Abstract
Methamphetamine use can produce psychotic symptoms almost indistinguishable from schizophrenia (SCZ). Variation in DNA methylation may be closely implicated in the etiology and longitudinal development of psychiatric disorders. However, the relationship between psychotic symptoms, functional disability, and DNA methylation is still unclear. This study consists of three periods: discovery, validation, and follow-up. In the discovery stage, we employed genome-wide DNA methylation profiling (Illumina 450K) in peripheral blood mononuclear cells to test whether DNA methylation associates with psychotic symptoms and function state in representative SCZ and methamphetamine-induced psychotic disorder (MIP) patients. Then, we found seven differentially methylated regions/genes (DMRs, in UBA6, APOL3, KIF17, MLLT3, GRM8, CSNK1E, SETDB1) overlapping with genetic variants reported in previous studies of psychosis. In the validation stage, we compared the above-mentioned seven genes by MethLight qPCR method in Chinese Han males (N = 109 SCZ patients, N = 99 methamphetamine use disorder with MIP patients, N = 150 methamphetamine use disorder without MIP patients, N = 282 normal controls, age range: 18–50 years). GRM8 showed robustly altered methylation, which has passed rigorous filtration in subsequent validation, suggesting a remarkable contribution to SCZ and MIP. In addition, hypermethylation of GRM8 showed a significant association with the total scores of the Positive Negative Syndrome Scale and WHO disability assessment schedule II in both baseline and follow-up periods. Our findings suggest that increased CpG methylation in the promoter of GRM8 is a potential candidate epigenetic biomarker of psychotic symptoms in transdiagnostic samples of SCZ and MIP.
Similar content being viewed by others
Introduction
Epidemiological studies place amphetamine-type stimulant (ATS) as the most widely used illicit drug in the world after cannabis1,2, with up to 51 million users globally between 15 and 64 years old3. Methamphetamine, a kind of ATS, has become the primarily used illicit drug in China, with 1.19 million registered users (55.2% of all registered drug users)4. Chronic, high dose and continuous use of methamphetamine can produce transient psychotic symptoms almost indistinguishable from schizophrenia (SCZ), which is a severe, highly heritable and socially disabling mental disorder called methamphetamine-induced psychotic disorder (MIP)5.
In addition to the clinical similarities, such as age of onset6, predisposed personality traits7 and cognitive function impairment, there are also shared susceptibility factors in terms of neurochemistry, neuroimaging5,8 and genetic pathway9,10 between the two conditions. Moreover, cohort studies indicate that a substantial proportion of MIP diagnoses develop into persistent psychotic conditions in the long run11,12. Unlike SCZ, the presence of MIP is generally transient and releases typically in a few hours or days13 and rarely lasts for more than one month14. However, the psychotic symptoms of the susceptible population could be prolonged for months or years after methamphetamine abstinence. After following up for 6–8 years, 30–38% of individuals with MIP were re-diagnosed as SCZ or SCZ spectrum disorders12,15. We speculate these two highly comorbid disorders that may have shared neurobiological underpinnings.
Growing evidence has suggested that epigenetic dysregulation might be implicated in the etiology and pathogenesis of psychotic disorders16,17. DNA methylation is a major epigenetic mechanism that effectively regulates gene expression profiles in response to environmental challenge18. This process involves the addition of an extra methyl group on the cytosine of CpG dinucleotides without altering the DNA sequence. Because of the nature of being inheritable and mitotically retainable, DNA methylation has been postulated to promote the onset of many complex diseases19. Hence, we speculate DNA methylation alterations may regulate gene expression leading to the resemblant phenotype of psychotic disorders and functional disability. Considering the heterogeneity of mental disorders, it is now recognized that mapping cross-diagnostic symptoms to potential neurobiology may have a translational impact than trying to identify the target of classification disorders20,21,22.
We, therefore, adopted a transdiagnostic and symptom-based approach to explore shared candidate epigenetic biomarkers that could underlie the symptomatic overlap between both primary and substance-induced psychotic symptoms. In the present study, we first investigated the DNA methylation profiles of peripheral blood mononuclear cells (PBMCs) from male Chinese Han patients with SCZ, methamphetamine use disorder with MIP (MUD-MIP), methamphetamine use disorder without MIP (MUD-only) and normal controls (NC) using Infinium Human Methylation 450 BeadChip Array.
Then, we quantitatively evaluated the candidate differentially methylation genes with MethLight qPCR and analyzed their correlation with clinical variables (Positive and Negative Syndrome Scale-PANSS, Clinical Global Impressions-Severity Scale-CGI-SI, and WHO disability assessment schedule II-WHODAS 2.0). A follow-up examination for the methylation changes after a 5-week effective antipsychotic treatment was subsequently conducted. By comparing the methylation status of an individual at different time points, longitudinal design has advantages in analyzing potential sources of methylation variation.
Patients and methods
Study samples
We recruited SCZ patients (n = 109) from the Institute of Mental Health at the Second Xiangya Hospital. MUD-MIP patients (n = 99) and MUD-only patients (n = 150) from the Kangda, Xinkaipu and Bainihu rehabilitation centers and the Second People’s Hospital of Hunan Province. NC (n = 282) were recruited by word of mouth from hospital staff or University undergraduates documented to be free from any psychiatric disorders. All participants were Chinese Han nationals of the male gender and 18–50 years old. The diagnosis was decided according to the Diagnostic and Statistical Manual of Mental Disorders, 5 Edition (DSM-5) criteria by at least three experienced psychiatrists on the basis of extensive clinical interviews and reviews of the patient’s medical records.
The clinical symptoms of patients with SCZ and MUD-MIP were assessed by using PANSS, the CGI-S scale (Guy, 1976) and WHODAS 2.0. The PANSS provides a total score and separate scores for three subscales, namely the 7-item Positive Symptoms subscale (P1–P7), the 7-item Negative Symptoms subscale (N1–N7), and the 16-item General Psychopathology subscale (G1–G16). The WHODAS 2.0 is a 36-item measure that assesses disability across six domains, including understanding and communicating, getting around, self-care, getting along with people, life activities (i.e., household, work, and/or school activities), and participation in society. Higher scores indicated more severe disability (0–4 indicates no difficulty, 5–24 indicates mild difficulty, 25–49 indicates moderate difficulty, 50–95 indicates severe difficulty and 96–100 indicates extreme difficulty). The Chinese version of the PANSS and WHODAS 2.0 questionnaire have satisfactory validity and reliability23,24.
The diagnosis of SCZ and MIP were based on the Diagnostic and Statistical Manual of Mental Disorders, 5th Edition (DSM-5) criteria25. Participants satisfied the following criteria: (1) at least one of three items (P1. Delusions, P3. Hallucinatory behavior, P6. Suspiciousness) of Positive Scale in PANSS ≥ 5, a minimum CGI-S scale rating of 4 (moderately ill) or greater for SCZ and MUD-MIP patients; (2) The improved follow-up SCZ and MUD-MIP patients defined a priori as ≥50% reduction in PANSS total score after 4 weeks of antipsychotic treatment; (3) NC did not meet the criteria set by Criteria of Prodromal Syndromes (COPS) or DSM-5 and had no documented family history of psychiatric disorders or medications; and (4) All participants had not received any psychiatric treatment in the past 6 months. The exclusion criteria included: (1) met DSM-5 criteria for any other psychotic disorder, or had delirium, dementia, amnesia, or other severe cognitive impairment in the past or intellectual developmental disabilities before current diagnosis were made; (2) subjects with a documented history of brain injury, epilepsy, or other known diseases of central nervous system; and (3) subjects who were high cigarette smokers (>1 pack/day), because of its effects on some epigenetic markers26,27,28.
This study was conducted in accordance with the principles expressed in the Declaration of Helsinki and was approved by the Ethics Committee of the Second Xiangya Hospital. Prior to the study, the procedure was fully explained, and written informed consent was obtained from each subject. All the subjects were free to participate in or abstain from this study, and free to withdraw from this study anytime without threat of punishment.
All participants were recruited between February 2014 and May 2016.
Study design
Study Period I (discovery): Whole genome methylation profiling using Infinium Methylation 450 BeadChip
During Study Period I, genomic DNA was isolated from PBMCs and then bisulfite-converted following standard procedures. The bisulfite-converted DNA was used for the whole genome amplification reaction, enzymatic fragmentation, precipitation, and resuspension in a hybridization buffer. Subsequent steps were carried out according to the standard Infinium HD assay methylation protocol guide. Methylomic profiling read out by Illumina iScan System. The Illumina 450K BeadChip simultaneously profiles 482,421 CpG and 3091 non-CpG sites, covering 99% of RefSeq genes with multiple sites in annotated promoters (1500 or 200 bp upstream of the transcription start site), 5′-untranslated regions (UTRs), first exons, gene body, and 3′-UTRs29. The array incorporates two different chemical assays: Infinium I, which includes 135,501 probes, and Infinium II, which includes 350,076 probes. Each CpG site of Infinium I is targeted by two probes that, respectively, detect methylated (M) and unmethylated (U) signal intensities, while each CpG site of Infinium II is targeted by a single probe with two different dye colors (green and red) that distinguish M and U signal intensities. The genome-wide methylation analysis was performed on SCZ (n = 8), MUD-MIP (n = 8), MUD-only (n = 9), and NC (n = 8) after age and education matching.
Study Period II (validation) and Study Period III (follow-up): sequence validation of selected probes using MethLight qPCR
During Study Period II, the methylomic characteristics (candidate methylated genetic loci relevant to mental disorders or neural systems) discovered in Study Period I were examined in all the SCZ (n = 109), MUD-MIP (n = 99), MUD-only (n = 150) and NC (n = 282). The follow-up cohort enrolled SCZ (n = 16) and MUD-MIP (n = 15), who exhibited a positive response to antipsychotic treatment (defined as reduction ≥50% in PANSS) after 5 weeks. Every follow-up subject has received monotherapy of olanzapine (15–20 mg/day) or risperidone (4–6 mg/day) as clinically indicated after being diagnosed. We chose olanzapine and risperidone because they have no indirect effect or interaction with candidate genes. MethLight qPCR was used to verify the methylation status of candidate genes, based on the principle of fluorescence-based Real-time PCR with double probes. We prepared standard gene products of selected differentially methylated positions (DMPs) by amplification of target DNA fragments and purification of agarose gel DNA fragments using a Gel Extraction Kit and DNA Methylation Kit (CWBiotech, Beijing, China). MethLight qPCR was performed on a PikoReal 96 Real-Time PCR System (Thermo Fisher Scientific Inc., USA) using bisulfite-treated DNA and a GoldStar TaqMan Mixture (CWBiotech, Beijing, China). The flowchart of participants who were screened and participated in the study is shown in Fig. 1.
Sample storage and processing
PBMCs samples were collected and stored at −80 °C in the ultra-low temperature freezer of the Psychiatry Laboratory at the Second Xiangya Hospital, Central South University. Following the collection of all samples, DNA was extracted concurrently in a unified batch, facilitating subsequent integrated analysis.
Statistical analysis
For methylation analysis, raw Illumina IDAT files were run in the R environment and underwent background correction, probe type I/II correction, and normalization. The probes with low-quality signals (p-value > 0.05) or those overlapped with single nucleotide polymorphisms were discarded. The methylation status of each individual CpG locus was determined by calculating the Beta value, which represents the ratio of methylated signal intensity to the sum of methylated (M) and unmethylated (U) signals after background subtraction. This value ranged from 0 (indicating no methylation) to 1 (representing complete methylation). The original data of the chip was preprocessed using the minfi package of the R software, and then the methylation sites and methylation regions of the samples were differentiated using the IMA package of the R software. Probes were considered to be differentially methylated if the resulting adjusted |beta.difference|>0.14 by Empirical Bayes method t-test analysis and a Benjamini-Hochberg corrected p-value was <0.05.
Their primers and probes of candidate genes were designed by Beacon Designer 7.0. In MethLight qPCR test, according to the slope and intercept of the standard curve and the cycle number of the sample to be tested, the copy number ‘M’ of candidate gene methylation and the copy number ‘U’ of gene unmethylation of the sample to be tested are calculated. The formula Beta = M/(M + U + 100). The Beta-value statistic results in a number between 0 and 1; under ideal conditions, a value of zero indicates that all copies of the CpG site in the sample were completely unmethylated (no methylated molecules were measured), and a value of one indicates that every copy of the site was methylated30. The remaining data were analyzed using SPSS 19.0 software with a t-test, chi-squared test, and Pearson correlation analysis. The Mann–Whitney U test was employed to compare the differences between two independent samples that deviated from a normal distribution. For paired samples with non-normality distribution, the Wilcoxon rank-sum test was utilized. The false discovery rate was <0.05 for all statistical analyses.
Results
Demographic and clinical data
Demographic and PANSS, WHODAS 2.0, CGI, and GAF scores of SCZ (n = 109) and MUD-MIP (n = 99) before treatment are shown in Table 1. The MUD-MIP group was older, had poorer marital status than the SCZ group (ps < 0.001), and had no statistically significant difference in educational level between the two groups. No significant difference was found in whether it was the first onset psychosis (χ2 = 0.895, p = 0.405), PANSS total score (t = 1.548, p = 0.123), WHODAS2.0 total score (t = 1.161, p = 0.247), WHODAS 2.0 classification (χ2 = 1.409, p = 0.703), CGI (SI) scores (t = 0.833, p = 0.406). SCZ patients had higher scores in PANSS negative score (t = 4.542, P < 0.001) and cognition ___domain in WHODAS2.0 (t = 2.631, p = 0.009) than MUD-MIP patients. The average age of MUD-only (n = 150) was 30.31 ± 6.83 years, and the average age of NC (n = 282) was 32.66 ± 7.66 years. No differences were found in frequency, average daily dose, administration route, and duration of METH use between MUD-MIP and MUD-only.
Whole genome methylation profiling data
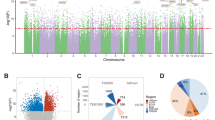
Whole-genome methylation profiling was conducted within a meticulously selected cohort, encompassing subjects with SCZ (n = 8), MUD-MIP (n = 8), MUD-only (n = 9), and NC (n = 8) following stringent matching for age and educational attainment to control for potential confounding variables. Demographic data of these samples analyzed in this study are presented in the Supplementary Document Table S1. There was no significant difference in age among the four groups (F = 0.746, p = 0.533). No significant difference was found in the age of initial methamphetamine use (t = 0.603, p = 0.144) and duration of methamphetamine use (t = 0.067, p = 0.948) between MUD-MIP and MIP-only. To identify probes with differential methylation respectively between SCZ and NC, or MUD-MIP and MUD-only, we run epigenome-wide methylation analysis in representative samples. In comparison with NC samples, 125 of these DMPs were significantly hypomethylated, whereas 406 DMPs were hypermethylated in SCZ patients. 963 DMPs were found between MUD-MIP and MUD-only, with 797 hypomethylated and 166 hypermethylated (see Fig. 2). Finally, 79 hypermethylated DMPs were the intersection of two groups (Supplemental document Table S2).
The DMPs are clustered in the y-row, and the samples are listed in the x-column. The map was generated using the normalized methylation scores (β-values). The higher of the blue signals represents a decreased level of DNA methylation, and the higher of yellow signals indicates an increased DNA methylation level.
Identification of psychotic disorder-associated DNA methylation genes
To screen for candidate genes from the 79 DMPs and eliminate the impact of confounding factors, the following screening strategies were performed: (1) the aberrant methylation of genes should correspond to the specific gene name or UCSC_REFGENE_ACCESSION number. (2) Gene Ontology analysis (GO, http://www.geneontology.org) should confirm the function of the methylation abnormality genes related to the nervous system or psychotic disorders.
The top-ranked DMPs corresponded to the following genes: CSNK1E (casein kinase I isoform epsilon), APOL3 (apolipoprotein L3), UBA6 (ubiquitin-like modifier activating enzyme 6), KIF17 (kinesin superfamily motor proteins17), SETDB1 (SET ___domain bifurcated 1), MLLT3 (mixed lineage leukemia translocated to, 3) and GRM8 (metabotropic glutamate receptor 8). Their primers and probes were designed by Beacon Designer 7.0, as shown in Table 2. These seven genes were selected to perform the MethLight qPCR verification. The epigenetic regulation of these genes in SCZ has been reported in previous studies.
Identification of psychotic disorders-associated DNA methylation pattern
Mann–Whitney U test method was used to analyze methylated Beta values in patients with MUD-MIP (n = 99), MUD-only (n = 150), SCZ (n = 109), and NC (n = 282). Patients with SCZ had higher Beta values in UBA6 (p < 0.05), KIF17 (p < 0.05), and GRM8 (p < 0.001) than NC. Patients with SCZ had higher GRM8 Beta values (p < 0.001) than patients with MUD-only. Patients with MUD-MIP had higher Beta values of APOL3 (p < 0.001), UBA6 (p < 0.001), KIF17 (p < 0.001), MLLT3 (p < 0.001), and GRM8 (p < 0.001) than patients with MUD without MIP. Patients with MUD-MIP had higher Beta values of APOL3 (p < 0.001), UBA6 (p < 0.001), KIF17 (p < 0.001), MLLT3 (p < 0.001), and GRM8 (p < 0.001) than NC (Table 3 and Fig. 2).
The Wilcoxon rank-sum test was used to analyze methylated Beta values in SCZ and MUD-MIP patients before and after treatment. After antipsychotic treatment, Beta values of CSNKE1 (p = 0.007), APOL3 (p = 0.002), and GRM8 (p = 0.001) were significantly decreased in the following-up patients with SCZ (n = 16). Beta values of APOL3 (p = 0.001), UBA6 (p = 0.001), KIF17 (p = 0.001), SETDB1 (p = 0.005), MLLT3 (p = 0.001) and GRM8 (p = 0.001) were significantly decreased in the following up patients with MUD-MIP (n = 15) (Table 4, Figs. 3 and 4).
GRM8 is the only gene whose beta values are significantly higher in patients with SCZ and patients with MUD-MIP than in patients with MUD-only and NC groups. And GRM8 beta values decreased after antipsychotic treatment. Beta values of the promoter region (TS1500) of GRM8 of patients with MUD-MIP or SCZ were significantly higher than patients with MUD-only and NC. Pearson correlation and linear regression analysis showed Beta values of GRM8 were significantly correlated with PANSS total scores (R = 0.404, R2 = 0.164, p < 0.001) and WHODAS2.0 total scores (R = 0.476, R2 = 0.021, p < 0.05) in all patients with SCZ and MUD-MIP (n = 208) in the baseline. In the follow-up patients with SCZ and MUD-MIP (n = 31), Pearson correlation and linear regression analysis showed alternation of Beta values of GRM8 were significantly correlated with reduction of PANSS scores (R = 0.476, R2 = 0.200, p < 0.001) and reduction of WHODAS 2.0 (R = 0.080, R2 = 0.07, p < 0.001) (Table 5, Fig. 5).
Discussion
In the present study, we test whether there were DNA methylation changes related to psychotic symptoms. Our research findings show a correlation between psychotic symptoms and increased CpG methylation in the promoter of GRM8, and remission of psychotic symptoms under the treatment of antipsychotics (risperidone, olanzapine) would be associated with a reduction in hypermethylation of GRM8 in patients with SCZ and MIP. To our knowledge, the present study is one of the largest two-stage epigenome-wide DNA methylation investigations in both primary and substance-induced psychotic disorders and the first to investigate levels of GRM8 methylation in this context. GRM8 is a metabotropic glutamate receptor that maps to chromosome 7q31.3-q32, spanning over 800 kb, and is composed of 11 exons31. Group III metabotropic glutamate receptors (including mGluR8) expressed in the broad region of CNS, predominantly located in the active zone close to the glutamate release site in pre-synapse, and act as autoreceptors mediating the inhibition of glutamate release32,33. mGluR8s has been considered involved in modulating the glutamate level within a normal range to prevent excessive glutamate in synaptic space34, which will produce excitotoxic effects, causing cell damage, neuron amyotrophy, and loss.
Growing evidence suggests that the glutamatergic system plays a major role in the pathophysiology of psychiatric disorders, and glutamate receptors are seen as promising therapeutic targets35,36. Chen et al.37 found three SNPs in intron 6 of GRM8 (rs1361995, rs10487457, rs10487459) in 1049 individual samples from 209 families was significantly correlated with alcohol dependence. In addition, the rs886003 and rs17862325 SNPs loci of GRM8 were associated with alcohol dependence in a European-American population (age 18–26 years). A GWAS study found that rs2237781 genotype type of GRM8 was associated with smoking onset and current smoking38. A study that included Americans (n = 839) versus Italians (n = 3972) subjects found the (rs17864092) mutation in GRM8 to be significant with depressive traits39. Moreover, rs1361995 mutation on GRM8 was found to be associated with major depressive disorder in Han Chinese by Li et al.40. Other lines of research showed GRM8 deletion or copy number variants were identified in attention deficit and hyperactivity disorder41 or autism spectrum disorder42,43.
The dynamic changes of GRM8 methylation in the present study are concordant with the mutation of GRM8 observed in SCZ patients or animal models of SCZ. For example, Takaki et al.44 selected 22 SNPs loci with an average length of 40.3 kb on the GRM8 gene and found statistically significant differences in genotype distribution frequencies between the two groups in 100 Japanese schizophrenic patients and 100 controls (rs2237748, rs2299472), but no significance after Bonferroni correction. Among the four loci constitutive haplotypes on GRM8, rs2237797-rs1361963-rs2283094, and rs1361963-rs2283094-rs2402851 were associated with SCZ in Japanese44. Zhang et al.45 found that the genotype distribution of rs2299472 of GRM8 gene polymorphism was statistically associated with the onset of SCZ in the Uyghur population in China. Later, Li et al.40 found that the rs2237781 mutation on GRM8 was associated with SCZ in Han Chinese. In the Iranian population, rs712723 in GRM8 may play an important role in the pathogenesis of SCZ46. However, an early report of an SNP locus (2846-C/T polymorphism) in GRM8 was not associated with SCZ in British Caucasians47. It is thus clear that the positive association of SCZ with multiple haplotypes in the GRM8 region needs to be confirmed in different ethnic populations.
Abnormally elevated glutamate levels in the striatum or PFC are also involved in the biological process of MIP48, while clinical studies of the relationship between metabotropic glutamate receptors and MIP are relatively rare. Tsunoka T et al.10 found that GRM2 may play a role in the pathophysiology of MIP but not SCZ in the Japanese population. They did not find any correlation between MIP and rs6465084 of GRM3, but this study also pointed out that no further linkage disequilibrium analysis and GRM3 mutation scanning were carried out49. At present, there is no research on whether GRM8 mutation or the function of mGluR8 can influence MIP susceptibility. However, animal experiments have shown that GRM8 or mGluR8 is associated with numerous behavioral paradigms in rodents. The absence of mGluR8 leads to reduced locomotor activity in a novel enclosed environment, delayed response to electric shock, transient delay in response to scene fear and increased measures of anxiety in the open field and elevated plus maze50,51. Intraperitoneal injection of (R,S)-3,4-dicarboxyphenylglycine (80 mg/kg), which is a mGluR8 selective agonist and antagonist of AMPA receptors, could decrease the amphetamine (2.5 mg/kg s.c.) induced hyperactivity52. Continuous chronic intraperitoneal injection of amphetamine (12 days), the expression of GRM8 mRNA in the caudate nucleus and nucleus accumbent, cerebral cortex increased 1 day after withdrawal and continued until 21 days after withdrawal in rat53. The similar reduction of GRM8 mRNA levels were also found in prefrontal lobe and striatum 25 min after rapid cocaine administration54. In this study, the level of GRM8 methylation decreased in PBL after the remission of psychotic symptoms. In fact, methylation modification of cytosine in the promoter region was once considered a highly stable and robust epigenetic mark that ensures a steady state of gene expression and maintains the phenotypic characteristics of cells55. However, subsequent studies found that this methylation modification is actually a highly dynamic process, and the DNA methylation process will not only be inhibited but also regulated by the demethylation process56. Guo et al.57 quantitatively compared the CpG methylation landscape of adult mouse dentate granule neurons in vivo before and after synchronous neuronal activation. About 1.4% of 219,991 CpGs measured showed rapid active demethylation or de novo methylation. By performing an in-depth DNA methylation profiling of two genomic loci surrounding the transcription start site of 2 genes in 51 samples, Giulia et al.58 confirmed a dynamic balance of DNA methylation and demethylation in the same genomic region. Antipsychotics can change the DNA methylation level of different neural pathways in specific neurotransmitter-associated candidate genes59,60,61,62 or at the genome-wide level63,64. Global methylation results revealed a highly significant hypomethylation in SCZ patients, and antipsychotic treatment (haloperidol) was associated with higher methylation variance65. However, very few have assessed clinical outcomes and the potential of differential DNA methylation profiles as predictors of antipsychotic response.
Nevertheless, our results should be considered in light of a number of limitations. First, our analyses were constrained by the technical limitations of the Illumina 450K arrays, which assay only ~3% of CpG sites in the genome. Due to the small sample size in SP I, the whole genome sequencing analysis results need further validation. These May increase the possibility of false negative results. Second, this is a longitudinal study, but we failed to follow up with all the subjects on baseline except the patients who effectively improved the outcome after 5 weeks. We did not evaluate the long-term effects of the treatments after a 5-week treatment period, although most patients visited the outpatient clinic regularly. Third, our study has analyzed DNA methylation in peripheral blood, which is an easily accessible tissue, so it can only provide limited information on disease-related primary tissue variation and potential pathogenic processes. Fourth, we have explored the potential effects of olanzapine or risperidone on DNA methylation by assessing a sub-group of patients with SCZ and MIP. We didn’t strictly standardize the intervention program of the corresponding subjects. Fifth, only male patients from Huan, China, were included in the present study. Considering the possible regional variation, further validation in large and more diverse populations is needed. Finally, although we found some evidence that psychotic symptoms-associated DMPs, we did not detect the expression of candidate genes. Future work will aim to explore more interactions of GRM8 and psychotic symptoms both in animal studies and clinical research.
Conclusion
This study investigated the genome-wide DNA methylation status, which could be used to identify potential biomarkers for psychotic disorders in Chinese male patients with SCZ and MUD-MIP. Seven DMPs associated with psychosis were identified, which were verified in large sample sizes by the MethLight qPCR method. GRM8 was the only gene screened from the genome methylation chip data that showed significant differences in methylation levels in pairwise comparisons of the four groups (SCZ, NC, MUD-MIP, and MUD-only). Meanwhile hypermethylation of GRM8 showed a significant association with the total scores of the Positive Negative Syndrome Scale and WHO disability assessment schedule II in both baseline and follow-up. Further investigations on the function and mechanism of GRM8 will help us to better understand the pathogenesis and prognostic biomarkers of psychotic symptoms as well as therapeutic options that target causative epigenetic alterations.
References
UNODC. World Drug Report (UNODC, New York, NY, 2009).
UNODC. Global Amphetamine-type Stimulant Assessment: Amphetamine and Ecstasy (UNODC, New York, NY, 2011).
UNODC. World Drug Report (UNODC, New York, NY, 2014).
Annual Report on Drug Control in China (Ministry of Public Security of China, Beijing, 2019).
Srisurapanont, M. et al. Comparisons of methamphetamine psychotic and schizophrenic symptoms: a differential item functioning analysis. Prog. Neuropsychopharmacol. Biol. Psychiatry 35, 959–964 (2011).
Hermens, D. F., Lubman, D. I., Ward, P. B., Naismith, S. L. & Hickie, I. B. Amphetamine psychosis: a model for studying the onset and course of psychosis. Med. J. Aust. 190, S22–S25 (2009).
Shelly, J. et al. First-rank symptoms in methamphetamine psychosis and schizophrenia. Psychopathology 49, 429–435 (2016).
Okada, N. et al. Characterizing prefrontal cortical activity during inhibition task in methamphetamine-associated psychosis versus schizophrenia: a multi-channel near-infrared spectroscopy study. Addict. Biol. 21, 489–503 (2016).
Ikeda, M. et al. Evidence for shared genetic risk between methamphetamine-induced psychosis and schizophrenia. Neuropsychopharmacology 38, 1864–1870 (2013).
Tsunoka, T. et al. Association analysis of GRM2 and HTR2A with methamphetamine-induced psychosis and schizophrenia in the Japanese population. Prog. Neuropsychopharmacol. Biol. Psychiatry 34, 639–644 (2010).
Callaghan, R. C. et al. Methamphetamine use and schizophrenia: a population-based cohort study in California. Am. J. Psychiatry 169, 389–396 (2012).
Kittirattanapaiboon, P. et al. Long-term outcomes in methamphetamine psychosis patients after first hospitalisation. Drug Alcohol Rev. 29, 456–461 (2010).
Ujike, H. & Sato, M. Clinical features of sensitization to methamphetamine observed in patients with methamphetamine dependence and psychosis. Ann. N. Y. Acad. Sci. 1025, 279–287 (2004).
A. P. Association. Diagnostic and Statistical Manual of Mental Disorders 5th edn (American Psychiatric Association, Arlington, VA, 2013).
Niemi-Pynttari, J. A. et al. Substance-induced psychoses converting into schizophrenia: a register-based study of 18,478 Finnish inpatient cases. J. Clin. Psychiatry 74, e94–e99 (2013).
Smigielski, L., Jagannath, V., Rossler, W., Walitza, S. & Grunblatt, E. Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings. Mol. Psychiatry 25, 1718–1748 (2020).
Kebir, O., Chaumette, B. & Krebs, M. O. Epigenetic variability in conversion to psychosis: novel findings from an innovative longitudinal methylomic analysis. Transl. Psychiatry 8, 93 (2018).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33(Suppl.), 245–254 (2003).
Kalayasiri, R., Kraijak, K., Mutirangura, A. & Maes, M. Paranoid schizophrenia and methamphetamine-induced paranoia are both characterized by a similar LINE-1 partial methylation profile, which is more pronounced in paranoid schizophrenia. Schizophr. Res. 208, 221–227 (2019).
Lotan, A. et al. Neuroinformatic analyses of common and distinct genetic components associated with major neuropsychiatric disorders. Front. Neurosci. 8, 331 (2014).
Cross-Disorder Group of the Psychiatric Genomics, C. et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat. Genet. 45, 984–994 (2013).
Cross-Disorder Group of the Psychiatric Genomics, C. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet 381, 1371–1379 (2013).
Chen, R. et al. Assessment of functioning and disability in patients with schizophrenia using the WHO Disability Assessment Schedule 2.0 in a large-scale database. Eur. Arch. Psychiatry Clin. Neurosci. 268, 65–75 (2018).
Yen, C. F. et al. Validity and reliability of the Functioning Disability Evaluation Scale-Adult Version based on the WHODAS 2.0-36 items. J. Formos. Med. Assoc. 113, 839–849 (2014).
A. P. Association, Diagnostic and Statistical Manual of Mental Disorders, 5th ed., (American Psychiatric Association, Arlington, VA, 2013).
Satta, R. et al. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc. Natl Acad. Sci. USA 105, 16356–16361 (2008).
Guidotti, A. et al. Epigenetic GABAergic targets in schizophrenia and bipolar disorder. Neuropharmacology 60, 1007–1016 (2011).
Smith, R. C. et al. Varenicline treatment decreases DNMT1 mRNA expression in lymphocytes of schizophrenic patients who are cigarette smokers. Schizophr. Res. 119, 269–270 (2010).
Bibikova, M. et al. High density DNA methylation array with single CpG site resolution. Genomics 98, 288–295 (2011).
Du, P. et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 11, 587 (2010).
Scherer, S. W. et al. Localization of two metabotropic glutamate receptor genes, GRM3 and GRM8, to human chromosome 7q. Genomics 31, 230–233 (1996).
Schoepp, D. D. Unveiling the functions of presynaptic metabotropic glutamate receptors in the central nervous system. J. Pharmacol. Exp. Ther. 299, 12–20 (2001).
Shigemoto, R. et al. Differential presynaptic localization of metabotropic glutamate receptor subtypes in the rat hippocampus. J. Neurosci. 17, 7503–7522 (1997).
Mao, L. M., Mathur, N., Shah, K. & Wang, J. Q. Roles of metabotropic glutamate receptor 8 in neuropsychiatric and neurological disorders. Int. Rev. Neurobiol. 168, 349–366 (2023).
Sanacora, G., Zarate, C. A., Krystal, J. H. & Manji, H. K. Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nat. Rev. Drug Discov. 7, 426–437 (2008).
Lavreysen, H. & Dautzenberg, F. M. Therapeutic potential of group III metabotropic glutamate receptors. Curr. Med. Chem. 15, 671–684 (2008).
Chen, A. C. et al. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am. J. Med. Genet. B Neuropsychiatr. Genet. 150B, 359–368 (2009).
Vink, J. M. et al. Genome-wide association study of smoking initiation and current smoking. Am. J. Hum. Genet. 84, 367–379 (2009).
Terracciano, A. et al. Genome-wide association scan of trait depression. Biol. Psychiatry 68, 811–817 (2010).
Li, W. et al. Significant association of GRM7 and GRM8 genes with schizophrenia and major depressive disorder in the Han Chinese population. Eur. Neuropsychopharmacol. 26, 136–146 (2016).
Sangu, N. et al. A 7q31.33q32.1 microdeletion including LRRC4 and GRM8 is associated with severe intellectual disability and characteristics of autism. Hum. Genome Var. 4, 17001 (2017).
Prasad, A. et al. A discovery resource of rare copy number variations in individuals with autism spectrum disorder. G3 (Bethesda) 2, 1665–1685 (2012).
Serajee, F. J., Zhong, H., Nabi, R. & Huq, A. H. The metabotropic glutamate receptor 8 gene at 7q31: partial duplication and possible association with autism. J. Med. Genet. 40, e42 (2003).
Takaki, H. et al. Positive associations of polymorphisms in the metabotropic glutamate receptor type 8 gene (GRM8) with schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 128B, 6–14 (2004).
Zhang, L. et al. Association analysis of the GRM8 gene with schizophrenia in the Uygur Chinese population. Hereditas 151, 140–144 (2014).
Tavakkoly-Bazzaz, J. et al. TCF4 and GRM8 gene polymorphisms and risk of schizophrenia in an Iranian population: a case-control study. Mol. Biol. Rep. 45, 2403–2409 (2018).
Bolonna, A. A., Kerwin, R. W., Munro, J., Arranz, M. J. & Makoff, A. J. Polymorphisms in the genes for mGluR types 7 and 8: association studies with schizophrenia. Schizophr. Res. 47, 99–103 (2001).
Hsieh, J. H., Stein, D. J. & Howells, F. M. The neurobiology of methamphetamine induced psychosis. Front. Hum. Neurosci. 8, 537 (2014).
Tsunoka, T. et al. No association between GRM3 and Japanese methamphetamine-induced psychosis. Curr. Neuropharmacol. 9, 160–162 (2011).
Duvoisin, R. M. et al. Increased measures of anxiety and weight gain in mice lacking the group III metabotropic glutamate receptor mGluR8. Eur. J. Neurosci. 22, 425–436 (2005).
Gerlai, R., Adams, B., Fitch, T., Chaney, S. & Baez, M. Performance deficits of mGluR8 knockout mice in learning tasks: the effects of null mutation and the background genotype. Neuropharmacology 43, 235–249 (2002).
Ossowska, K., Pietraszek, M., Wardas, J. & Wolfarth, S. Potential antipsychotic and extrapyramidal effects of (R,S)-3,4-dicarboxyphenylglycine [(R,S)-3,4-DCPG], a mixed AMPA antagonist/mGluR8 agonist. Pol. J. Pharmacol. 56, 295–304 (2004).
Parelkar, N. K. & Wang, J. Q. Upregulation of metabotropic glutamate receptor 8 mRNA expression in the rat forebrain after repeated amphetamine administration. Neurosci. Lett. 433, 250–254 (2008).
Zhang, G. C. et al. Acute administration of cocaine reduces metabotropic glutamate receptor 8 protein expression in the rat striatum in vivo. Neurosci. Lett. 449, 224–227 (2009).
Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007).
Moore, L. D., Le, T. & Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 38, 23–38 (2013).
Guo, J. U. et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351 (2011).
De Riso, G. et al. Modeling DNA methylation profiles through a dynamic equilibrium between methylation and demethylation. Biomolecules 10, 1271 (2020).
Abdolmaleky, H. M. et al. DNA hypermethylation of serotonin transporter gene promoter in drug naive patients with schizophrenia. Schizophr. Res. 152, 373–380 (2014).
Tang, H., Dalton, C. F., Srisawat, U., Zhang, Z. J. & Reynolds, G. P. Methylation at a transcription factor-binding site on the 5-HT1A receptor gene correlates with negative symptom treatment response in first episode schizophrenia. Int. J. Neuropsychopharmacol. 17, 645–649 (2014).
Ghadirivasfi, M. et al. Hypomethylation of the serotonin receptor type-2A Gene (HTR2A) at T102C polymorphic site in DNA derived from the saliva of patients with schizophrenia and bipolar disorder. Am. J. Med. Genet. B Neuropsychiatr. Genet. 156B, 536–545 (2011).
Bonsch, D. et al. Methylation matters? Decreased methylation status of genomic DNA in the blood of schizophrenic twins. Psychiatry Res. 198, 533–537 (2012).
Melka, M. G. et al. The effects of olanzapine on genome-wide DNA methylation in the hippocampus and cerebellum. Clin. Epigenet. 6, 1 (2014).
Murata, Y. et al. Comprehensive DNA methylation analysis of human neuroblastoma cells treated with blonanserin. Neurosci. Lett. 563, 123–128 (2014).
Melas, P. A. et al. Epigenetic aberrations in leukocytes of patients with schizophrenia: association of global DNA methylation with antipsychotic drug treatment and disease onset. FASEB J. 26, 2712–2718 (2012).
Acknowledgements
This paper is dedicated to the memory of Dr. Liang Liu, who made significant contributions to the early stages of this research. He was an excellent psychiatrist whose insights and expertise were invaluable to our work. We would like to express our gratitude to Dr. Yuanyuan Dai from the Department of Psychiatry at the Second People’s Hospital of Hunan Province and Dr. Wei Zhou from the Department of Psychiatry at Hunan Kangda Voluntary Drug Rehabilitation Center for their assistance in subject recruitment. We would also like to thank all the volunteers who understood our study purpose and participated in this study.
Funding
This work was supported by the Key Program of the National Natural Science of China [Grant Number 81130020] and the National 973 Program [Grant Number 2015CB553500] to Wei Hao; the National Natural Science Foundation of China [Grant Number 82001405] to and Natural Science Foundation of Hunan Province [Grant Number 2021JJ40979] to Huixi Dong.
Author information
Authors and Affiliations
Contributions
H.D. collected the majority of the data and wrote the manuscript. T.L., C.Y., and M.L. helped with data collection. H.D. and Y.S. performed all data analyses. Y.S. and W.H. designed the study. All authors contributed to and have approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dong, H., Luo, T., Yang, C. et al. Psychotic symptoms associated increased CpG methylation of metabotropic glutamate receptor 8 gene in Chinese Han males with schizophrenia and methamphetamine induced psychotic disorder: a longitudinal study. Schizophr 10, 91 (2024). https://doi.org/10.1038/s41537-024-00506-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41537-024-00506-9