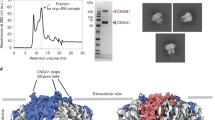
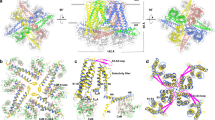
Abstract
Plant cyclic nucleotide-gated channels (CNGCs) belong to the cyclic nucleotide-binding ___domain (CNBD) channel family, but are phylogenetically classified in a distinct branch. In contrast to their animal counterparts of K+-selective or non-selective cation channels, plant CNGCs mainly mediate Ca2+ influx and are involved in various physiological processes, such as stomatal movements, pollen-tube growth and immune responses. Here, we present the cryo-EM structure and electrophysiological analysis of plant CNGC representatives, Arabidopsis CNGC1 and CNGC5. We found that CNGC1 and CNGC5 contain a unique extracellular ___domain featuring disulfide bonds that is essential for channel gating via coupling of the voltage-sensing ___domain with the pore ___domain. The pore ___domain selectivity filter possesses a Gln residue at the constriction site that determines the Ca2+ selectivity. Replacement of this Gln with Glu, typically observed in CNBD-type non-selective cation channels, could convert CNGC1 and CNGC5 from Ca2+-selective channels to non-selective cation channels permeable to Ca2+, Na+ or K+. In addition, we found that the CNGC1 and CNGC5 CNBD homology ___domain contains intrinsic-ligand-like interactions, which may devoid the binding of cyclic nucleotides and lead to gating independent of cAMP or cGMP. This research not only provides a mechanistic understanding of plant CNGCs’ function, but also adds to the comprehensive knowledge of the CNBD channels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The 3D cryo-EM density maps of CNGC1Ca, CNGC5 and CNGC5Ca have been deposited in the Electron Microscopy Data Bank under the accession numbers EMD-61105, EMD-61106 and EMD-61107. Coordinates for structure models have been deposited in the Protein Data Bank (PDB) under the accession codes 9J34, 9J35 and 9J36. Source data are provided with this paper.
References
Demidchik, V., Shabala, S., Isayenkov, S., Cuin, T. A. & Pottosin, I. Calcium transport across plant membranes: mechanisms and functions. New Phytol. 220, 49–69 (2018).
Chiasson, D. M. et al. A quantitative hypermorphic CNGC allele confers ectopic calcium flux and impairs cellular development. eLife 6, e25012 (2017).
Huang, J., Pan, X. & Yan, N. Structural biology and molecular pharmacology of voltage-gated ion channels. Nat. Rev. Mol. Cell Biol. 25, 904–925 (2024).
James, Z. M. & Zagotta, W. N. Structural insights into the mechanisms of CNBD channel function. J. Gen. Physiol. 150, 225–244 (2018).
Clark, M. D., Contreras, G. F., Shen, R. & Perozo, E. Electromechanical coupling in the hyperpolarization-activated K+ channel KAT1. Nature 583, 145–149 (2020).
Kaupp, U. B. & Seifert, R. Cyclic nucleotide-gated ion channels. Physiol. Rev. 82, 769–824 (2002).
Dzeja, C., Hagen, V., Kaupp, U. B. & Frings, S. Ca2+ permeation in cyclic nucleotide-gated channels. EMBO J. 18, 131–144 (1999).
Gauss, R., Seifert, R. & Kaupp, U. B. Molecular identification of a hyperpolarization-activated channel in sea urchin sperm. Nature 393, 583–587 (1998).
Sanguinetti, M. C. & Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 440, 463–469 (2006).
Xue, J., Han, Y., Zeng, W. Z., Wang, Y. & Jiang, Y. X. Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron 109, 1302–1313 (2021).
Wang, W. W. & MacKinnon, R. Cryo-EM structure of the open human Ether-à-gogo-related K+ channel hERG. Cell 169, 422–430 (2017).
Li, M. H. et al. Structure of a eukaryotic cyclic-nucleotide-gated channel. Nature 542, 60–65 (2017).
Lee, C. H. & MacKinnon, R. Structures of the human HCN1 hyperpolarization-activated channel. Cell 168, 111–120 (2017).
Whicher, J. R. & MacKinnon, R. Structure of the voltage-gated K+ channel Eag1 reveals an alternative voltage sensing mechanism. Science 353, 664–669 (2016).
Köhler, C., Merkle, T. & Neuhaus, G. Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana. Plant J. 18, 97–104 (1999).
Leng, Q., Mercier, R. W., Yao, W. Z. & Berkowitz, G. A. Cloning and first functional characterization of a plant cyclic nucleotide-gated cation channel. Plant Physiol. 121, 753–761 (1999).
Hua, B. G., Mercier, R. W., Leng, Q. & Berkowitz, G. A. Plants do it differently. A new basis for potassium/sodium selectivity in the pore of an ion channel. Plant Physiol. 132, 1353–1361 (2003).
Wang, Y. F. et al. Identification of cyclic GMP-activated nonselective Ca2+-permeable cation channels and associated CNGC5 and CNGC6 genes in Arabidopsis guard cells. Plant Physiol. 163, 578–590 (2013).
Gao, Q. F., Fei, C. F., Dong, J. Y., Gu, L. L. & Wang, Y. F. Arabidopsis CNGC18 is a Ca2+-permeable channel. Mol. Plant 7, 739–743 (2014).
Gao, Q. F. et al. Cyclic nucleotide-gated channel 18 is an essential Ca2+ channel in pollen tube tips for pollen tube guidance to ovules in Arabidopsis. Proc. Natl Acad. Sci. USA 113, 3096–3101 (2016).
Wang, Y. et al. CNGC2 is a Ca2+ influx channel that prevents accumulation of apoplastic Ca2+ in the leaf. Plant Physiol. 173, 1342–1354 (2017).
Zhang, Z., Hou, C., Tian, W., Li, L. & Zhu, H. Electrophysiological studies revealed CaM1-mediated regulation of the Arabidopsis calcium channel CNGC12. Front. Plant Sci. 10, 1090 (2019).
Tan, Y. Q. et al. Three CNGC family members, CNGC5, CNGC6, and CNGC9, are required for constitutive growth of Arabidopsis root hairs as Ca2+-permeable channels. Plant Commun. 1, 100001 (2020).
Tan, Y.-Q. et al. Multiple cyclic nucleotide-gated channels function as ABA-activated Ca2+ channels required for ABA-induced stomatal closure in Arabidopsis. Plant Cell 35, 239–259 (2023).
Yang, Y. et al. OPEN STOMATA 1 phosphorylates cyclic nucleotide-gated channels to trigger Ca2+ signaling for abscisic acid-induced stomatal closure in Arabidopsis. Plant Cell 36, 2328–2358 (2024).
Stael, S. et al. Plant organellar calcium signalling: an emerging field. J. Exp. Bot. 63, 1525–1542 (2012).
Rudd, J. J. & Franklin-Tong, V. E. Calcium signaling in plants. Cell. Mol. Life Sci. 55, 214–232 (1999).
Gilroy, S., Fricker, M. D., Read, N. D. & Trewayas, A. J. Role of calcium in signal transduction of commelina guard-cells. Plant Cell 3, 333–344 (1991).
Hetherington, A. M. & Brownlee, C. The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 55, 401–427 (2004).
Clapham, D. E. Calcium signaling. Cell 131, 1047–1058 (2007).
Dodd, A. N., Kudla, J. & Sanders, D. The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620 (2010).
Yuan, P. G., Jauregui, E., Du, L. Q., Tanaka, K. & Poovaiah, B. W. Calcium signatures and signaling events orchestrate plant–microbe interactions. Curr. Opin. Plant Biol. 38, 173–183 (2017).
Kudla, J. et al. Advances and current challenges in calcium signaling. New Phytol. 218, 414–431 (2018).
Tian, W., Wang, C., Gao, Q. F., Li, L. G. & Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 6, 750–759 (2020).
Siegel, R. S. et al. Calcium elevation-dependent and attenuated resting calcium-dependent abscisic acid induction of stomatal closure and abscisic acid-induced enhancement of calcium sensitivities of S-type anion and inward-rectifying K+ channels in Arabidopsis guard cells. Plant J. 59, 207–220 (2009).
Brost, C. et al. Multiple cyclic nucleotide-gated channels coordinate calcium oscillations and polar growth of root hairs. Plant J. 99, 910–923 (2019).
Zhang, S. S. et al. CNGC14 mediates calcium influx required for tip growth in root hairs. Mol. Plant 10, 1004–1006 (2017).
Tian, W. et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135 (2019).
Wang, J. C. et al. A cyclic nucleotide-gated channel mediates cytoplasmic calcium elevation and disease resistance in rice. Cell Res. 29, 820–831 (2019).
Frietsch, S. et al. A cyclic nucleotide-gated channel is essential for polarized tip growth of pollen. Proc. Natl Acad. Sci. USA 104, 14531–14536 (2007).
Tunc-Ozdemir, M. et al. Cyclic nucleotide gated channels 7 and 8 are essential for male reproductive fertility. PLoS ONE 8, e55277 (2013).
Charpentier, M. et al. Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105 (2016).
Wang, J. C. et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 14, 315–329 (2021).
Peng, Y. et al. Differential phosphorylation of Ca2+-permeable channel CNGC20 modulates calcium-mediated freezing tolerance in Arabidopsis. Plant Cell 36, 4356–4371 (2024).
Saponaro, A. et al. Gating movements and ion permeation in HCN4 pacemaker channels. Mol. Cell 81, 2929–2943 (2021).
Bucher, D., Guidoni, L., Carloni, P. & Rothlisberger, U. Coordination numbers of K+ and Na+ ions inside the selectivity filter of the KcsA potassium channel: insights from first principles molecular dynamics. Biophys. J. 98, L47–L49 (2010).
Wu, J. P. et al. Structure of the voltage-gated calcium channel Cav1.1 at 3.6 Å resolution. Nature 537, 191–196 (2016).
Zhao, Y. Y. et al. Cryo-EM structures of apo and antagonist-bound human Cav3.1. Nature 576, 492–496 (2019).
He, L. L. et al. Structure, gating, and pharmacology of human Cav3.3 channel. Nat. Commun. 13, 2084 (2022).
Lu, Y. et al. Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis. Nat. Commun. 13, 5682 (2022).
Köhler, C. & Neuhaus, G. Characterisation of calmodulin binding to cyclic nucleotide-gated ion channels from Arabidopsis thaliana. FEBS Lett. 471, 133–136 (2000).
Pan, Y. et al. Dynamic interactions of plant CNGC subunits and calmodulins drive oscillatory Ca2+ channel activities. Dev. Cell 48, 710–725 (2019).
Fischer, C. et al. Calmodulin as a Ca2+-sensing subunit of arabidopsis cyclic nucleotide-gated channel complexes. Plant Cell Physiol. 58, 1208–1221 (2017).
Fischer, C., Kugler, A., Hoth, S. & Dietrich, P. An IQ ___domain mediates the interaction with calmodulin in a plant cyclic nucleotide-gated channel. Plant Cell Physiol. 54, 573–584 (2013).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Pravda, L. et al. MOLEonline: a web-based tool for analyzing channels, tunnels and pores. Nucleic Acids Res. 46, W368–W373 (2018).
Acknowledgements
We thank M. Zhang, L. Zhu and W. Cai from the Center for Excellence in Molecular Plant Sciences core facility for diagnostic cryo-EM analysis and GFP fluorescence analysis; and A. Dong and H. Zhao from Fudan University for technical assistance in cryo-EM data collection. This work was supported by grants from the National Natural Science Foundation of China (grant no. 32025020 to P.Z. and grant no. 32270279 to Y.-F.W.), the Chinese Academy of Sciences (grant no. XDB0630100 to P.Z.) and the Shanghai Science and Technology Commission (grant no. 23310710100).
Author information
Authors and Affiliations
Contributions
J.W., B.D. and X.Z. designed and performed the bulk of the experiments. J.W. carried out protein expression and purification, and grid sample preparation. X.Z. and J.W. carried out cryo-EM data collection and structure determination, supervised by P.Z. B.D. carried out electrophysiological experiments guided by Y.-F.W. Y.Y. contributed to electrophysiological data analysis. X.Q. and Z.Y. contributed to protein purification and grid sample preparation. P.Z., Y.-F.W. and J.W. wrote the manuscript with inputs from other authors. P.Z. conceived the project.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Plants thanks Changlin Tian and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Cryo-EM analysis of CNGC5.
a, Gel filtration profile and Coomassie-blue-stained SDS-PAGE analysis of CNGC5 sample prepared in nanodiscs. Independent experiments have been repeated at least three times with similar results. b, Representative micrograph of cryo-EM. c, 2D class averages. d, cryo-EM data analysis pipeline. e, Local resolution estimation and gold-standard Fourier shell correlation (FSC) curves. The color represents the local resolution in Å.
Extended Data Fig. 2 Cryo-EM analysis of CNGC5Ca.
a, Gel filtration profile and Coomassie-blue-stained SDS-PAGE analysis of CNGC5 in buffer containing 0.01% GDN. Independent experiments have been repeated at least three times with similar results. b, Representative micrograph of cryo-EM. c, 2D class averages. d, cryo-EM data analysis pipeline. e, Local resolution estimation and gold-standard Fourier shell correlation (FSC) curves. The color represents the local resolution in Å.
Extended Data Fig. 3 Cryo-EM analysis of CNGC1Ca.
a, Gel filtration profile and Coomassie-blue-stained SDS-PAGE analysis of CNGC1 in buffer containing 0.01% GDN. Independent experiments have been repeated at least three times with similar results. b, Representative micrograph of cryo-EM. c, 2D class averages. d, cryo-EM data analysis pipeline. e, Local resolution estimation and gold-standard Fourier shell correlation (FSC) curves. The color represents the local resolution in Å.
Extended Data Fig. 4 Structure based sequence alignment of 20 CNGCs from Arabidopsis thaliana, showing exclusively on the segment of ECD.
In the illustration, the numbers highlighted in orange denote the cysteine residues that form the three corresponding disulfide bonds. Sequences are ordered by classification based on phylogenetic relationships.
Extended Data Fig. 5 VSDs of CNGC5 and CNGC1 in comparative perspective with other CNBD Channels.
a, Sequence alignment of S4 helix among 20 Arabidopsis CNGCs. Red asterisks indicate the conserved arginine site. b, VSDs of CNGC5, CNGC1, HCN1(PDB:5U6P) and KAT1(PDB:6V1X) are shown in side view, with the S1 helix omitted for clarity. The positive charged (or polar) residues on S4 and the gating charge transfer center residues on S2 and S3 are shown as sticks.
Extended Data Fig. 6 Ion selectivity of CNGC5 and CNGC1.
a, Sequence alignment of ion selectivity filter motifs in 20 Arabidopsis CNGCs. b-d, Density map at the selectivity filter of CNGC1Ca and CNGC5Ca. Both samples were prepared with a supplementation of 2 mM Ca2+. Selectivity filter regions of two diagonally opposed subunits are shown in sticks, the Ca2+ ions along the ion pathway are in green spheres and water molecules are in red spheres. b, The density at the selectivity filter of CNGC1Ca. Density map is obtained after C4 symmetry processing (contoured at 5.5 σ). c, The density at the selectivity filter of CNGC5Ca. Density map is obtained after C4 symmetry processing (contoured at 5.5 σ). d, The density at the selectivity filter of CNGC5Ca. Density map is obtained after C1 symmetry processing (contoured at 4.0 σ). Chain A, C and chain B, D exhibit distinct conformations due to their mobility or asymmetry, and are shown separately.
Extended Data Fig. 7 Comparison of the selectivity filter of CNGC5, CNGC1 and the selected CNBD channels.
a, Sequence alignment of ion selectivity filter motifs in CNGC5, CNGC1 and other CNBD channels. b, Structure comparison of the selectivity filter of CNGC5Ca, CNGC1Ca and the selected CNBD channels. For clarity, only two diagonally opposed subunits are shown. The PDB accession numbers of the comparison channels are KAT1, 6V1X; HCN1, 5U6P; TAX-4, 6WEK.
Extended Data Fig. 8 Electrophysiological studies on selectivity filter mutants.
Patch clamping experiments were performed in HEK293T cells for the analysis of ion permeability to diverse cations. a and c, The average current-voltage curves show the whole-cell currents of CNGC5Q383A and CNGC1Q371A in 1 mM Ca2+-based bath solution similar to that in 10 mM Ca2+-based bath solution, which are different from the mock control-like small currents of wild-type CNGC5 and CNGC1 in 1 mM Ca2+-based bath solution. b and d, The average current-voltage curves show the whole-cell currents of CNGC5Q383A and CNGC1Q371A in the bath solutions with 120 mM Na+ and 1 mM Ca2+, 8 mM Na+ and 1 mM Ca2+, 8 mM Na+ and 10 mM Ca2+, and 8 mM K+, and 1 mM Ca2+. The currents in different bath solutions show different reversal potentials (zoom-in view), which point to the ion selectivity of CNGC5Q383A and CNGC1Q371A. For the convenience of comparison, the data of mock control in panels a and c of this extended data figure are also shown as the mock control data in Fig. 4d and f, respectively; the data of CNGC5Q383A/CNGC1Q371A 1 mM Ca2+-based bath solution in panels a and c of this extended data figure are also shown as the data of CNGC5Q383A/CNGC1Q371A in 120 mM Na+ plus 1 mM Ca2+ condition in panels b and d, respectively. The letter n denotes the numbers of HEK293T cells tested. Data are presented as means ± SEM.
Extended Data Fig. 9 Representative fluorescent of CNGC5 wild-type, mutants and GFP in HEK 293T cells.
GFP tag was fused to the N terminus of CNGC5. GFP fluorescence was readily detected in cells expressed CNGC5 wild type (WT) and mutants with similar patterns. Independent experiments have been repeated at least three times with similar results. The varying lengths of the gray bars corresponded to 10 µm in all the indicated variant images.
Extended Data Fig. 10 Sequence alignment of cNMP-binding region.
a, Sequence alignment of cNMP binding region in Arabidopsis CNGC1/5 and other selected CNBD channels. b, Sequence alignment of 20 Arabidopsis CNGCs. Filled red triangles indicate the typical Arg residue required for cNMP binding in cNMP-regulated CNBD channels. Filled red circles indicate another distinctive Arg residue in Arabidopsis CNGCs that occupies the pseudo cNMP-binding pocket.
Supplementary information
Supplementary Information
Supplementary Figs. 1–3 and Table 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed gels.
Source Data Extended Data Fig. 8
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, J., Du, BY., Zhang, X. et al. Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity. Nat. Plants 11, 632–642 (2025). https://doi.org/10.1038/s41477-025-01923-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41477-025-01923-z
This article is cited by
-
Mechanisms of auxin action in plant growth and development
Nature Reviews Molecular Cell Biology (2025)