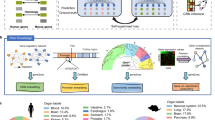
Abstract
Regulatory sequences encode crucial gene expression signals, yet the sequence characteristics that determine their functionality across species remain obscure. Deep generative models have demonstrated considerable potential in various inverse design applications, especially in engineering genetic elements. Here, we introduce DeepCROSS, a generative artificial intelligence framework for the inverse design of cross-species and species-preferred 5’ regulatory sequences in bacteria. DeepCROSS constructs a meta-representation using 1.8 million regulatory sequences from thousands of bacterial genomes to depict the general constraints of regulatory sequences, employs artificial intelligence-guided massively parallel reporter assay experiments in E. coli and P. aeruginosa to explore the potential sequence space, and performs multi-task optimization to obtain de novo regulatory sequences. The optimized regulatory sequences achieve similar or better performance to functional natural regulatory sequences, with high success rates and low sequence similarities with the natural genome. Collectively, DeepCROSS efficiently navigates the sequence-function landscape and enables the inverse design of cross-species and species-preferred 5’ regulatory sequences.
Similar content being viewed by others
Introduction
The evolution process fundamentally shapes the genomes of different organisms. The naturally-existing regulatory sequences (RSs) in different hosts thus generally adhere to related but different biophysical constraints. For example, despite the commonality of sigma factors between bacteria, each species exhibits specific sigma factors and transcription factor binding sites (TFBSs)1,2. Therefore, it is an interesting question whether one can break down the barriers between organisms to inverse design RSs that are species-preferred or functional across multiple species. Such RSs can help construct novel synthetic circuits in a wide range of microbial species for industrial and therapeutic applications3,4,5. Although a few natural RSs have been reported to be functional cross-species6,7, they generally exhibit low activities in heterologous species8,9,10. An effective strategy to obtain cross-species RSs with the desired function is still lacking.
Previous studies showed the existence of cross-species 5’ RSs in nature. For example, a recent work by Johns et al.7 randomly selected natural RSs from 180 bacteria species and tested their activities using massively parallel reporter assays (MPRA) in both E. coli and P. aeruginosa and identified several cross-species and species-preferred RSs (denoted as “Johns-Dataset”). The Johns-Dataset provided the paired information about the ‘sequence-activity’ landscape of a set of RSs across species7. However, the dataset has a limited number of functional cross-species RSs, and the activity and diversity of these RSs were severely restricted. These challenges restrict the understanding of sequence characteristics and impede efficient virtual exploration within the sequence space.
Here, we proposed DeepCROSS, a deep learning-empowered framework to inverse design cross-species and species-preferred 5’ RSs (Fig. 1a). The inverse design begins with defining desired functional characteristics, such as cross-species activity or species-preferred preferences, and works backward to create new RSs that meet these criteria. We demonstrated its effectiveness for designing gene promoters in E. coli and P. aeruginosa. DeepCROSS stands for Deep learning-empowered Cross-species regulatory sequence design based on Representation and Optimization in Sequence Space. By constructing a large-scale meta-representation of potential 5’ RSs derived from thousands of bacterial genomes and fine-tuning the model to target species using an adversarial autoencoder (AAE)11, DeepCROSS extracts the fundamental features of regulatory sequences into a compact deep statistical embedding. This representation, combined with a Dense-LSTM-based prediction model that predicts promoter activity of RSs, enables DeepCROSS to explore sequence space and generate cross-species RSs with high accuracy and sequence diversity. To address the issue of the limited number of functional cross-species RSs, we carried out an active exploration strategy, using DeepCROSS to guide in vivo MPRA experiments to fill the long-tail-distributed “sequence-activity” landscape. The DeepCROSS model refined by this strategy demonstrated a significant improvement in predicting RS activity. The experimental validation confirmed that the final DeepCROSS model achieved 90.0% and 93.3% accuracy in designing species-preferred and cross-species RSs respectively. These synthetic RSs exhibited high diversity and low sequence similarity to their natural counterparts within bacterial genomes. Our work provides a de novo strategy for effectively portraying the functional landscape within the vast DNA sequence space, positioning DeepCROSS as a valuable computational framework for the inverse design of cross-species and species-preferred RSs.
a In the meta-representation stage, tens of thousands of natural 5’ RSs were collected from the NCBI genome. The semi-supervised adversarial auto-encoder and Dense-LSTM predictor model, DeepCROSS, was applied to generate synthetic cross-species and species-preferred RSs. Then the AI-guided experimental quantification by MPRA experiments in E. coli and P. aeruginosa were conducted to measure the transcription activity of synthetic RSs. In the multi-task optimization stage, DeepCROSS-designed RSs were optimized, validated, and characterized. b The AAE model maps the one-hot encoded RSs to a 64-dimensional continuous vector. The encoder network (E-net) and two decoder networks (D-net-Gaussian and D-net-exp) are all based on groups of residual blocks (Supplementary Fig. 1). The three input datasets were ‘E. coli & P. aeruginosa’ (the RSs in the Johns-Dataset), ‘Enterobacterales and Pseudomonadales’ (RSs from Enterobacterales and Pseudomonadales bacteria), and RSs from more than 2000 broad species of bacteria.
Results
Inverse design of cross-species and species-preferred 5’ RSs by DeepCROSS
As reported in the Johns-Dataset which randomly selected natural RSs from broad species, though several sequences showed cross-species activities at some level, most of the randomly selected natural RSs are non-functional in target species7. Therefore, using randomly sampled 5’ RSs from other species to represent the functional subspace of the target species is ineffective12. Training a model directly on this kind of dataset tends to be ineffective in generating high-activity cross-species RSs. This ineffectiveness stems from two primary challenges: a limited number of functional samples with insufficient diversity13, and the long-tail distribution observed in the sequence activities of randomly selected samples12. Both of these challenges are prevalent issues in inverse design tasks across both biomolecular and material domains14,15,16.
To address these challenges, DeepCROSS was structured into three main steps (Fig. 1a): meta-representation, AI-guided experimental quantification by MPRA, and multi-task optimization. We demonstrated DeepCROSS with the inverse design task of cross-species and species-preferred 5’ RSs as promoters in E. coli and P. aeruginosa. The two widely used bacteria are both gram-negative proteobacteria but inhabit distinct environments and are equipped with diverse RSs.
For meta-representation, DeepCROSS employs an AAE11 to encode 5’ RSs into a lower-dimensional continuous vector, providing a compact and informative representation for the sequences. The AAE framework consists of one encoder, one decoder, and two adversarial discriminators (Fig. 1b). These two discriminators enforce constraints on the sequence representation, associating it with expression activity and aligning it with a Gaussian distribution. During training, DeepCROSS was first trained on 1.8 million 5’ RSs derived from 2621 bacteria genomes17 (Fig. 1a, Methods, Supplementary Fig. 1), effectively capturing critical sequence patterns. These RSs, sourced from diverse bacterial strains, endowed DeepCROSS with a broad understanding of the physicochemical constraints that are shared across different bacteria. This extensive training helped in framing a meta-representation of the potential functional sequence space. DeepCROSS was fine-tuned on a few bacteria species of the same genus as E. coli and P. aeruginosa (Enterobacterales and Pseudomonadales respectively). The pre-training and fine-tuning process can combine the sequence features of general constraints and the specific preferences of target hosts. We also built a quantitative prediction model (“Dense-LSTM” model), which aims to predict the transcriptional activity of the RSs based on their sequences, to guide the exploration of functional RSs based on the Johns-Dataset7 (Supplementary Methods).
To deal with the long-tail distribution observed in the sequence activities of randomly selected samples, we then used DeepCROSS to sample from the subspace of interest to generate candidate RSs and performed MPRA experiments18 to quantify the activities of these RSs (Methods, Supplementary Data 2). To guide the exploration of the potential sequence space, these quantified RSs were appended to the training dataset to refine both the Dense-LSTM prediction and representation models. To achieve multi-task optimization for the cross-species in both E. coli and P. aeruginosa and species-preferred RSs, the refined prediction model was applied to guide the optimization direction in the representation space. These optimized RSs were the final inversely designed products, and their activities in both E. coli and P. aeruginosa were verified using sfgfp gene as a reporter.
Meta-representation captures essential constraints of RSs
To evaluate whether the DeepCROSS-generated RSs captured essential constraints of RSs, we examined the representation of RSs from various bacteria genomes to assess DeepCROSS’s ability to derive meaningful features. As shown in Fig. 2a, although the edit distance between RSs within the same taxonomic class is indistinguishable from that between different taxonomic classes (Supplementary Fig. 2a), the two-dimensional principal component analysis (PCA) projection of the large-scale representation of DeepCROSS distinctly clusters RSs from different organisms based on their phylogenetic levels. Specifically, RSs from the target species E. coli and P. aeruginosa positioned themselves within their species-preferred territories. We found that the previously reported cross-species RSs between E. coli and P. aeruginosa7 are predominantly located between the two species-preferred subspace, suggesting that the cross-species RSs are constrained by the sequence features of both species. We assumed that cross-species RSs and species-preferred RSs could be generated by sampling from the intersectant region and the corresponding non-overlapping region, respectively (Supplementary Methods). In addition, we investigated whether the similarity between two RSs corresponded to the Euclidean distance between their embeddings in the representation space (Supplementary Methods). We analyzed the averaged embedding of the generated RSs at each step and found a gradual decline in sequence similarity to the starting sequence (Supplementary Fig. 2b). These results suggest that the representations generated by the AAE model preserve the inherent similarities within sequences, enabling clustering for the functional properties of sequences and thereby improving the efficiency of sampling and optimization.
a The PCA result of the statistic representation of natural cross-species RSs (the host-preferred genera are annotated by color. E.g., the RSs from Actinobacteria are in color red). The enlarged part is the local territory of RSs from our targeted species E. coli and P. aeruginosa in the testing dataset of the Johns-Dataset. b Left: comparison of PCC of 6-mer base frequency between DeepCROSS-generated RSs and reported functional RSs under different representation approaches (Supplementary Methods). Right: comparison of JS divergence of GC content between DeepCROSS-generated RSs and reported functional RSs under different representation approaches. Bar height represents the mean of n = 3 independent experiments per group, with black dots indicating individual experiment results. The p-values were determined by a two-tailed t-test, where ns represents not significant. c Comparison of the measured MPRA activity of cross-species RSs generated by the full DeepCROSS framework (AAE & Dense-LSTM model) under different representation approaches. The relative MPRA activity of RSs was calculated using the BBa_J23119 as the control sequence. Johns-DeepCROSS (n = 491), genus-DeepCROSS (n = 492), Motif (n = 395), Random RSs (n = 391), Johns-Dataset (n = 1470), Meta-DeepCROSS (n = 485). n represents the number of RSs. Box plots depict the median (center line), interquartile range (box limits), whiskers (1.5×IQR), and outliers (points beyond whiskers). Source data are provided as a Source Data file.
Considering the diversity in GC content and k-mer preferences of RSs across different organisms19,20, and the importance of conserved −10 and −35 elements across bacteria for promoter function21, we computationally evaluated the feature extraction ability of AAE model from three perspectives: 6-mer frequencies, GC-contents, sigma70 factor match score (Fig. 2b and Supplementary Fig. 3). Besides DeepCROSS trained with meta-representation which has been described above (meta-DeepCROSS in short), we also considered two versions of DeepCROSS trained using only the Johns-Dataset (Johns-DeepCROSS in short), or using both the Johns-Dataset and RSs of representative Enterobacterales and Pseudomonadales bacteria which are in the same genus of E. coli and P. aeruginosa, respectively (genus-DeepCROSS in short). Regarding the 6-mer frequency, we calculated the Pearson’s Correlation Coefficient (PCC) for 6-mer base frequencies between the generated RSs and previously reported functional RSs. We found that in comparison with Johns-DeepCROSS and genus-DeepCROSS, RSs generated by meta-DeepCROSS exhibited a significantly higher PCC in both cross-species and E. coli-preferred generation tasks (Fig. 2b, two-tailed t-test). Hence, the model appears to more accurately mimic the 6-mer base frequency preference when utilizing the meta-representation. We examined the Jensen–Shannon (JS) divergence between the GC distribution of natural RSs in the training set and those generated by DeepCROSS. The GC content distribution of RSs generated by meta-DeepCROSS showed the lowest JS divergence with natural distribution (Fig. 2b, two-tailed t-test), which demonstrated that meta-representation can well learn the constraints of broad bacteria species and thus provides crucial sequence constraint information for DeepCROSS. To evaluate whether DeepCROSS-generated RSs contain sigma70 factor recognition sites, we calculated the cumulative match score of these RSs (Supplementary Methods and Supplementary Fig. 3). We observed that some DeepCROSS-generated RSs have higher sigma70 factor matching scores, which play a crucial role in initiating transcription by guiding RNA polymerase to specific promoter regions22. However, this feature has no significant difference in RSs designed by Johns-DeepCROSS, genus-DeepCROSS and meta-DeepCROSS methods, indicating that the model might have identified more specific features that dominate the regulatory effects of RSs.
We further experimentally measured the in vivo transcriptional activities of the DeepCROSS-generated RSs by MPRA experiment18 (Fig. 2c). The MPRA library contains a total of 24,000 candidate RSs, mainly including 6 groups of RSs: random RSs group, Motif group, Johns-Dataset group, Johns-DeepCROSS group, genus-DeepCROSS group, meta-DeepCROSS group, etc (see Supplementary Data 2 and Supplementary Methods for details). We used the DeepCROSS framework to generate cross-species, E. coli-preferred, and P. aeruginosa-preferred RSs as promoters and measured their in vivo activities (Methods). The measurements across three replicates within a given condition were highly correlated (average PCC stood at 0.9969 for E. coli and 0.9958 for P. aeruginosa, Supplementary Fig. 4a). As to the cross-species design task, the RSs designed by meta-DeepCROSS showed higher transcriptional activity than all the other aforementioned RS generation methods (Fig. 2c). Specifically, regarding the total transcriptional activity of RSs in both E. coli and P. aeruginosa, the meta-DeepCROSS group exhibited an average relative activity of 0.71, while other groups showed average relative activities ranging from 0.23 to 0.67.
Overall, both experimental and computational results demonstrated that meta-representation can serve as an efficient method to help the generative model better learn semantically rich representations, capture the essential constraints of RSs, and generate functional RSs with key sequence features. In addition to expanding the training dataset, incorporating a broader array of relevant samples and employing unsupervised learning steers the search away from non-functional sequences23, thereby effectively enabling inverse design.
DeepCROSS-guided exploration efficiently fills the sparse sequence-activity landscape
The performance of a prediction model is heavily influenced by the quality of its training dataset. A large proportion of low-activity sequences in the long tail can introduce noise, severely biasing the model and limiting its generalization ability. This issue was evident in the Johns-Dataset, which consists of RSs randomly selected from natural bacterial genomes. In our study, MPRA results included a subset of RSs from the Johns-Dataset, and experimental results from two studies showed a high correlation (PCC = 0.85 for E. coli and 0.71 for P. aeruginosa, Supplementary Fig. 5). We used this subset to correct the activity values of the remaining sequences in the Johns-Dataset. As shown in Fig. 3a, b, randomly selected natural RSs in the Johns-Dataset are over-abundant with zeros or near-zero activity values. In contrast, the activity distribution of the DeepCROSS-generated RSs mitigated the long tail effect and showed a significantly heavier tail. The ratio of functional RSs (normalized activities > 0.1) generated by DeepCROSS in E. coli and P. aeruginosa was 5.63 and 3.28 times higher, respectively, compared to that in the Johns-Dataset (Fig. 3a, b). These results suggest that DeepCROSS-generated RSs can effectively reshape the unbalanced activity distribution, providing a more diverse and functional dataset for model training.
a The distribution of RSs’ MPRA activity in E. coli. The activity of randomly selected natural RSs in the Johns-Dataset is shown in shallow red. The MPRA activity distribution of all RSs designed by DeepCROSS (AAE & Dense-LSTM model) is shown in dark red. b The distribution of RSs’ MPRA activity P. aeruginosa. The activity of randomly selected natural RSs in the Johns-Dataset is shown in shallow blue. The MPRA activity distribution of all RSs designed by DeepCROSS (AAE & Dense-LSTM model) is shown in dark blue. c The max activity achieved, mean activity achieved and NDCG are three evaluation metrics of inverse design efficiency. The design efficiency is evaluated under three expression situations: E. coli, P. aeruginosa, and cross-species expression. The cross-species expression activity in both E. coli and P. aeruginosa is calculated by the sum of MPRA activity (log2) in both species. n = 100 independent simulation experiments per group. The p-values were determined by a two-tailed t-test, where ns represents not significant. The outer violin curves represent the kernel density estimate of the data distribution. A box plot is embedded at the center, where the white dot indicates the median, the bounds of the box correspond to the 25th and 75th percentiles, and the whiskers extend to values within 1.5 times the interquartile range (IQR). Source data are provided as a Source Data file.
We then investigated whether the measurement of these DeepCROSS-generated RSs can enhance the accuracy of RS activity prediction models. We used RSs from the Johns-Dataset and DeepCROSS-generated RSs from our MPRA experiment as the training data to train seven state-of-the-art prediction models24,25,26,27,28. These models were then used to predict the activities of RSs in an independent test dataset (Methods). As shown in Supplementary Fig. 6, training on DeepCROSS-generated RSs can significantly improve PCCs between predicted and real activities and decrease root mean square errors (RMSEs) in all prediction models. The average PCC in the seven prediction models has been improved by 13.7% in E. coli and 46.5% in P. aeruginosa respectively. We finally adopted the Dense-LSTM model which was proved to show the highest accuracy as the prediction model in the following analysis.
After filling the sparse sequence-activity landscape with DeepCROSS-generated RSs validated by MPRA experiments, we obtained a more informative ‘RS-activity’ mapping landscape. To improve the designs of promising candidates, we employed transfer learning to integrate features from different datasets by pretraining the Dense-LSTM model on the Johns-Dataset and fine-tuning it on DeepCROSS-generated RSs. Given that sample sizes of functional and non-functional RSs were still unbalanced, which may dampen the generality of the model. We thus further investigated how the prediction accuracy of the Dense-LSTM model was affected by removing non-functional RSs. Since the goal of inverse design is to find functional RSs with high activity, the candidate sequences with high activities should rank at the top of the list and the non-functional RSs should rank at the bottom. We carried out three evaluation metrics, inspired by previous work in protein design13: the max and mean activity of the 100-highest-ranked RSs (“max activity achieved” and “mean activity achieved” in short) and the Normalized Discounted Cumulative Gain (NDCG). NDCG, a commonly used metric in information retrieval, evaluates the ranking accuracy of RSs with high activities29 (Supplementary Methods). All four models, each employing different performance enhancement strategies, were evaluated on an independent test dataset (Fig. 3c, Methods). The results showed that training on DeepCROSS-generated RSs, compared to Johns-Dataset, significantly improved the mean activity achieved and NDCG, suggesting DeepCROSS-generated RSs abundantly contain key functional features. Furthermore, fine-tuning on DeepCROSS-generated RSs and strategically removing non-functional RSs both significantly improved the prediction accuracy across E. coli-preferred, P. aeruginosa-preferred, and cross-species situations (Fig. 3c). These observations suggest that the abundance of non-functional sequences hampers the prediction model’s ability to learn functional features. By strategically employing transfer learning and removing these non-functional sequences, the model was able to better capture the sequence-activity landscape and more efficiently explore promising candidates.
DeepCROSS efficiently designs novel species-preferred and cross-species RSs
To design synthetic species-preferred RSs and cross-species RSs with higher activities, we applied a genetic algorithm (GA) to navigate in the representation space with the guidance of the prediction model (Methods). The Dense-LSTM prediction model was initially pre-trained on Johns-Dataset after removing non-functional RSs, and then fine-tuned using all DeepCROSS-generated RSs from our MPRA experiments. For the optimization task of species-preferred 5’ RSs, the objective function was set to maximize the fold-change of predicted activities between E. coli and P. aeruginosa. Each optimized RS was supposed to satisfy two properties at the same time: high activity in E. coli and low activity in P. aeruginosa, and vice versa. Similarly, the objective function of cross-species RSs was maximizing the product of the predicted activities of E. coli and P. aeruginosa (Methods). The GA-optimized RS group showed significantly higher predicted scores than that of the natural RS group in both species-preferred and cross-species situations (Fig. 4a–c).
a The natural RSs’ activities and the optimized RSs’ activities were scored by the prediction model in an cross-species RS design situation (EC and PA are short for E. coli and P. aeruginosa respectively). b The natural RSs’ activities and the optimized RSs’ activities were scored by the prediction model in the P. aeruginosa-preferred RS design situations. c The natural RSs’ activities and the optimized RSs’ activities were scored by the prediction model in E. coli-preferred RS design situations. Box plots depict the median (center line), interquartile range (box limits), whiskers (1.5×IQR), and outliers (points beyond whiskers). n = 2000 per group in a–c and n represents the number of RSs. d The relative activity of optimized cross-species and control 5’ RSs measured by sfGFP (BBa_J23119 as the control sequence). e The relative activity of optimized P. aeruginosa-preferred and control 5’ RSs. f The relative activity of optimized E. coli-preferred and control 5’ RSs. Bar height represents the mean of n = 3 biological replicates per group in d to f, with black dots indicating individual experiment results. Source data are provided as a Source Data file.
We selected in total 45 GA-optimized RSs with top predicted activity scores for E. coli-preferred, P. aeruginosa-preferred, and cross-species tasks. Additionally, we selected 6 RSs with high scores from the Johns-Dataset for each task after removing abnormal values, resulting in 18 RSs in total as positive controls (Supplementary Data 3). We measured the promoter activities of these RSs in both E. coli and P. aeruginosa by an sfgfp reporter gene (Supplementary Methods). In total, all GA-optimized RSs designed as cross-species RSs were validated to show significantly higher activities than negative control RSs in both species (P-value < 0.001, two-tailed t-test, Fig. 4d). Furthermore, 93.3% of these GA-optimized RSs exhibited higher activity levels than the native functional RS BBa_J23119 in both E. coli and P. aeruginosa. Besides, 90.0% of the GA-optimized RSs designed as species-preferred RSs showed activity fold-changes between the two species higher than 1.5 (Fig. 4e, f). Using the functional RS BBa_J23119 as control, the DeepCROSS-designed E. coli-preferred RSs showed an average fold-change of 4.14 and a maximum of 9.6 between E. coli and P. aeruginosa. Meanwhile, the DeepCROSS-designed P. aeruginosa -preferred RSs showed an average fold-change of 15.3 and a maximum of 51.0 between P. aeruginosa and E. coli. The PCCs of predicted and measured activities in E. coli and P. aeruginosa were 0.84 and 0.82, respectively (Supplementary Fig. 7). We also compared DeepCROSS with the promoter design methods proposed by Hossain et al.30. We experimentally characterized the top 6 promoters of their designs. DeepCROSS-generated RSs showed equivalent or superior promoter activity in both E. coli and P. aeruginosa (Supplementary Fig. 8). We further assessed the robustness of the DeepCROSS-generated RSs by testing them in two additional types of culture medium: M9 and EZ-rich (Methods). These RSs showed a very high correlation in promoter activity under diverse culture conditions in both E. coli and P. aeruginosa (Supplementary Fig. 9, Supplementary Data 3).
We also evaluated the sequence diversity of DeepCROSS-generated RSs. The average similarity of these optimized RSs calculated by multiple sequence alignments in ClustalW231 is 11.5, comparable to the similarity of random sequences with the same GC content (Supplementary Fig. 10a, Supplementary Methods). We also compared the similarity of DeepCROSS-generated RSs with functional natural RSs collected by Kosuri et al. (Supplementary Data 3)32. The average similarity calculated by ClustalW2 between DeepCROSS-generated RSs and functional natural RSs is 24.0, which is at a similar level of 24.3 observed between random sequences and functional natural RSs (Supplementary Methods). Furthermore, using BLAST33 to compare DeepCROSS-generated RSs against the bacterial genomes (E. coli and P. aeruginosa), we found that all e-values are higher than 0.25 (Supplementary Fig. 10b), indicating that DeepCROSS-generated RSs exhibit a strong dissimilarity to natural bacterial genomes.
Characteristics of DeepCROSS-generated cross-species and species-preferred 5’ RSs
To explore the characteristics of DeepCROSS-generated RSs, we generated 1 billion RSs from the representation space and used the prediction model to filter out the top 2000 sequences as E. coli-preferred, P. aeruginosa-preferred, and cross-species RSs, respectively. These DeepCROSS-generated RSs were characterized by three aspects: GC contents, k-mer frequencies, and DNA rigidity properties.
The top 2000 E. coli-preferred and P. aeruginosa-preferred RSs showed an average GC content of 62.2% and 66.7% respectively. The GC content distribution of cross-species RSs showed an average of 64.7%, lying between those of E. coli-preferred and P. aeruginosa-preferred RSs (Fig. 5a). Meanwhile, RS activities showed a weak negative correlation with GC contents for all three kinds of RSs (Supplementary Fig. 11). We then calculated the k-mer (k from 4 to 8) base frequencies of E. coli-preferred, P. aeruginosa-preferred, and cross-species RSs (Fig. 5b, Supplementary Fig. 12). Taking the 6-mer base frequency as an example, we found that about one-quarter of the 6-mers showed differential frequencies among the three kinds of RSs, forming six differentially occurred groups (Fig. 5b, Supplementary Methods). The P. aeruginosa-preferred RSs showed significantly higher frequencies of GC-rich 6-mers, such as GCCGCC and CCCGCC. The E. coli-preferred RSs showed significantly higher frequencies of some AT-rich 6-mers, such as TTTTTA and AATAAT. The cross-species RSs showed significantly higher frequencies of TATAAT and TTGACA, which are the conserved −10 and −35 elements in bacterial promoters, essential for RNA polymerase binding and transcription initiation. The results also showed similar k-mer frequencies between cross-species and E. coli-preferred RSs indicating that designing E. coli-preferred RSs is more challenging compared to P. aeruginosa-preferred RSs. This observation is consistent with the experimental results in multi-task optimization (Fig. 4e, f).
a The GC content distribution of species-preferred and cross-species RSs. b The heatmap illustrates the 6-mer frequency of species-preferred and cross-species RSs. The different preferred base 6-mer frequency encoding in host species. Differentially enriched groups are annotated above the heatmap with color bars. c The MGW, ProT, Roll, and HelT property characteristics of species-preferred and cross-species RSs. Source data are provided as a Source Data file.
We further investigated the DNA strand shape properties of these RSs. DNA shape plays an important role in transcription factor (TF) binding and is determined by local DNA sequences and some shape motifs, which could be missed by typical motif-finding approaches34. We calculated four crucial DNA shape property parameters, including Minor groove width (MGW), Helix twist (HelT), Propeller twist (ProT), and Roll reported by Zhou et al.35 through recently developed DeepDNAshape toolkit36. As shown in Fig. 5c, the HelT and Roll distributions of cross-species RSs were similar to those of P. aeruginosa-preferred RSs, and showed significant distribution differences compared to E. coli-preferred RSs. In contrast, the MGW distribution of cross-species RSs was similar to that of E. coli-preferred RSs and differed from P. aeruginosa-preferred RSs. For ProT, and the distributions of cross-species RSs located between the two distributions of E. coli-preferred and P. aeruginosa-preferred RSs (Fig. 5c). These results demonstrated that efficiently navigating the functional subspace in the representation space can help enrich and reveal transcription regulation constraints of these elements in both DNA sequence and DNA shape levels.
Discussion
The inverse design of cross-species and species-preferred RSs37,38 has the potential for building sophisticated gene circuits39,40 in metabolic engineering4,41 and biomedicine42. However, the limited number of functional samples with insufficient diversity and the long-tail distribution of sequence activities bring difficulty in extracting the functional features and exploring the sequence space efficiently.
Here, we presented DeepCROSS to achieve de novo cross-species and species-preferred regulatory element inverse design. DeepCROSS is trained on the characteristics of natural RSs from a broad range of bacteria and the DeepCROSS-designed 5’ RSs’ activities were validated in vivo. The inclusion of a wide variety of bacterial species enabled the generative model to learn general biophysical constraints and avoid the non-functional sequence features efficiently. The deep learning model distilled the fundamental features of RSs into a semantically rich representation and fine-tuned it to target species to form species-preferred subspace. The AI-guided MPRA experiments quantified the activities of DeepCROSS-designed RSs and filled the long-tail-distributed “sequence-activity” landscape. This AI-guided exploration strategy significantly improved the accuracy of RS activity predictions and enabled DeepCROSS to perform multi-task optimization within the representation space to generate cross-species RSs and species-preferred RSs.
Long-tail distribution is a common challenge in inverse design and engineering tasks14,15,16. A large number of sequences with low activities form the ‘hole’ in the sequence-activity landscape and often mislead the deep network-based model into learning inaccurate sequence patterns13. A small number of functional sequences (for example, cross-species and species-preferred RSs) exists in nature7, but they often show insufficient performance due to the lack of pressure to enhance activity or adapt to synthetic system. The combination of exploration procedures guided by AI and massive quantification experiments efficiently fills in the sequence-activity landscape and significantly improves the performance of predictive and generative models to inverse design functional sequence candidates. Furthermore, the strategy of meta-representation guided high-throughput exploration may have the potential to overcome data scarcity which is a common problem in inverse design and engineering tasks. In addition, researchers have been using machine learning (ML) algorithms to predict activities of eukaryotic RSs43,44. DeepCROSS has the potential to be applied to eukaryotic systems for designing cross-species RSs between eukaryotes, or even between prokaryotes and eukaryotes. This advancement represents a step toward constructing complex circuits for the synthetic biology community45.
Methods
Ethical statement
This research does not involve human participants, animals, or other ethical concerns requiring formal approval.
Overview of the inverse design procedure
From the perspective of statistical modeling, we used \(X\) to denote the regulatory sequences and \(Y\) to denote the target activities. The general sequence inverse design problem can be formulated to maximize the joint probability:
According to the chain rule, the following is obtained:
Supposing all the possible base combinations are in the set \(L\), there are in total \(L={4}^{N}\) possibilities with sequence length N. Directly analyzing mappings from one-hot encoded DNA sequences remains challenging. One key issue is the difficulty in inferring property similarity from sequence space, as sequences that are similar in their one-hot encodings may exhibit distinct functional properties. Additionally, the vastness of the sequence space further complicates this analysis. To address these challenges and map discrete candidate sequences to points in a representation space, DeepCROSS was used to map the one-hot encoded DNA sequences \(X\) into a low-dimensional continuous sequence representation \(Z\) (Fig. 1b). The training process of DeepCROSS is aimed at maximizing the reconstruction accuracy (Supplementary Methods), thus the sequences with highly similar transcriptional activities or the same species labels will be automatically clustered closely in the representation space. Therefore, after a well-established training process, DeepCROSS can better characterize the sequence properties in a low-dimensional representation, enabling visualization and statistical modelings. Thus, the objective can be rewritten as follows46:
To learn the species-preferred constraints, the sequence representation \(Z\) is divided into different sub-groups for each species: \(Z={Z}_{{EC}}\cup {Z}_{{PA}}\cup {Z}_{{species}}\) …, etc. The terms \({Z}_{{EC}}\) and \({Z}_{{PA}}\) denote the sub-group of sequence representation \(Z\) that prefers to be functional in E. coli and P. aeruginosa respectively. Each sub-group has its own constraints, making its RSs form a functional subspace in the whole sequence space. Using the E. coli sub-group as an example, the objective of designing E. coli-preferred RSs can be rewritten as follows:
Sampling the vectors from \({Z}_{{EC}}\) instead of the other sequence space, can help avoid nonfunctional regions and provide better generalization conditions. The large-scale representation helps the model to capture general biophysical constraints which helps to better capture the sequence features like GC content distribution and k-mer frequency (See the section ‘Meta-representation captures essential constraints of RSs’). Similarly, the sub-group of \({Z}_{{PA}}\) was calculated to design P. aeruginosa. The cross-species situation is not natural in the wild. However, using the sequence representation by generative model, one can sample from the cross-species subregion \({Z}_{{cross}}=\left\{{z|z}\in {Z}_{{EC}}\bigwedge z\in {Z}_{{PA}}\right\}\) and optimize the product of activities in two species, and generate novel cross-species sequences.
The DeepCROSS strategy was designed based on this formulation, which contained three main steps (Fig. 1a): meta-representation and generation, MPRA quantification, and multi-task optimization. Metagenomic mining offered 1.8 million putative 5’ RSs as a training dataset to estimate \(P(x)\), where \(x\) is a one-hot-coded DNA sequence. Then the deep representation and sampling by DeepCROSS used the training dataset to learn the continuous representation space \(P(z)\) and generated novel 5’ RSs, which corresponds to the term \(P(x,z)\). The DeepCROSS-guided quantification by MPRA experiments provides RSs’ activity to fill the long-tail-distributed ‘sequence-activity’ distribution and refines the prediction model by improving the prediction accuracy. The optimization procedure under the guidance of the prediction model navigates the continuous representation space \(P(z)\) to maximize the term \(P(y,x,z)\) and inverse design synthetic cross-species and species-preferred RSs.
Meta-representaiton of RS space
DeepCROSS was built to map the 165nt RSs to low-dimensional representation space and sample novel cross-species and species-preferred RSs from the corresponding subspace. DeepCROSS contains four sub-networks: the encoder, decoder, discriminator for Gaussian distribution in representation space, and discriminator for expression bin distribution. The input 5’ RSs were one-hot encoded and passed through a 64-dimensional embedding. The encoder network (E-net) and decoder network (D-net) are based on groups of residual blocks (Fig. 1b, Supplementary Fig. 1).
The DeepCROSS was trained by the following four steps:
-
1.
Large-scale unsupervised training on 1.8 million 5’ RSs from 2621 representative bacteria genomes, to learn the general constraints of functional RSs.
-
2.
Unsupervised fine-tuning of the DeepCROSS model on representative Enterobacterales and Pseudomonadales bacteria species to learn the features of E. coli and P. aeruginosa respectively. The closely related species of E. coli selected in this study are E. coli str. K-12 substr. DH10B, Salmonella typhimurium LT2, Erwinia amylovora ATCC 49946, Klebsiella pneumoniae subsp. pneumoniae MGH 78578 and Enterobacter sp. 63847. The closely related species of P. aeruginosa selected in this study are Pseudomonas entomophila L48, Pseudomonas putida KT2440, Pseudomonas syringae pv. tomato str. DC3000, Pseudomonas fluorescens Pf-547. DeepCROSS would combine features from both the large-scale species and closely related sequence landscape.
-
3.
Supervised learning on functional 5’ RSs characterized by MPRA7. The paired ‘RS-activity’ data was provided to model every 5 iterations.
For robust prediction in the generative model, the RSs’ transcription activity is coded into three bins to represent the high, middle, and low expression levels48. Each RS was in total encoded in 6 bins for the two target species.
Prediction of RSs’ expression activity
The prediction models were trained to predict the transcription activity of input RSs. The sequences were encoded in one-hot format and the output transcription activity was normalized after taking the logarithm (log2). To systematically compare the natural training set and the training set after AI-based sampling, these two datasets were compared under seven state-of-the-art model structures for representation and prediction: DeepGOPlus24, ResNet25, Basset-based network26, CNN connected with attention layers27, DenseNet28, Dense-connected LSTM, Dense-LSTM. The independent test dataset used in Fig. 3c and Supplementary Fig. 6 was derived from the Lib-1 MPRA experiment results, containing 10,311 unique RSs. The Lib-2 MPRA experiment results, which contained 10,282 unique RSs, were used for training models in Supplementary Fig. 6. Additionally, the 9400 DeepCROSS-generated RSs from Lib-2 results were used to train the models in Fig. 3c. Finally, the Dense-LSTM model was initially pre-trained on Johns-Dataset after removing non-functional RSs, and then fine-tuned using all DeepCROSS-generated RSs from our MPRA experiments and was used as the scoring model in the genetic algorithm.
Optimization in continuous representation space
The genetic algorithm (GA)49 is used to optimize the AI-generated sequences to become species-preferred or cross-species RSs. The Dense-LSTM model as the scoring model in GA. The transcriptional activity of RSs in E. coli and P. aeruginosa were trained in two different prediction models respectively. For the E. coli-preferred RSs, the optimization target is the difference in the predicted activity between E. coli (described as \({P}_{{EC}}\)) and P. aeruginosa (described as \({P}_{{PA}}\)):
The optimization target of P. aeruginosa-preferred is the difference in the predicted activity between P. aeruginosa and E. coli:
The optimization target of cross-species RSs is the sum of the predicted activity of E. coli and P. aeruginosa:
Based on the MPRA results, we set \(\varepsilon\) = 2−5 to mitigate the impact of small values in the prediction model. Moreover, to prevent potential overfitting in the predictive models, we filtered out RSs with predicted values that significantly exceed the upper bound achievable within the training data when designing species-preferred RSs.
The pool size in GA was set to 2000 and selected half ordinary and half high-scored representation vectors for the next epoch in each optimization epoch. The decoder (D-net) then mapped the representation vector into RSs. The maximum epoch of GA optimization was set to 5000 for each task and the optimization stopped when the target objective converged. To contain sequence diversity, the sequences were ordered by hamming distance, and the adjacent sequences with lower than 25nt hamming distance (whole length=165nt) were deleted. After approximately 2000 epochs, fitness scores plateaued, confirming the robustness of the optimized RSs against overfitting (Supplementary Fig. 13). The GA process for the cross-species task is shown in Supplementary Fig. 13a, and the species-preferred tasks are shown in Supplementary Fig. 13b, c. Considering the challenge of designing E. coli-preferred RSs, we also selected some from the early epochs for experimental validation.
Characterization of DeepCROSS-designed 5’ RSs
The AI-designed 5’ RSs were evaluated in the following four aspects: 1) GC content distribution; 2) k-mer frequency; 3) DNA rigidity; 4) The similarity of sequences within designed ones, and the similarity between the designed ones and natural 5’ RSs.
Measuring the 5’ RSs activity by massive parallel reporter assay
Oligo synthesis and cloning
The library was synthesized by TWIST Bioscience (www.twistbioscience.com) and then resuspended in 100 μL of elution buffer before cloning into plasmid pMPRA2 (Supplementary Fig. 14). Our MPRA experiment procedure referred to Yu et al.50 and Tewhey et al.18. The replicon of the pMPRA1 plasmid (Addgene ID no. 49349) was replaced with pBBR1 to generate the plasmid pMPRA2, which has a broad host range and is compatible for replication in both E. coli and P. aeruginosa. A total of 24,000 oligos were synthesized as 195 bp sequences containing 165 bp of designed 5’ RSs and 15 bp of adapter sequence at both 5’ and 3’ end (5’ [165 bp oligo] 3’). These oligos were synthesized as two separate libraries named Lib-1 and Lib-2 (Supplementary Data 2).
The 20 bp barcodes were added by performing 6 emulsion PCR reactions by DNA emulsion and purification Kit (EURX, E3600). Each reaction was a 50 μL water phase in the volume containing approximately 109 copies of oligo, 25 μl NEBNext® Ultra™ II Q5® Master Mix (NEB, M0544S), 0.2 μM Library_F and Barcode_R primers, and 0.5 ng acetylated BSA. The oil surfactant phase was prepared following the manufacturer’s protocol (EURX, E3600). The 50 μl precooled PCR Water Phase was added to the 300 μl precooled oil surfactant mixture, and then vortexed in a cold room using a vortexer for 5 min. The 350 μL of Emulsion mixture was equally divided into 4 PCR tubes and cycled with the following conditions; 95 °C for 30 sec, 20 cycles (95 °C for 20 sec, 67 °C for 10 sec, 72 °C for 15 sec), 72 °C for 5 min. Amplified emulsion mixture was broken and purified by adding 1 mL of butanol followed by vortexing at room temperature. The broken emulsion/butanol mixture was conducted by adding orange DX buffer in the DNA emulsion and purification Kit and gently agitating the mixture on a rotator for 2 min. The mixture was centrifuged at 16,000 g for 2 min for phase separation. Most of the yellow-colored organic phase was removed and the water phase and interphase were used for spin column purification. In total 100 μl of Elution buffer was added to elute the final bound DNA. The product was then cleaned with Zymo Clean and Concentrator Kit (Zymo Research, D4014), eluting into 10 μL of Ultra-pure H2O.
To clone the RiboJ-RBS-sfGFP sequence into the plasmid, the sfGFP was first amplified with NEBNext® Ultra™ II Q5® Master Mix (NEB, M0544S) for 35 cycles using primers sfGFP_F and sfGFP_R at 10 μM. The plasmid was digested with AsiSI (NEB, R0630S) for 3.5 h at 37 °C. The digested plasmid and PCR product were run on a 1% agarose gel and then purified by a Multifunctional DNA purification Kit (Aidlab Biotech, DR0303). Both the purified plasmid and sfGFP PCR product were concentrated by Zymo Clean and Concentrator Kit (Zymo Research, D4014) and the final concentration is 148 ng/μL and 222 ng/μL respectively. The sfGFP PCR product was inserted into the digested pMPRA2 plasmid by Gibson assembly (NEB, E2611L) using 3 μg of oligos and 0.8 μg of digested vector in a 40 μl reaction incubated for 60 min at 50 °C. The Gibson assembly product was concentrated by Zymo Clean and Concentrator Kit and eluted into 40 μL of Ultra-pure H2O. To remove the remaining uncut plasmids, the 20 U of AsiSI, 5 U of RecBCD (NEB, M0345), 5 μL 1 mM ATP, and 5 μL 1x NEB Buffer 4 were incubated in a 50 μl reaction for 4 hours at 37 °C. The product was concentrated by Zymo Clean and Concentrator Kit and eluted into 6 μL of Ultra-pure H2O. The product was next transformed into 10-beta electrocompetent E. coli cells and competent P. aeruginosa cells.
Transformation into E. coli and P. aeruginosa
The ligated plasmid was transformed into 10-beta electrocompetent E. coli cells (DH10B) and competent P. aeruginosa cells (PAO1). The ~500 ng ligation product was transferred into 100 ml competent cells by electronic transformation at 2.1 kV with a Bio-Rad Micropulser. The transfected cells were recovered in 1 mL of SOC for 1 hour at 37 °C. The serial dilutions (10−1, 10−2, 10−3, and 10−4) of 100 μl cell culture were plated after SOC recovery and counted ~102 CFUs in 10−4 plate. The estimated transformation efficiency is >107 CFU. The 1 ml cell culture in 5 mL of Luria−Bertani (LB) was inoculated with Ampicillin (50 μg/mL) and 107 cells were grown overnight.
Barcode mapping
The RSs and barcode region from pMPRA2 plasmid were extracted and purified by QIAGEN Plasmid Maxi Kit (QIAGEN, 12162) for sequencing and downstream mapping of the barcodes to their respective 5’ RSs.
The library was amplified by two PCR reactions. The library was amplified with NEBNext® Ultra™ II Q5® Master Mix (NEB, M0544S) at 6 cycles using primers MPRA_v3_Amp2Sa_Illumina_F and Illumina_Universal_Adapter_R at 5 μM as well in triplicate. Replicates were pooled, and then cleaned with Zymo Clean and Concentrator Kit (Zymo Research, D4014), eluting into 10 μL of Ultra-pure H2O.
For the second PCR, primers with Illumina adapters P7, P5, and a unique sample index were used. The product was amplified with NEBNext® Ultra™ II Q5® Master Mix (NEB, M0544S) at 5 cycles using primers Illumina_Multiplex and Illumina_Universal_Adapter_R at 5 μM.
Ultimately, the PCR product was cleaned with Zymo Clean and Concentrator Kit (Zymo Research, D4014) and ran on a 1.0% agarose gel for final confirmation. After quality assessment, samples were sequenced on an Illumina 2 × 150 bp chemistry on Illumina NovaSeq instruments through the sequencing service by Azenta company. Barcodes were mapped to their respective 5’ RSs using the pipeline from MPRAflow51. Paired-end reads are merged into a 239nt sequence. Then the first 150 bp of each read was extracted, which encodes the RS variant, as well as the last 20 bp encoding the barcode, and generated a list of barcode-5’ RSs associations. The statistical distribution of barcodes corresponding to each sequence in Lib-1 and Lib-2 is shown in Supplementary Fig. 15a, d. Finally, the additional filtering steps were performed for quality control purposes.
Library growth, final plasmids, and cDNA synthesis
Bacteria in glycerol stocks (20% glycerol) were picked out and inoculated in 100 mL of LB + Carbenicillin (50 µg/mL for E. coli and 150 µg/ml for P. aeruginosa) at 37 °C for 16 h overnight. The overnight culture was diluted into approximately OD 0.01, inoculated into 200 mL of LB + Carbenicillin, and grown at 37 °C to OD 0.3–0.4 (~2.5 h).
To harvest the final plasmids library, which consists of plasmids with the RiboJ-RBS-sfGFP sequence inserted, we extracted separately the plasmids from 50 ml culture medium of E. coli and P. aeruginosa using the QIAGEN Plasmid Maxi Kit (QIAGEN, 12162).
To harvest RNA pellets, the culture was first cooled for two minutes in an ice slurry while periodically swirling. For each sample, three 50 mL aliquots of culture were poured into pre-chilled tubes and spun for two minutes at 13,000 g at 4 °C. The supernatant was carefully poured off. RNA was extracted from P. aeruginosa and E. coli pellets using a Qiagen RNEasy Midiprep kit (QIAGEN, 75142). The technical replicates of this extraction in biological was performed. Subsequent wash steps concentrated isolated RNA with Qiagen Minelute Cleanup Kit (QIAGEN, 74204). Next, isolated RNA was converted to cDNA with Transcript II one-step gDNA Removal and cDNA Synthesis SuperMix (TransGen biotech, AH311) following the supplementary protocol. The cDNA reaction mixture was cleaned with Zymo Clean and Concentrator Kit (Zymo research, D4014).
The final plasmid library was amplified by two PCR reactions, following the same procedure as the Barcode mapping, except that the primer MPRA_v3_Amp2Sa_Illumina_F was replaced with MPRA_v3_Illumina_GFP_F. The statistical distribution of barcodes corresponding to the sequence of the final plasmid library in Lib-1 and Lib-2 is shown in Supplementary Fig. 15b, c, and e, f. The barcoded cDNA was amplified with NEBNext® Ultra™ II Q5® Master Mix (NEB, M0544S) by perfectly matched primer MPRA_v3_Amp2Sc_R at 5 μM for 10 cycles. The product was cleaned with Zymo Clean and Concentrator Kit (Zymo Research, D4014). Then the product was amplified with primers MPRA_v3_Illumina_GFP_F and Illumina_Universal_Adapter_R at 5 μM for 6 cycles. Both prepared DNA and RNA library samples were pooled and sequenced using 2 × 150 bp chemistry on Illumina NovaSeq instruments through the sequencing service by Azenta company. A detailed list of all primers and plasmids used in this paper can be found in Supplementary Data 1.
Measuring the activity of 5’ RSs by sfgfp gene expression
The strains containing the targeted plasmid were stored as 20% glycerol stocks in sterile centrifuge tubes (1.5 ml). E. coli and P. aeruginosa with target plasmid were picked out using a sterilized metal pinner and grown on plates containing 5 ml of LB medium with 50 μg/ml and 150 μg/ml carbenicillin respectively. Monoclonal selections were performed overnight (16 hours) in a 5 mL shaker tube at 37 °C with shaking on an orbital shaker. Then the overnight cultures were diluted 1:100 into a final volume of 1 ml of fresh LB medium with the appropriate carbenicillin concentration and grown for another 6~8 h. Then, the 150 μl of culture was added to each well of clear bottom black plates, and repeated measurements of the optical density at 600 nm (OD600) and fluorescence (relative fluorescence units [RFU]; excitation at 485 nm and emission at 520 nm) were performed with a microplate reader (EnVision 2105). 96 well microplates were analyzed using EnVision Workstation (version 1.14.3049.1534). The process of evaluating promoter activity in the M9 (Coolaber, SL0060) and EZ-rich (Coolaber, MK0100) culture medium was performed in the same way as in the LB medium. All experiments were conducted in three biological replicates.
Quantifying RSs’ transcription activity levels
For the transcription activity measured by the MPRA experiment, the overall data processing method is mainly divided into two steps according to the MPRAflow processing protocol51. The first step is to calculate the correspondence between the RSs and the barcodes. The upstream sequencing result information was compared with the input sequence library and the corresponding barcode in the downstream file through the unique tags in the paired file. Since a sequence often contains multiple barcodes, a sequence-barcode dictionary can be obtained by comparing and splicing reads. The second step is to calculate the activity of RSs. The RNA amount corresponding to each piece of cDNA was calculated. The final splicing result is a 239 bp sequence, of which the last 20 bp is a barcode sequence. Finally, the first 20 bp of the “sequence barcode” dictionary was used in the first step, and the reverse complementation sequence to obtain a positive chain barcode as the barcode identifier. After this filtering step, the transcription activity of each regulatory element was calculated using the following formula:
where each barcode is represented by j, ∑j∊p RNAj is the sum of the number of reads of each cDNA barcode sequence combined with the regulatory element p in the cDNA library sequencing results. The term ∑j∊p DNAj is the sum of the number of reads of each DNA barcode sequence combined with the regulatory element p in the final plasmids library sequencing results. The term Nbc_nonzero is the non-zero barcode number corresponding to the RNA expression of the regulatory element p. The CRNA and CDNA are normalization coefficients for the size of the cDNA library and RNA library, respectively. RNA libraries and DNA libraries were normalized to the size of 106, eliminating the impact of sequencing depth on the transcription activity results.
The activity of RSs optimized by the GA was measured based on the expression of the sfgfp gene. The activities of RSs are calculated as follows52:
where F refers to the fluorescence (relative fluorescence units [RFU]; excitation at 485 nm and emission at 520 nm) and the control plasmid used the BBa_J23119 RS from Biobrick53. The blank control refers to a plasmid without a promoter sequence. The final RSs’ activities were calculated by taking the average of three independent biological repeated experiments. The activity of sequence measured by MPRA and the sfgfp gene showed a highly correlation, wth Pearson r2 values of 0.765 for E. coli and 0.840 for P. aeruginosa (Supplementary Fig. 4b, Supplementary Data 3).
Statistical analysis
Comparisons between two groups of data in this study were analyzed by two-tailed t-test. Multiple comparisons among three or more groups were analyzed by pairwise comparison and were corrected for multiple comparisons. All statistical analyses were performed using SciPy package in Python. GraphPad Prism (version 9.0.0) and Adobe Illustrator (version 25.0.1) were used to generate and prepare all figures.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The primers and plasmids used in this study are available in Supplementary Data 1. The Lib-1 and Lib-2 MPRA data generated in this study are available in Supplementary Data 2. The RSs tested using the sfgfp gene, as well as the natural sequences and random sequences used for ClustalW2 analysis in this study, are available in Supplementary Data 3. The validated natural 5’RSs in bacteria proposed by Johns et al. used in this study are available at https://static-content.springer.com/esm/art%3A10.1038%2Fnmeth.4633/MediaObjects/41592_2018_BFnmeth4633_MOESM4_ESM.xlsx. The metagenome species proposed by Fahimipour et al. used in this study are available at https://figshare.com/articles/software/Diffusion_mapping_bacterial_metabolic_traits/12864011/4. The datasets used for pretraining and fine-tuning in this study are available in the Zenodo repository under accession code 1459856754. The raw sequence data in this study have been deposited in the Genome Sequence Archive (GSA)55 in National Genomics Data Center56, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences under accession codes: CRA018904. Source data are provided with this paper.
Code availability
The source code used to develop the DeepCROSS in this study is publicly available and has been deposited in GitHub repository at https://github.com/WangLabTHU/DeepCROSS, under an MIT license. The specific version of the code associated with this publication is archived in Zenodo repository and is accessible via https://doi.org/10.5281/zenodo.1460029557.
References
Hu, E.-Z., Lan, X.-R., Liu, Z.-L., Gao, J. & Niu, D.-K. A positive correlation between GC content and growth temperature in prokaryotes. BMC Genomics 23, 110 (2022).
Pishdadian, K., Fimlaid, K. A. & Shen, A. SpoIIID-mediated regulation of σK function during Clostridium difficile sporulation. Mol. Microbiol. 95, 189–208 (2015).
Lynch, S. A. & Gill, R. T. Synthetic biology: new strategies for directing design. Metab. Eng. 14, 205–211 (2012).
Sheth, R. U., Cabral, V., Chen, S. P. & Wang, H. H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 32, 189–200 (2016).
Kotula, J. W. et al. Programmable bacteria detect and record an environmental signal in the mammalian gut. Proc. Natl. Acad. Sci. USA 111, 4838–4843 (2014).
Patel, J. R., Oh, J., Wang, S., Crawford, J. M. & Isaacs, F. J. Cross-kingdom expression of synthetic genetic elements promotes discovery of metabolites in the human microbiome. Cell 185, 1487–1505.e14 (2022).
Johns, N. I. et al. Metagenomic mining of regulatory elements enables programmable species-selective gene expression. Nat. Methods 15, 323–329 (2018).
Kolber, N. S., Fattal, R., Bratulic, S., Carver, G. D. & Badran, A. H. Orthogonal translation enables heterologous ribosome engineering in E. coli. Nat. Commun. 12, 599 (2021).
Kang, M.K. et al. Synthetic biology platform of CoryneBrick vectors for gene expression in Corynebacterium glutamicumand its application to xylose utilization. Applied Microbiology & Biotechnology 98, 5991–6002 (2014).
Song, Y. et al. Promoter screening from Bacillus subtilis in various conditions hunting for synthetic biology and industrial applications. PLoS One 11, e0158447 (2016).
Makhzani, A., Shlens, J., Jaitly, N., Goodfellow, I. & Frey, B. Adversarial Autoencoders. Preprint at https://arxiv.org/abs/1511.05644 (2015).
Hie, B., Bryson, B. D. & Berger, B. Leveraging uncertainty in machine learning accelerates biological discovery and design. Cell Syst. 11, 461–477.e9 (2020).
Wittmann, B. J., Yue, Y. & Arnold, F. H. Informed training set design enables efficient machine learning-assisted directed protein evolution. Cell Syst. 12, 1026–1045.e7 (2021).
Arnold, F. H. The library of Maynard-Smith: My search for meaning in the protein universe. Microbe Wash. DC 6, 316–318 (2011).
Romero, P. A., Krause, A. & Arnold, F. H. Navigating the protein fitness landscape with Gaussian processes. Proc. Natl. Acad. Sci. USA 110, E193–E201 (2013).
Qiu, C. et al. Nanophotonic inverse design with deep neural networks based on knowledge transfer using imbalanced datasets. Opt. Express 29, 28406–28415 (2021).
Fahimipour, A. K. & Gross, T. Mapping the bacterial metabolic niche space. Nat. Commun. 11, 4887 (2020).
Tewhey, R. et al. Direct identification of hundreds of expression-modulating variants using a multiplexed reporter assay. Cell 172, 1132–1134 (2018).
Foerstner, K. U., von Mering, C., Hooper, S. D. & Bork, P. Environments shape the nucleotide composition of genomes. EMBO Rep. 6, 1208–1213 (2005).
Vogl, T., Ruth, C., Pitzer, J., Kickenweiz, T. & Glieder, A. Synthetic core promoters for Pichia pastoris. ACS Synth. Biol. 3, 188–191 (2014).
Feklístov, A., Sharon, B. D., Darst, S. A. & Gross, C. A. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 68, 357–376 (2014).
Paget, M. S. B. & Helmann, J. The σ 70 family of sigma factors. Genome Biology 4, 1–6 (2003).
Biswas, S., Khimulya, G., Alley, E. C., Esvelt, K. M. & Church, G. M. Low-N protein engineering with data-efficient deep learning. Nat. Methods 18, 389–396 (2021).
Kulmanov, M. & Hoehndorf, R. DeepGOPlus: improved protein function prediction from sequence. Bioinformatics 37, 1187 (2021).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770–778 (2016).
Kelley, D. R., Snoek, J. & Rinn, J. L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 26, 990–999 (2016).
Vaswani, A. et al. Attention is all you need. Preprint at https://arxiv.org/abs/1706.03762 (2017).
Huang, G., Liu, Z., Van Der Maaten, L. & Weinberger, K. Q. Densely connected convolutional networks. In IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 4700–4708 (2017).
Järvelin, K. & Kekäläinen, J. Cumulated gain-based evaluation of IR techniques. ACM Trans. Inf. Syst. 20, 422–446 (2002).
Hossain, A. et al. Automated design of thousands of nonrepetitive parts for engineering stable genetic systems. Nat. Biotechnol. 38, 1466–1475 (2020).
Patel, S., Panchal, H. & Anjaria, K. Phylogenetic analysis of some leguminous trees using CLUSTALW2 bioinformatics tool. In IEEE International Conference on Bioinformatics and Biomedicine Workshops, 917–921 (2012).
Kosuri, S. et al. Composability of regulatory sequences controlling transcription and translation in Escherichia coli. Proc. Natl. Acad. Sci. USA 110, 14024–14029 (2013).
Gilchrist, C. L. M. et al. Cblaster: A remote search tool for rapid identification and visualization of homologous gene clusters. Bioinform. Adv. 1, vbab016 (2021).
Samee, M. A. H., Bruneau, B. G. & Pollard, K. S. A DE Novo shape motif discovery algorithm reveals preferences of transcription factors for DNA shape beyond sequence motifs. Cell Syst. 8, 27–42.e6 (2019).
Zhou, T. et al. DNAshape: a method for the high-throughput prediction of DNA structural features on a genomic scale. Nucleic Acids Res. 41, W56–W62 (2013).
Li, J., Chiu, T.-P. & Rohs, R. Predicting DNA structure using a deep learning method. Nat. Commun. 15, 1243 (2024).
Kushwaha, M. & Salis, H. M. A portable expression resource for engineering cross-species genetic circuits and pathways. Nat. Commun. 6, 7832 (2015).
Gaida, S. M. et al. Expression of heterologous sigma factors enables functional screening of metagenomic and heterologous genomic libraries. Nat. Commun. 6, 7045 (2015).
Sharon, E. et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat. Biotechnol. 30, 521–530 (2012).
Tian, C., Li, J. & Wang, Y. From qualitative to quantitative: the state of the art and challenges for plant synthetic biology. Quant. Biol. 11, 214–230 (2023).
Bayer, T. S. et al. Synthesis of methyl halides from biomass using engineered microbes. J. Am. Chem. Soc. 131, 6508–6515 (2009).
Fukazawa, T., Maeda, Y., Sladek, F. M. & Owen-Schaub, L. B. Development of a cancer-targeted tissue-specific promoter system. Cancer Res. 64, 363–369 (2004).
Hong, J., Gao, R. & Yang, Y. CrepHAN: cross-species prediction of enhancers by using hierarchical attention networks. Bioinformatics 37, 3436–3443 (2021).
MacPhillamy, C., Alinejad-Rokny, H., Pitchford, W. S. & Low, W. Y. Cross-species enhancer prediction using machine learning. Genomics 114, 110454 (2022).
Peng, S., Huang, H., Wei, P. & Xie, Z. Synthetic gene circuits moving into the clinic. Quant. Biol. 9, 100–103 (2021).
Brookes, D. H., Park, H. & Listgarten, J. Conditioning by adaptive sampling for robust design. Proceedings of the 36th International Conference on Machine Learning, PMLR 97, 773–782 (2019).
Leyn, S. A. et al. Comparative genomics and evolution of transcriptional regulons in Proteobacteria. Microb. Genom. 2, e000061 (2016).
Senior, A. W. et al. Improved protein structure prediction using potentials from deep learning. Nature 577, 706–710 (2020).
Whitley, D. A genetic algorithm tutorial. Stat. Comput. 4, 65–85 (1994).
Yu, T. C. et al. Multiplexed characterization of rationally designed promoter architectures deconstructs combinatorial logic for IPTG-inducible systems. Nat. Commun. 12, 325 (2021).
Gordon, M. G. et al. Author Correction: lentiMPRA and MPRAflow for high-throughput functional characterization of gene regulatory elements. Nat. Protoc. 16, 3736 (2021).
Meng, H. et al. Quantitative design of regulatory elements based on high-precision strength prediction using artificial neural network. PLoS One 8, e60288 (2013).
Smolke, C. D. Building outside of the box: iGEM and the BioBricks foundation. Nat. Biotechnol. 27, 1099–1102 (2009).
Zhang, P., Du, Q., Wang, Y. & WangLabTHU. (Datasets) Systematic representation and optimization enable the inverse design of cross-species regulatory sequences in bacteria. Zenodo https://doi.org/10.5281/ZENODO.14598567 (2025).
Chen, T. et al. The Genome Sequence Archive family: Toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics 19, 578–583 (2021).
CNCB-NGDC Members and Partners. Database resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 50, D27–D38 (2022).
Zhang, P., Du, Q., Wang, Y. & WangLabTHU. (Toolkits) Systematic representation and optimization enable the inverse design of cross-species regulatory sequences in bacteria. Zenodo https://doi.org/10.5281/ZENODO.14600295 (2025).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 62250007, 62225307 to X.W.W.), and the grant from the Guoqiang Institute, Tsinghua University (2021GQG1023 to X.W.W.). We also thank Zheng Wei, Guanhua Hu, Haochen Wang, and Wenbo Guo for the positive discussion.
Author information
Authors and Affiliations
Contributions
X.W.W., Y.W., P.C.Z., and Q.X.D. conceived the study. Y.W. and Q.X.D. implemented in silico designs. P.C.Z. and Y.W. designed the experiments and performed the experiments. P.C.Z., Q.X.D., Y.W., and X.W.W. analyzed the data. Q.X.D. built the statistical modeling method for sequence optimization. P.C.Z., Y.W., Q.X.D., W.L., and X.W.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Wai Low, Yi Wu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, P., Du, Q., Wang, Y. et al. Systematic representation and optimization enable the inverse design of cross-species regulatory sequences in bacteria. Nat Commun 16, 1763 (2025). https://doi.org/10.1038/s41467-025-57031-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-57031-1