Abstract
Deep learning (DL) enabled liquid-based cytology has potential for cervical cancer screening or triage. Here, we develop a DL model using whole cytology slides from 17,397 women and test it on 10,826 additional cases through a three-stage process. The DL model achieves robust performance across nine hospitals. In a multi-reader, multi-case study, it outperforms cytopathologists’ sensitivity by 9%. Reading time significantly decreases with DL assistance (218s vs 30s; p < 0.0001). In community-based organized screening, the DL model’s sensitivity matches that of senior cytopathologists (0.878 vs 0.854; p > 0.999), yet it has reduced specificity (0.831 vs 0.901; p < 0.0001). Notably, hospital-based opportunistic screening shows that junior cytopathologists with DL assistance significantly improve both their sensitivity and specificity (0.857 vs 0.657, 0.840 vs 0.737; both p < 0.0001). When triaging human papillomavirus-positive cases, DL assistance exhibits better performance than junior cytopathologists alone. These findings support using the DL model as an assistance tool in cervical screening and case triage.
Similar content being viewed by others
Introduction
Cervical cancer is one of the most common gynecologic tumors, with 662,301 new cases and 348,874 deaths worldwide in 20221. Cytology-based screening is associated with significant declines in mortality, but this is limited by low sensitivity and significant heterogeneity across different providers, particularly in low-middle income countries2,3. Human papillomavirus (HPV) testing has been recommended for primary cervical screening due to high sensitivity and long-term reassurance after a negative test result4. However, HPV infections can be transient, which influences specificity. In high-resource countries, the screening strategy is often based on co-testing for HPV and cytology every five years to identify those at increased risk5. However, HPV testing is not yet widely available across all regions, creating barriers to screening coverage6. Countries with quality-assured cytology programs can justifiably continue to utilize cytology as a primary screening test for the general population and even if HPV testing is available, cytology is preferred for triaging HPV-positive women7. The widespread use of cytology over the past several decades is due to utility and applicability, making it a crucial component of national screening programs in 109 countries8. However, this approach relies on cytopathologists’ subjective judgments, making it both labor-intensive and time-consuming.
Traditional microscopy is gradually transitioning to a digital process which has facilitated the development of artificial intelligence (AI), specifically deep learning (DL) in cytopathology (see Supplementary Note 1 for literature review). Recently, the U.S. Food and Drug Administration approved a Hologic Genius Digital Diagnostic System with DL assistance to identify abnormal cells9. This system uses automated scanning to digitize slides and generate high-resolution cell images. This has the potential to improve both diagnostic accuracy and efficiency by reducing reading time. Several studies have presented supportive findings related to the technical feasibility of DL models for abnormal cell detection10,11,12,13. However, there is insufficient evidence around the clinical impact of DL models on disease outcomes and it remains necessary to conduct further investigations with histological outcomes as the reference standard. In addition, it is also unlikely that these AI tools will be used independently until more is known about the effects, safety, clinical acceptance, or the implications for public trust. Therefore, we do not yet know whether DL tools can help cytopathologists improve diagnostic performance and few have analysed the integration of DL-based cytology solutions in different scenarios.
In this study, we developed a DL model to detect cervical intraepithelial neoplasia grade 2 or worse (CIN2+ ). Initially, we assessed the generalizability of this DL model using liquid-based cytology (LBC) slides from several different hospitals across China, which had not been previously analyzed with this model. The Findings were directly compared with those from human cytopathologists. Then, a multi-reader multi-case (MRMC) study was performed to investigate whether cytopathologists improve their diagnostic performance with DL assistance. Finally, we applied the DL model in two different screening scenarios to validate its performance and to assess its influence on clinical practice.
Results
Study population
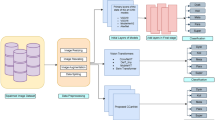
In total, 19,318 slides were obtained for the training set from two independent pathology archives. After quality control, 1921 slides were excluded. 17,397 slides remained and were used to develop the DL model. For the three-stage stepwise validation, we obtained four datasets, which included 5803 slides (test set A), 550 slides (test set B), 3001 slides (test set C) and 1472 slides (test set D). An overview of the study design is presented in Fig. 1. Details of the characteristics of the modules and each dataset are provided in Supplementary Tables 1-4 and Supplementary Fig. 1.
This DL model was trained to classify LBC digital slides as positive or negative cases using data from two independent pathology archives. By histological reference standards, the DL model was then tested in a stepwise validation study which had three stages (test sets A–D). First, a multi-institutional validation study was performed to assess the generalizability across nine hospitals. Second, diagnostic performance and efficiency of cytopathologists before and after DL assistance was assessed in a multi-reader multi-case study. Third, diagnostic performance and referral efficiency between senior cytopathologists and DL alone were evaluated using a cloud platform in a community-based organized screening population. Diagnostic performance and referral efficiency of junior cytopathologists before and after DL assistance was assessed locally in a hospital-based opportunistic screening population. DL Deep learning, LBC Liquid-based cytology.
Multi-institutional validation study
As part of the multi-institutional validation study (stage 1), we analysed the diagnostic performance of our DL model at CIN2+ detection (Table 1). DL alone achieved a pooled accuracy of 0.832, a sensitivity of 0.861 and a specificity of 0.829, when generalized across nine different hospitals. DL accuracy varied between 0.798 to 0.857, as did sensitivity (ranging from 0.833 to 0.883) and specificity (ranging from 0.794 to 0.856). Area under the curve (AUC) values ranged from 0.814 to 0.868 (Fig. 2A). The most common false positives and false negatives are presented in Supplementary Fig. 2.
Multi-institutional validation study (A–C): A ROC curves for diagnostic performance of DL alone across nine different hospitals. B Diagnostic performance of DL model compared with each junior cytopathologist. Blue dots indicate diagnostic sensitivities and specificities for individual junior cytopathologists. Blue rhombus indicates the average sensitivity and specificity for all junior cytopathologists. C Diagnostic performance of DL model compared with each senior cytopathologist. Orange dots indicate sensitivities and specificities for individual senior cytopathologists. Orange rhombus indicates the average sensitivity and specificity of all senior cytopathologists. Multi-reader multi-case study (D–F): D Diagnostic sensitivities and specificities of individual junior and senior cytopathologists before and after DL assistance. Blue dots indicate sensitivities and specificities of individual cytopathologists without DL assistance. Orange triangles indicate sensitivities and specificities of individual cytopathologists with the use of DL assistance. E ROC curves for diagnostic performance of DL alone, junior and senior cytopathologists with and without DL assistance. Blue and orange dots indicate sensitivities and specificities of individual junior cytopathologists without and with DL assistance, respectively. Blue and orange rhombi indicate sensitivities and specificities of individual senior cytopathologists without and with DL assistance, respectively. Blue and orange rhombi indicate average sensitivities and specificities of all junior cytopathologists without and with DL assistance, respectively. Blue and orange stars indicate average sensitivities and specificities of all senior cytopathologists without and with DL assistance, respectively. F The average reading time is measured for all cytopathologists (n = 28), junior cytopathologists (n = 16), and senior cytopathologists (n = 12) with and without DL assistance. The data represent independent assessments by each group, with biological replicates defined as separate assessments by different cytopathologists with and without DL assistance. The upper and lower bounds of the box represent the 75th percentile (Q3) and 25th (Q1) percentile, respectively. The line within the box indicates the median. The rhombus outside of whiskers refer to outliers. The time of review per case is described as the median and interquartile range (IQR). CIN2+ Cervical intraepithelial neoplasia grade 2 or worse, AUC Area under the receiver operating characteristic curves, DL Deep learning, APH Anhui Provincial Hospital, GZPH Guangxi Zhuang Autonomous Region People’s Hospital, HHMU The First Affiliated Hospital of Hainan Medical University, WCSUH West China Second University Hospital, GHPLA The 7th Medical Center, General Hospital of PLA, SMCHH Shenzhen Maternity and Child Healthcare Hospital, NWCH Northwest Women’s and Children’s Hospital, XH Xijing Hospital, ZCH Zhejiang Cancer Hospital, IQR Interquartile Range.
Additionally, we stratified the 5803 slides (test set A) from nine hospitals based on the level of expertise of each cytopathologist who conducted initial diagnoses. Specifically, the subset comprising 2147 slides was evaluated by junior cytopathologists at three hospitals, whereas the remaining 3656 slides were diagnosed by senior cytopathologists at six different hospitals. Within this independent subset of 2147 slides, the DL model had higher sensitivity and specificity than the average of three junior cytopathologists (0.850 vs 0.719, 0.822 vs 0.746, both p < 0.0001). See Fig. 2B and Supplementary Table 5. In another independent subset of 3656 slides, the DL model had comparable sensitivity (0.867 vs 0.854, p = 0.572) but lower specificity (0.834 vs 0.888, p < 0.0001) than the average of six senior cytopathologists. See Fig. 2C and Supplementary Table 6 for further details.
Multi-reader multi-case study
In the second stage of the MRMC study, we compared the DL model to cytopathologists’ readings of 550 digitized slides. The sensitivity and specificity of DL significantly exceeded the average level of all 28 cytopathologists (0.870 vs 0.780, 0.831 vs 0.813, both p < 0.0001). However, with DL assistance, these cytopathologists had higher average sensitivity and specificity (0.874 vs 0.780, 0.852 vs 0.813, both p < 0.0001; Table 2). Individual cytopathologists’ sensitivity without DL varied from 0.609 to 0.913, and specificity varied from 0.677 to 0.948. After adding DL assistance, sensitivity varied from 0.804 to 0.935, and specificity varied from 0.762 to 0.972, but the greatest improvement was observed in junior cytopathologists (Fig. 2D and Supplementary Table 7).
The average sensitivity and specificity of junior cytopathologists significantly improved (0.717 vs 0.858, 0.750 vs 0.815, both p < 0.0001), but there was no difference for senior cytopathologists before and after adding DL assistance (0.864 vs 0.897, p = 0.073; 0.898 vs 0.903, p = 0.300). Receiver operating characteristic (ROC) curves are provided in Fig. 2E. In addition, the average review time was reduced with the use of DL compared to without (218s vs 30s per case, p < 0.0001, Fig. 2F). Similar results were observed in subgroups of junior and senior cytopathologists (Supplementary Table 8).
The application validation study
In the two application validation studies (stage 3), we investigated influence of the DL model on cytopathologists’ performances in the community-based organized screening and hospital-based opportunistic screening. Both patterns of local service and the cloud-based platform were made available to allow cytopathologists to upload digital slides for DL analysis (Supplementary Fig. 3). The community-based organized screening study (test set C) represented a general population with a CIN2+ prevalence (1.366 cases per 100 women), and consisted of 41 CIN2+ cases and 2960 <CIN2 (Supplementary Table 3). Compared to senior cytopathologists, the specificity of DL alone was lower (0.831 vs 0.901, p < 0.0001), but sensitivity was comparable (0.878 vs 0.854, p > 0.999, Fig. 3A, B).
The application validation study 1 was conducted in a community-based organized screening population (A–C): A ROC curves for diagnostic performance of DL alone and senior cytopathologists. Blue triangles indicate the average sensitivities and specificities of senior cytopathologists. B Diagnostic accuracy, sensitivity and specificity between DL alone and the average of senior cytopathologists. c Referral efficiency includes colposcopy referrals and NNR between DL alone and the average of senior cytopathologists. The application validation study 2 was conducted in a hospital-based opportunistic screening population (D–F): D ROC curves for diagnostic performance of DL alone and junior cytopathologists with and without DL assistance. Blue and orange triangles indicate the average sensitivities and specificities of junior cytopathologists with and without DL assistance, respectively. E Diagnostic accuracy, sensitivity and specificity, for DL alone and junior cytopathologists with and without DL assistance. F Referral efficiency including colposcopy referrals and NNR for DL alone and junior cytopathologists with and without DL assistance. CIN2+ Cervical intraepithelial neoplasia grade 2 or worse, AUC Area under the receiver operating characteristic curves, DL Deep learning, NNR Number of colposcopies required to yield one CIN2+.
Positive results initiated colposcopy referrals, and senior cytopathologists had a lower rate of colposcopy referrals (0.109 vs 0.179, p < 0.0001) with few number of colposcopies required to yield one CIN2+ (NNR, 9.371 vs 14.889, respectively), compared to the DL model alone (Fig. 3C). To extend our analysis and simulate HPV testing as a primary screening method, we compared triage performance of DL alone to seniors for HPV-positive or non-HPV16/18-positive women. Similar to the overall assessment, DL alone was equally sensitive but had lower specificity and a higher rate of referrals compared to seniors alone (Supplementary Table 9).
The hospital-based opportunistic screening study (test set D) represented an opportunistically screened population with a CIN2+ prevalence of 4.755 per 100 women, and included 70 CIN2+ cases and 1402 <CIN2 (Supplementary Table 3). Through initial diagnoses, juniors without DL assistance had a sensitivity of 0.843 and a specificity of 0.830 (Fig. 3D, E). However, sensitivity associated with juniors significantly improved from 0.657 to 0.857, and specificity from 0.737 to 0.840 (both p < 0.0001).
In terms of clinical efficiency, DL assistance reduced the overall colposcopy referral rate by one-third, from 0.282 to 0.193 (p < 0.0001) compared to juniors alone (Fig. 3F). This resulted in a lower number of colposcopies required to detect one CIN2+ case (NNR, 4.733 vs 9.022, respectively). Similar to the overall assessment observed in HPV-positive or non-HPV16/18-positive women, DL assistance yielded higher sensitivity, specificity and a lower referral rate than juniors alone (Supplementary Table 10).
Discussion
We developed and validated a DL model using LBC slides for detecting CIN2+. DL alone was effective and robust in analyzing data from geographically different hospitals across China, exceeding the average performance of cytopathologists. Notably evidence from clinical practice suggested juniors can significantly improve their diagnoses with DL assistance. Of course, efficiency would also be improved in relation to the reading time required but also by reducing the number of unnecessary colposcopy referrals. Similar results were observed when DL model was used as a triage method for HPV-positive women. These findings support the use of DL to assist cytopathologists, especially junior practitioners.
The DL model proved both effective and robust. It was developed using slides from 17,397 women, and was externally validated across different geographical, independent and heterogeneous datasets. This is distinguished from other available DL models14,15 based on LBC slides for detecting CIN2+ which have been developed using single center datasets for training and conducted without external validation. To improve upon the existing evidence base, we included a wider variety of slides including negative, squamous and those with glandular cells abnormalities, which were created by specialists with different professional statuses using different devices. Including a greater number of scanners (developed by different manufacturers) ensures the DL model is more generalizable to a diverse range of settings. In this study, we applied the DL model to previously unseen data from nine hospitals in China. We found that the DL alone can achieve high diagnostic performance although it is yet to be seen whether this model will prove sufficiently sensitive and specific for populations outside the Chinese mainland.
This model has the potential to improve cytological practice by assisting in diagnostic processes. Cytopathologists using conventional microscopy spend a long time looking for suspicious lesions among thousands of cells or clusters within a single slide. This prolonged process is prone to distractions which can result in missed diagnoses16. Our DL model can analyze whole slides within milliseconds, providing cytopathologists with a series of the most diagnostically important regions of interest (ROIs). Evidence demonstrates that some DL models can match and even exceed human cytopathologists17,18,19. Therefore, some suggest that these technologies will replace cytopathologists as independent screeners15. However, these advanced technologies can also mismatch, omit key subimages and can generate errors. These issues are forcing this field of research to consider AI models as supplementary tools rather than being replacements for human practitioners. Our MRMC study findings support this assertion suggesting the most important role of this DL model is to improve the diagnostic abilities and reading efficiencies by assisting cytopathologists, which could alleviate the burden on health-care systems, especially in large-scale population screening. This proposition may seem unduly tentative given the reading time reduction observed in this study; however, we do not want cytopathologists to become completely reliant on DL tools. Human-in-the-loop AI has not been fully tested and should enable cytopathologists to focus on more obscure cases with a higher risk of misdiagnosis, rather than spending time on clearer cases.
The application validation studies conducted in both the community-based organized screening and hospital-based opportunistic screening enabled us to consider changes in the disease spectrum. Previous studies in rural Kenya have shown the feasibility of DL models for pap smear20; however, the approach implemented had methodological weaknesses including limited training and testing samples, and the researchers used cytological outcomes as opposed to histological outcomes, which hindered the application of their DL model into clinical practice. This study was designed to assess the application of this tool and we found the DL model to be both reliable and effective in practice. In the community-based organized screening, which had a low CIN2+ prevalence, of prospectively collected 3001 LBC slides, the DL model detected four new CIN2+ cases which were missed by senior cytopathologists. However, specificity was lower than that of seniors, although sensitivity was comparable. Further investigation showed that most false positives were due to DL misclassifying true negatives as atypical squamous cells of undetermined significance or worse (ASC-US), and false negatives were due to DL misclassifying ASC-US as true negatives. We suggest this could be improved in the future with ASC-US training based upon histological outcomes although this would require further research. Sensitivity or specificity could also be improved by tweaking algorithmic configurations based on refined cut-off values to match requirements. However, this again raises questions around the positioning of AI assistance and the implications generally. It may be necessary to develop a safety mechanism, where cytopathologists can easily and quickly find ROIs with potential abnormal cells identified by DL.
In the hospital-based opportunistic screening, which had a CIN2+ prevalence of 4.755 per 100 women, we found that junior cytopathologists benefit most from DL support. There were significant increases in both sensitivity from 0.657 to 0.857 and specificity from 0.737 to 0.840, which were comparable to that reported in a meta-analytical study21. This consistent finding suggests that junior cytopathologists will derive significant benefit, particularly in county hospitals across China but also across the world. This assistance becomes crucial in environments where experienced professionals are few and far between, enabling more accurate and timely diagnoses for patients, who previously had to wait weeks before receiving their screening results. This makes our DL model an affordable solution to improve practice.
Several challenges exist when integrating the DL model into clinical workflow. First, unsatisfactory sample processing, staining and preparation including deep dyeing, random dots and speckles, blurring and the influence of untrained operators may impact scanning quality, which could be mistaken by DL. However, these processes were performed by experienced cytotechnologists in a standardized laboratory setting which strictly controlled for interferences in our study. This is not always possible in community hospitals. Second, the absence of automatic high-volume, specialized scanners slows DL model-based screening processes. Currently, the cost of scanners can be prohibitive and a possible solution would be to transport slides to a nearby ___location which possesses a whole slide scanner22. This analytical centralization may lower labour costs and help standardize identification but it will also come with logistics challenges. Therefore, it may be necessary to develop a portable scanner which can be shared among facilities23. Third, despite cloud servers, slow internet connectivity may result in locally delayed image uploading and diagnostic result generation. The provision of portable wireless networks at each site mitigated internet reliability issues. However, augmenting data management in high-volume community hospitals and outpatient clinics requires an integrated image compression algorithm or an offline DL model for optimization. Fourth, introducing the DL model into new environments necessitates optimization and validation to ensure it is feasible. This step is crucial for adapting the model to different datasets and clinical settings, ensuring that it performs reliably across various populations and regions. Fifth, ensuring the DL model is economical in different health systems requires a cost-effectiveness analysis that balances implementation costs against potential savings resulting from reduced false positives and negatives. Additionally, the model must be scalable and integrate seamlessly with existing workflows to be economically viable in both well-funded or resource-limited environments. In practice, we also need to foster trust in DL models among cytotechnologists as this is crucial for effective integration. As the model is introduced, it will impact cytopathology practices, potentially altering job roles and necessitating retraining to ensure professionals can effectively interpret and act on AI-generated insights. This transition should be managed to maintain job satisfaction by positioning the DL model as a supportive tool rather than a replacement24.
We also found that using DL model as the cytology-based screening method alone resulted in a higher referral rate for colposcopy, but as evidenced by screening of 25,404 Chinese women in 13 population-based pooled studies25,26, this referral rate is common in China and is influenced by the specific screening population and regional practices. To address this, we propose the DL model could be suitable for triaging HPV-positive women, where it improved junior cytopathologists’ triage performance by significantly reducing unnecessary colposcopies. This was consistent with the findings from our previous study which highlighted a reduction in colposcopic referrals by approximately 10%27. This aligns with the World Health Organization recommendation that HPV testing be the mainstream primary screening method with triaging HPV-positive women. Moreover, self-sampling for HPV screening combined with AI triage is appealing in middle-to-high resource settings, but currently it’s limited as it doesn’t provide material for cytological analysis. Future developments are needed to enhance self-sampling techniques for cytological use. However again, the positioning of DL model will depend upon the allocation of health resources. There is clearly a need for further health economics research and to assess the duration of assurance of negative DL results against precancer to determine a safe screening interval. It is also necessary to understand its effectiveness through longer-term surveillance.
This study has some limitations. First, we did not have biopsies from every woman who provided slides, and therefore could not ascertain disease status for all. Women with negative screening results for both hr-HPV and cytology were assumed to be at very low risk of developing high-grade cervical lesions, which may have affected our results. Second, the validated cohorts in our study exhibited a higher prevalence of CIN2+, which may not reflect the characteristics of the general screening population. Therefore, while these metrics provide valuable insights within the context of our specific study cohorts, their applicability to the general population may be limited and should be interpreted cautiously. Third, poor images were excluded to ensure data quality. However, the main reason is their susceptibility to fading over time, which is primarily attributed to factors such as prolonged light exposure, inadequate storage conditions, suboptimal mounting medium quality, inconsistencies in the staining process, and issues with chemical reagents. Therefore, it is recommended that cytology laboratory staff ensure the quality of slide preparation and staining, and that slides are scanned promptly to preserve the integrity of the cellular details. Fourth, while the cell-level performance of DL model was not shown, we directly assessed its slide-level performance, which may offer greater utility than focusing on individual cells or clusters. Finally, the presence of atypical glandular cell (AGC) could help identify adenocarcinoma precursors, but this was not measured due to the limited number of cases. This aspect requires further investigation.
In conclusion, cytopathologists’ diagnostic accuracy and efficiency were substantially improved when detecting CIN2+ with DL assistance. DL has potential for practice and is suitable for both screening and triage. With a cloud platform, this DL model could be globally accessible and would serve as an ‘assistant’ which would be particularly beneficial in areas where experienced cytopathologists are scarce. Furthermore, regardless of a country’s transitional status or advancement in cytology-based screening, the implementation of DL model in combination with point-of-care testing could lead to timely detection and intervention for cervical cancer, thereby improving screening or triage performance.
Methods
Ethical approval
This study was approved by the institutional review board of the Chinese Academy of Medical Sciences and Peking Union Medical College (IEC-2021-023; IEC-2022-022) and Peking Union Medical College Hospital (KS2023129). The need for informed consent was waived because this study involved the retrospective LBC slide and text data collection, as these were anonymized. However, written informed consent was obtained from patients whose LBC slides and text data were prospectively collected. All data were de-identified prior to model training and testing.
Study design and datasets
This was a multicentre, diagnostic study using LBC slides from 13 clinical hospitals in mainland China. The study comprised a training phase and a testing phase with separate sets of LBC slides (one slide per woman). The training set, consisting of 17,397 LBC slides with their cytological results, was retrospectively extracted from two independent datasets from the institutes of pathology in Chongqing and Guangzhou between February 2020 and August 2021. The testing phase was conducted in three stepwise stages with distinct test sets: Stage 1 involved a multi-institutional validation study where we retrospectively collected 5803 LBC slides (test set A) from nine different hospitals between October 2021 and November 2022 to evaluate the model’s generalizability, ensuring its reliability for broader clinical use. Stage 2 featured the MRMC study, where we randomly selected 550 slides (test set B) from stage 1 between December 2022 and January 2023 to compare the diagnostic performance and efficiency of a total of 28 cytopathologists with and without DL assistance. Stage 3 encompassed two application validation studies: one with 3001 prospectively collected slides (test set C) from a community-based organized screening population between December 2022 and April 2023, and the other with 1472 slides (test set D) prospectively obtained from four independent tertiary hospitals between April and September 2023. All slides were collected and scanned within three months after cytological samples were prepared to prevent fading and there was no overlap between the training set and any test sets, ensuring an independent evaluation.
Women aged 18 years or older with intact uteri and no history of pelvic radiation or hysterectomy were considered eligible. All participants provided LBC samples taken using one of the three widely used products (ThinPrep, Hologic, USA; SurePath, BD, USA; LBP, Guangzhou LBP, China). Whole slides were processed and digitized at a 0.18 μm/pixel resolution using ×20 magnification with one of three scanners (IBL-300 Scanner, Bingli, China; SQ-600 Scanner, Shengqiang, China; EasyScan NFC-60 Scanner, Xiamen Motic, China). All slides were initially screened and the low-quality images were removed, including unsatisfactory, blurred, and defocused images, as well as those with poor staining. Details in relation to eligibility criteria, labeling, and clinical information are provided in Supplementary Note 2-3, Supplementary Fig. 1, and Supplementary Tables 2-3.
A total of 56 cytopathologists participated. Of these, 12 were involved in the development of the DL model, the remaining 44 participated in validation processes (stages 1-3). Each cytopathologist participated voluntarily and was involved in only one stage. Cytopathologists were categorized as either junior or senior based on their experience. The juniors were essentially beginners who were accredited trainees and had 1-3 years of cytological experience. The seniors had specialist qualifications related to cytological diagnosis and more than eight years of experience. Further details about the participating cytopathologists are provided in Supplementary Table 4.
Clinical outcomes
All women adhered to recommended testing and care at their respective hospitals. Cytological results were reported as negative or squamous and/or glandular cells abnormalities: ASC-US; atypical squamous cells cannot exclude high-grade squamous intraepithelial lesion (ASC-H); low-grade squamous intraepithelial lesion (LSIL); high-grade squamous intraepithelial lesion (HSIL); squamous cell carcinoma (SCC); AGC; adenocarcinoma in situ (AIS) or adenocarcinoma (ADC), according to the Bethesda System28. The threshold for cytological positivity was ASC-US or worse. HPV testing was performed (if available) according to manufacturer’s instructions (using either HC2 hr-HPV test or clinically validated genotype PCR assays). Those who cytologically tested positive and/or had hr-HPV received a local colposcopy and biopsy within six months. Decisions to biopsy were taken in accordance with the International Federation for Cervical Pathology and Colposcopy Standards29,30. Patients with biopsies were classified based on the most severe histological diagnosis. Biopsies were used to determine disease status according to cervical intraepithelial neoplasia (CIN) nomenclature: normal/benign, CIN1, CIN2, CIN3, or cancer. The disease endpoint was a histologically confirmed CIN2+ diagnosis, which is the threshold for treatment. However, when a biopsy-based diagnosis was not available, a combination of secondary test results was used to verify the final disease status. According to previous studies on risk assessment in a Chinese population26,31,32, women without biopsy but with negative or ASC-US cytology and hr-HPV-negative results were deemed to be histologically normal/benign. Women without biopsy were also considered histologically normal/benign if they were hr-HPV positive but had negative cytology and a negative colposcopic finding. Those without biopsy were considered incomplete but were still assigned to to one of the following groups: ASC-US and hr-HPV positive; LSIL or worse; hr-HPV positive, negative cytology, and missing or positive colposcopy groups.
Procedures
The DL model was engineered through a two-stage process which comprised cell-level detection and slide-level classification, tailored for cytological diagnosis. Cell-level detection was trained to identify abnormal squamous and glandular cells using 1,011,322 expert-annotated cell image tiles, including 225,970 ASC-US, 121,404 LSIL, 318,241 ASC-H, 232,199 HSIL and SCC, 113,508 AGC, AIS or ADC. Heatmaps were generated to visualize the outputs of the cell-level detector and have been provided in Supplementary Fig. 4. We then trained the slide-level classifier based on a multiple instance learning strategy using 17,397 slides including 10,601 negative, 2314 ASC-US, 1833 LSIL, 1657 ASC-H, 693 HSIL and SCC, as well as 299 AGC, AIS or ADC. This strategy aggregated individual cell outputs generated by the cell-level detector to slide-level classifier in order to distinguish between whole slides that are most likely negative or positive, and generated DL-suggested ROIs, which were most likely to contain abnormal cells (Supplementary Note 4, Supplementary Figs. 5-7 and Supplementary Table 1).
This model was tested through a three-stage stepwise process. In the first stage, we validated its generalizability across nine different hospitals, each with a proportion of women diagnosed with CIN2+. We then compared diagnostic performance of the DL model to original cytological results using histology as the gold standard. The original findings were provided by six senior and three junior cytopathologists. All cytopathologists analyzed each slide and relied on their experience to make decisions.
In the second stage, we performed the MRMC study involving 28 recruited cytopathologists, 12 seniors and 16 juniors. Each cytopathologist was asked to independently read all digital slides and provide initial diagnoses via a web-based platform. For cytopathologists without DL assistance, only the digital slides were displayed. After a 4-week interval, the slides were shuffled and presented randomly, and then the slide-level results (negative or positive) of the DL prediction, along with the indication of the ROIs with potential abnormal cells, were displayed. Each cytopathologist made their second diagnosis with the DL assistance. Initial diagnoses, final assisted diagnoses, and reading times were automatically recorded for each slide.
In the third stage, we conducted two application validation studies, where our DL model was applied in a community screening site and an outpatient clinic. For community-based organized screening, which involved a general population with a relatively low prevalence of CIN2+, we validated the diagnostic performance and referral efficiency of the DL model. Manual diagnoses were provided by senior cytopathologists and results were compared to those generated through the cloud platform by the DL. In the hospital-based opportunistic screening, the DL model was prospectively validated across a sample of women who participated in hospital screening. Initial cytological diagnoses were provided by junior cytopathologists. The slides were digitized and transferred to a locally deployed DL model to generate outputs. If two results were inconsistent and junior cytopathologists had to choose whether to rely on their diagnosis or adopt the DL model’s determination. Final diagnoses were made with DL assistance.
Positive results triggered colposcopy referrals and biopsies, in accordance with the standard of care. This enabled us to measure both the diagnostic performance and the rate of colposcopy referrals for DL alone, and juniors with and without DL assistance. As primary screening is switching to HPV testing, we also simulated scenario using DL alone, and juniors with and without DL assistance as a triaging method, restricted to HPV-positive women. These results were recorded and statistically analyzed.
Statistical analysis
A pre-study sample size was calculated for the DL model validation (Supplementary Note 5, Supplementary Figs. 8-9). The diagnostic performances of the DL model alone, and cytopathologists with and without DL assistance for CIN2+ detection were assessed using diagnostic accuracy, sensitivity, specificity, positive predictive value and negative predictive value with respective confidence intervals (95% CI). ROC curves were generated to calculate the AUC values using the ROCR package (version 1.18.5). McNemar’s and a standard Chi-squared test were used to compare diagnostic performances, while Wilcoxon’s signed rank test determined differences in reading times. For referral efficiency, we counted colposcopy referrals and considered NNR. Subgroup analyses were conducted to assess performance and referral efficiency using DL alone, and cytopathologists with and without DL assistance as a triaging method for HPV-positive women. All statistical tests were two-sided, with statistical significance set at a threshold of 0.05. R software (version 4.4.0) was used for all analyses. This study is reported according to the DECIDE-AI reporting guidelines in Supplementary Note 633.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
In this study, we developed a DL model using whole cytology slides for detection of cervical precancer and cancer. Due to personal information protection, patient privacy regulations, and medical institutional data policies, slide images have not been publicly deposited. However, the authors have made every effort to make the resources publicly available, such as the source code, software methods, and supporting information to reproduce the technical pipeline and analyses. All data supporting the findings of this work are available unconditionally for accredited scientific researchers for the purpose of reproducing the results and/or academic activities from the corresponding authors upon request within 14 working days.
Code availability
The codes used in this study are available online https://github.com/LuZWCHA/LBC_WSI_Classification.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Habbema, D., De Kok, I. M. & Brown, M. L. Cervical cancer screening in the United States and the Netherlands: a tale of two countries. Milbank Q 90, 5–37 (2012).
Denny, L., Kuhn, L., Hu, C. C., Tsai, W. Y. & Wright, T. C. Jr. Human papillomavirus-based cervical cancer prevention: long-term results of a randomized screening trial. J. Natl Cancer Inst. 102, 1557–1567 (2010).
Zhang, J. et al. Effectiveness of High-risk Human Papillomavirus Testing for Cervical Cancer Screening in China: A Multicenter, Open-label, Randomized Clinical Trial. JAMA Oncol. 7, 263–270 (2021).
Bouvard, V. et al. The IARC Perspective on Cervical Cancer Screening. N. Engl. J. Med. 385, 1908–1918 (2021).
Asgary, R., Beideck, E. & Naderi, R. Comparative assessment of test characteristics of cervical cancer screening methods for implementation in low-resource settings. Prev. Med. 154, 106883 (2022).
Simms, K. T. et al. Benefits, harms and cost-effectiveness of cervical screening, triage and treatment strategies for women in the general population. Nat. Med. 29, 3050–3058 (2023).
Bruni, L. et al. Cervical cancer screening programmes and age-specific coverage estimates for 202 countries and territories worldwide: a review and synthetic analysis. Lancet Glob. Health 10, e1115–e1127 (2022).
Ikenberg, H. et al. Comparison of the Hologic Genius Digital Diagnostics System with the ThinPrep Imaging System-A retrospective assessment. Cancer Cytopathol. 131, 424–432 (2023).
Cheng, S. et al. Robust whole slide image analysis for cervical cancer screening using deep learning. Nat. Commun. 12, 5639 (2021).
Xue, P. et al. Improving the Accuracy and Efficiency of Abnormal Cervical Squamous Cell Detection With Cytologist-in-the-Loop Artificial Intelligence. Mod. Pathol. 36, 100186 (2023).
Wang, J. et al. Artificial intelligence enables precision diagnosis of cervical cytology grades and cervical cancer. Nat. Commun. 15, 4369 (2024).
Zhu, X. et al. Hybrid AI-assistive diagnostic model permits rapid TBS classification of cervical liquid-based thin-layer cell smears. Nat. Commun. 12, 3541 (2021).
Bao, H. et al. Artificial intelligence-assisted cytology for detection of cervical intraepithelial neoplasia or invasive cancer: A multicenter, clinical-based, observational study. Gynecol. Oncol. 159, 171–178 (2020).
Bai, X. et al. Assessment of Efficacy and Accuracy of Cervical Cytology Screening With Artificial Intelligence Assistive System. Mod. Pathol. 37, 100486 (2024).
Lew, M., Wilbur, D. C. & Pantanowitz, L. Computational Cytology: Lessons Learned from Pap Test Computer-Assisted Screening. Acta Cytol. 65, 286–300 (2021).
Xue, P. et al. Deep learning in image-based breast and cervical cancer detection: a systematic review and meta-analysis. NPJ Digit Med 5, 19 (2022).
Wentzensen, N. et al. Accuracy and Efficiency of Deep-Learning-Based Automation of Dual Stain Cytology in Cervical Cancer Screening. J. Natl Cancer Inst. 113, 72–79 (2021).
Liu, X. et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 1, e271–e297 (2019).
Holmström, O. et al. Point-of-Care Digital Cytology With Artificial Intelligence for Cervical Cancer Screening in a Resource-Limited Setting. JAMA Netw. Open 4, e211740 (2021).
Koliopoulos, G. et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database Syst. Rev. 8, Cd008587 (2017).
Lahrmann, B., Valous, N. A., Eisenmann, U., Wentzensen, N. & Grabe, N. Semantic focusing allows fully automated single-layer slide scanning of cervical cytology slides. PLoS One 8, e61441 (2013).
Jiang, P. et al. Development of Automatic Portable Pathology Scanner and Its Evaluation for Clinical Practice. J. Digit Imaging 36, 1110–1122 (2023).
Wang, H. et al. Chinese colposcopists’ attitudes toward the colposcopic artificial intelligence auxiliary diagnostic system (CAIADS): A nation-wide, multi-center survey. Digit Health 10, 20552076241279952 (2024).
Pan, Q. J. et al. Liquid-based cytology and human papillomavirus testing: a pooled analysis using the data from 13 population-based cervical cancer screening studies from China. Gynecol. Oncol. 133, 172–179 (2014).
Zhao, F. H. et al. Performance of high-risk human papillomavirus DNA testing as a primary screen for cervical cancer: a pooled analysis of individual patient data from 17 population-based studies from China. Lancet Oncol. 11, 1160–1171 (2010).
Xue, P. et al. Assessing artificial intelligence enabled liquid-based cytology for triaging HPV-positive women: a population-based cross-sectional study. Acta Obstet. Gynecol. Scand. 102, 1026–1033 (2023).
Nayar, R. & Wilbur, D. C. The Pap test and Bethesda 2014. Cancer Cytopathol. 123, 271–281 (2015).
Bornstein, J. et al. 2011 colposcopic terminology of the International Federation for Cervical Pathology and Colposcopy. Obstet. Gynecol. 120, 166–172 (2012).
Qin, D. et al. Colposcopic accuracy in diagnosing squamous intraepithelial lesions: a systematic review and meta-analysis of the International Federation of Cervical Pathology and Colposcopy 2011 terminology. BMC Cancer 23, 187 (2023).
Belinson, J. et al. Shanxi Province Cervical Cancer Screening Study: a cross-sectional comparative trial of multiple techniques to detect cervical neoplasia. Gynecol. Oncol. 83, 439–444 (2001).
Shi, J. F. et al. Human papillomavirus testing for cervical cancer screening: results from a 6-year prospective study in rural China. Am. J. Epidemiol. 170, 708–716 (2009).
Vasey, B. et al. Reporting guideline for the early stage clinical evaluation of decision support systems driven by artificial intelligence: DECIDE-AI. Bmj 377, e070904 (2022).
Acknowledgements
This study was funded by China Postdoctoral Science Foundation (GZB20230076, 2023M740323, and 2024T170072), and the ownership of these grants is attributed to P.X. This study was funded by Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (CIFMS 2021-I2M-1-004), National High Level Hospital Clinical Research Funding (2022-PUMCH-D-003), and the ownership of these grants is attributed to L.Zhu. This study was supported by Sanming Project of Medicine in Shenzhen (SZSM202211032) and the ownership of these grants is attributed to Y.LQ. We thank for LBP Medical Science and Technology Inc, Guangzhou, China for support on the development of DL model.
Author information
Authors and Affiliations
Contributions
P.X., L.D., Y.LQ., L.Zhu., and F.HZ. contributed to the study design. P.X., L.HK., H.PT., H.MX., H.YW., Z.W., R.GW., L.X., H.XL., H.YN., M.JW., Z.FL., W.C., Q.JP., and X.Z. contributed to sample collection and experiment or data interpretation. P.X., Z.CY., Y.LQ., L.Zhu., and F.HZ. were the core team for data analysis and manuscript preparation. All the authors had access to a summary of all the study data. P.X., R.R., L.Zhang., and Y.J. reviewed and verified the data in the study. The corresponding author and equally contributing joint senior authors had final responsibility for the decision to submit for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks William Lotter, Andrzej Nowakowski, Shaokai Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xue, P., Dang, L., Kong, LH. et al. Deep learning enabled liquid-based cytology model for cervical precancer and cancer detection. Nat Commun 16, 3506 (2025). https://doi.org/10.1038/s41467-025-58883-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-58883-3