Abstract
Cell state dynamics underlying successful tissue regeneration are undercharacterized. In the intestine, damage prompts epithelial reprogramming into revival stem cells (revSCs) that reconstitute Lgr5+ intestinal stem cells (ISCs). Here single-nuclear multi-omics of mouse crypts regenerating from irradiation shows revSC chromatin accessibility overlaps with ISCs and differentiated lineages. While revSC genes themselves are accessible throughout homeostatic epithelia, damage-induced remodelling of chromatin in the crypt converges on Hippo and the transforming growth factor-beta (TGFβ) signalling pathway, which we show is transiently activated and directly induces functional revSCs. Combinatorial gene expression analysis further suggests multiple sources of revSCs, and we demonstrate TGFβ can reprogramme enterocytes, goblet and paneth cells into revSCs and show individual revSCs form organoids. Despite this, loss of TGFβ signalling yields mild regenerative defects, whereas interference in both Hippo and TGFβ leads to profound defects and death. Intestinal regeneration is thus poised for activation by a compensatory system of crypt-localized, transient morphogen cues that support epithelial reprogramming and robust intestinal repair.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Raw data for 10x multi-modal RNA-seq and ATAC-seq of 0 and 2 dpi samples, and RNA-seq of 1 dpi sample, along with processed Seurat R objects associated with 0 and 2 dpi data, and 0, 1 and 2 dpi data are available at Gene Expression Omnibus (GEO) (accession GSE230766). The JASPAR database is available at www.bioconductor.org/packages/release/data/annotation/html/JASPAR2022.html. Source data are provided with this paper.
Code availability
The R and Python code used to process, analyse and produce the panels for the various different figures in this study are available via GitHub at https://github.com/wrana-lab/revSC_tgfb. Intermediate files needed to run the example code are available via Zenodo at https://doi.org/10.5281/zenodo.13315436 (ref. 47).
References
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Tao, S. et al. Wnt activity and basal niche position sensitize intestinal stem and progenitor cells to DNA damage. EMBO J. 36, 2920–2921 (2017).
Nusse, Y. M. et al. Parasitic helminths induce fetal-like reversion in the intestinal stem cell niche. Nature 562, 109–113 (2018).
Ayyaz, A. et al. Single-cell transcriptomes of the regenerating intestine reveal a revival stem cell. Nature 569, 121–125 (2019).
de Sousa e Melo, F. & de Sauvage, F. J. Cellular plasticity in intestinal homeostasis and disease. Cell Stem Cell 24, 54–64 (2019).
Ohara, T. E., Colonna, M. & Stappenbeck, T. S. Adaptive differentiation promotes intestinal villus recovery. Dev. Cell 57, 166–179.e6 (2022).
Karo-Atar, D. et al. Helminth-induced reprogramming of the stem cell compartment inhibits type 2 immunity. J. Exp. Med. 219, e20212311 (2022).
Goldsmith, J. R. et al. TNFAIP8 controls murine intestinal stem cell homeostasis and regeneration by regulating microbiome-induced Akt signaling. Nat. Commun. 11, 2591 (2020).
Andersen, C. L. et al. Clusterin expression in normal mucosa and colorectal cancer. Mol. Cell Proteom. 6, 1039–1048 (2007).
Chen, X. et al. Clusterin as a biomarker in murine and human intestinal neoplasia. Proc. Natl Acad. Sci. USA 100, 9530–9535 (2003).
Yui, S. et al. YAP/TAZ-dependent reprogramming of colonic epithelium links ECM remodeling to tissue regeneration. Cell Stem Cell 22, 35–49.e7 (2018).
Morral, C. et al. p53 promotes revival stem cells in the regenerating intestine after severe radiation injury. Nat. Commun. 15, 3018 (2024).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Heuberger, J. et al. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc. Natl Acad. Sci. USA 111, 3472–3477 (2014).
Wallaeys, C., Garcia‐Gonzalez, N. & Libert, C. Paneth cells as the cornerstones of intestinal and organismal health: a primer. EMBO Mol. Med. 15, e16427 (2023).
Treveil, A. et al. Regulatory network analysis of paneth cell and goblet cell enriched gut organoids using transcriptomics approaches. Mol. Omics 16, 39–58 (2020).
Kim, C. K. et al. Krüppel-like factor 5 regulates stemness, lineage specification, and regeneration of intestinal epithelial stem cells. Cell Mol. Gastroenterol. Hepatol. 9, 587–609 (2020).
van Es, J. H. et al. Wnt signalling induces maturation of paneth cells in intestinal crypts. Nat. Cell Biol. 7, 381–386 (2005).
Ishibashi, F. et al. Contribution of ATOH1+ cells to the homeostasis, repair, and tumorigenesis of the colonic epithelium. Stem Cell Rep. 10, 27–42 (2018).
Tomic, G. et al. Phospho-regulation of ATOH1 Is required for plasticity of secretory progenitors and tissue regeneration. Cell Stem Cell 23, 27–42 (2018).
Wester, R. A. et al. Retinoic acid signaling drives differentiation toward the absorptive lineage in colorectal cancer. iScience 24, 103444 (2021).
Hickey, J. W. et al. Organization of the human intestine at single-cell resolution. Nature 619, 572–584 (2023).
Lopez, R. et al. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMG Genomics 19, 477 (2018).
Moor, A. E. et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell 175, 1156–1167.e15 (2018).
Chen, L. et al. TGFB1 induces fetal reprogramming and enhances intestinal regeneration. Cell Stem Cell 30, 1520–1537.e8 (2023).
Hayashi, S., Lewis, P., Pevny, L. & McMahon, A. P. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119, 93–97 (2002).
Hayashi, H. et al. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 89, 1165–1173 (1997).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Serra, D. et al. Self-organization and symmetry breaking in intestinal organoid development. Nature 569, 66–72 (2019).
Ko, T. C., Beauchamp, R. D., Townsend, C. M., Thompson, E. A. & Thompson, J. C. Transforming growth factor-β inhibits rat intestinal cell growth by regulating cell cycle specific gene expression. Am. J. Surg. 167, 14–20 (1994).
Yin, X. et al. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112 (2014).
El Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Levéen, P. et al. Induced disruption of the transforming growth factor beta type II receptor gene in mice causes a lethal inflammatory disorder that is transplantable. Blood 100, 560–568 (2002).
Li, M. et al. Mothers against decapentaplegic-related protein 2 expression in avian granulosa cells is up-regulated by transforming growth factor β during ovarian follicular development. Endocrinology 138, 3659–3665 (1997).
Guillermin, O. et al. Wnt and Src signals converge on YAP‐TEAD to drive intestinal regeneration. EMBO J. 40, e105770 (2021).
Gregorieff, A., Liu, Y., Inanlou, M. R., Khomchuk, Y. & Wrana, J. L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 526, 715–718 (2015).
Barry, E. R. et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature 493, 106–110 (2013).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
Cai, J. et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 24, 2383–2388 (2010).
Sanman, L. E. et al. Transit-amplifying cells coordinate changes in intestinal epithelial cell-type composition. Dev. Cell 56, 356–365.e9 (2021).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e4 (2019).
Van den Berge, K. et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 11, 1201 (2020).
Sato, T. et al. Single Lgr5 stem cells build crypt–villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009).
Gregorieff, A. et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology 129, 626–638 (2005).
Caldwell, L. & Patel, S. Custom revSC analysis files for Fink et al. 2024, in Nature Cell Biology. Zenodo https://doi.org/10.5281/zenodo.13315436 (2024).
Acknowledgements
We thank L. Attisano (University of Toronto) for critical comments, L. Caldwell for managing data and code availability and the Network Biology Collaborative Centre (nbcc.lunenfeld.ca) for single-cell profiling and sequencing service. The Network Biology Collaborative Centre is a facility supported by Canada Foundation for Innovation, the Ontarian Government, Genome Canada and Ontario Genomics (OGI-139). This research is supported by the Canadian Cancer Society (grant no. 706715), the Terry Fox Research Institute (programme project grant no. 1107) and the Canadian Institutes of Health Research (CIHR grant no. PJT-191835) to J.L.W. D.C. and A.R. are supported by a CIHR postdoctoral fellowship award (FRN 193957). Schematics throughout the manuscript were created with Biorender.com.
Author information
Authors and Affiliations
Contributions
A.A., M.F. and J.L.W. co-conceived the study. A.A. and D.T designed and executed the multi-omics experiment. K.C. performed 10x genomics. S.J.P., D.P.C. and A.O. performed all multi-omics data processing and analysis. D.P.C. applied scVI data integration methods. S.J.P. developed Combinator and performed the associated analyses. M.J.P. maintained mouse colonies for various genotypes involved, along with D.T. and K.N. M.F. performed experiments pertaining to Tgfbr2−/− mice. M.F. and K.N. performed experiments pertaining to YTT mice; this included TAM and irradiation treatments, tissue collection, preparation and characterizations by H&E, IHC, IF staining and RNAscope. M.N. developed and applied Smad7 FISH. A.R. performed lineage-enriched organoid experiments. V.M. and F.R. aided in additional tissue preparation and processing. M.F. performed in vitro organoid experiments. M.F., S.J.P, D.P.C., K.N. and J.L.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Cell Biology thanks Nick Barker and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Temporal and transcriptional analysis of multiomics data set.
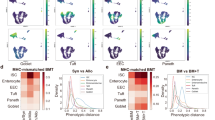
a) WNN embedding of cells (1b) colored based on time point origin. b) Proportion of the number of cells within each identified WNN cluster relative to total number of cells from 0 dpi or 2 dpi across the full embedding (top). Relative proportion of number of cells within each identified WNN cluster from either 0 dpi or 2 dpi relative to total number of cells from both timepoints within each cluster (bottom). c) UMAP plot visualizing intestinal cell types generated using unsupervised clustering of single nuclei RNA-Seq data at 0 and 2 dpi. Clusters identified at higher resolutions (colour coded) were collapsed into groups corresponding to the indicated major lineages. d) Clusterwise median of the weighting of ATAC (red bar) and RNA (blue bar) data on the nuclei embedding for each of the indicated WNN clusters (x axis) identified in Fig. 1b. e) Relative proportion of nuclei (legend) from each WNN cluster (x axis) that mapped to the indicated ATAC cluster is shown as a heatmap. f) Proportion of the number of cells within each identified ATAC cluster relative to total number of cells from 0 dpi (blue) or 2 dpi (red) across the full embedding (top). Relative proportion of number of cells within each identified ATAC cluster from either 0 dpi (blue) or 2 dpi (red) relative to total number of cells from both timepoints within each cluster (bottom). g) Dot plot of the average ChromVAR Z-scores of each cell type for the indicated motifs. Size of the dot corresponds to the percentage of nuclei from any given cell type that that has >0 or <0 ChromVAR z-scores depending on if the average is positive or negative, respectively. Multiomics source data available at GEO: GSE230766.
Extended Data Fig. 2 Accurate reconstruction of zonation patterns by trajectory inference.
a) scVI-integrated UMAP embedding of 0 and 2 dpi cells, discretized into five bins based on their position along the inferred trajectory. b) Expression levels of zonation markers defined by Moor et al. in each of the five discrete regions. To assess features at homeostasis, only 0 dpi cells were evaluated. Boxes range between the 25th and 75th percentiles, bars reflect the median, and whiskers extend 1.5x the interquartile range. c) Chromatin accessibility and expression evaluated at 0dpi for Tfrc, Slc2a2 and Slc15a1. Pseudo-bulk profiles of snATAC-Seq data associated with the indicated loci are plotted as genomic traces, along with the corresponding gene structure and peak identifications (left panels). Light grey bars highlight peaks with zonation-associated accessibility. Corresponding expression is shown to the right and colour coded according to pseudotime levels. For each gene, the height of ATAC traces was set to a common range and RNA expression quantified as log-transformed UMI counts per 10k counts (log CP10k + 1).
Extended Data Fig. 3 RevSC features along the crypt-villus axis.
a) scVI-integrated UMAP embeddings showing expression of revSC associated genes. b-d) Chromatin accessibility and expression along the CV axis for the indicated genes at 0 and 2 dpi. Pseudo-bulk profiles of snATAC-Seq data associated with the indicated loci are plotted as genomic traces, along with the corresponding gene structure and peak identifications (left panels). Grey bars highlight injury-associated peaks. Corresponding expression is also shown, and colour coded according to levels. Genomic coordinates are shown and for each gene, the height of ATAC traces were set to a common range and RNA expression is quantified as log-transformed UMI counts per 10k counts (log CP10k + 1).
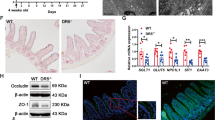
Extended Data Fig. 4 TGFβ is transiently activated in the regenerating intestine.
a) ChromVAR activity scores for TEAD motifs in 0 and 2 dpi cells. For each cell, scores for TEAD1-4 were averaged given the similarity of their binding motif. p-value calculated using a linear model to compare chromVar scores between 0 and 2 dpi cells within Level 1 zonation bin (P = 1.62E-13). b) Pseudo-bulk profiles of snATAC-Seq data of Tgfbr2 in 0 and 2 dpi cells along the CV axis, plotted as genomic traces with corresponding gene structure and peak identifications (see ED3b-d). Grey bars highlight injury-associated peaks. c) Dot plot of median gene expression of Smad2 and Smad3 in 0 and 2 dpi nuclei along the CV axis. P-values calculated via Wilcoxon rank sum test comparing 0 and 2 dpi cells within each level, adjusted for multiple comparisons using the Benjamini-Hochberg method *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. Smad3: P= Level 5, 9.4E-3; Level 4, 9.9E-2; Level 3, 4.2E-4; Level 2, 6.0E-6; Level 1, 1.2E-2. Smad2: P=Level 5, 0.97; Level 4, 0.29; Level 3, 0.16; Level 2, 0.0017; Level 1, 0.055. d) Representative RNAscope images of Tgfb1 expression with T-cell marker (CD3E), Macrophage marker (Adgre1) or Stromal marker (Pdgfra) at 2 dpi (12 Gy). White arrows denote co-localization. Scale bar, 50 µm. e) Representative RNAscope images for Tgfbr2 and Clu in wild-type intestines at 0 and 2 dpi (12 Gy). Scale bar, 100 µm. f) Immunohistochemistry staining of Smad2/3 protein in wild-type mice following intestinal injury at 0-, 1-, 2-, 3-, 5-, and 7- dpi (12 Gy). Boxes denote enlarged regions. Red and white arrows denote cells with or without nuclear accumulation of Smad2/3, respectively. Scale bar, 100 µm. g) Representative immunofluorescence staining of Smad7 (red) expression in wild type mice following intestinal injury at 0-, 2-, 3-, 5-, and 7 dpi (12 Gy). Dotted lines denote intestinal crypts. Scale bar, 100 µm. h) Fluorescent in situ hybridization of Smad7 in wild type mice at 0-,3-,5-, and-7 dpi (12 Gy). Scale bar 50 µm. N = 3 mice per condition (c-d, f-h).
Extended Data Fig. 5 Characterizing the in vitro response of intestinal organoids to TGFβ pathway stimulation.
a) Representative immunofluorescent images of Clu-GFP organoids treated with TGFβ1 or SB431542 (10 µM) for 24 hours, as indicated. Staining shows EpCam, Clu-GFP, and Smad2/3. Nuclear Smad2/3 is indicated by solid arrows, nuclear holes indicated by open arrows. Scale bar,10 µm. N = 3 replicates. b) Gating strategy identifying GFP+ cells isolated from organoids via flow cytometry. Plots indicate cell populations that were included based on size (singlets) and viability (DAPI-) (left). Representative plots indicating the GFP+ fraction in (1) GFP negative organoids or (2) Clu-GFP organoids treated with SB431542 (10 µM) or induced with TGFβ1(200 pM) (left). SS, side scatter; FS, forward scatter. c) Organoids expressing Clu-GFP were treated with TGFβ1 (200 pM; 24 or 48 h) and the percent GFP+ cells quantified by flow cytometry. Bars represent the mean ( ± SEM) of N = 3 individual experiments performed in triplicate, N = 5 for control and 48 h treatment groups (dots). Unpaired, two-tailed t-test; SB P = 0.25, 24 h TGFβ1 P = 0.04, 48 h TGFβ1 P = 0.01. d) Representative IF images of Lgr5-GFP-expressing organoids treated +/- TGFβ1 (50 or 200pM; 48 h, as indicated). Scale bar, 50 µm (left, middle), Scale bar, 10 µm (right). e) Quantification of the number of organoids in culture following 24 h exposure. Organoids were treated for 24 h after 3 days of growth and the total number of organoids/well quantified. N = 3 independent experiments performed in triplicate (dots). All data shown as mean ± SEM. One-way ANOVA, P = 0.54. Source numerical data available in source data.
Extended Data Fig. 6 Flow cytometry analysis strategy of lineage-traced organoids.
a) Schematic of experimental design accompanying Fig. 4d. Clu-CreERT2;lsl-tdTom;Lgr5-GFP organoids were treated with a pulse of TGFβ (TGFβ1; 24 h) and 4-OHT to induce recombination prior to passaging and flow cytometry. b) Gating strategy used to quantify cell populations isolated from organoids via flow cytometry. Plots indicate cell populations that were included based on size (singlets) and viability (DAPI-). c) Representative plots indicating the proportion of (1) GFP-/tdTom- (2) GFP + /tdTom-; (3) GFP + /tdTom+ and (4) GFP-/tdTom+ populations in Clu-CreERT2;lsl-tdTom;Lgr5-GFP organoids treated +/-4-OHT to induce lineage tracing and +/-TGFβ1 (1, 50 or 200 pM), as indicated (bottom panels). GFP-/tdTom- and GFP-/tdTom+ organoids were used for gating controls.
Extended Data Fig. 7 Organoids employ revSCs for functional regeneration following damage.
a) Representative images of Clu-tdTom;Lgr5-GFP organoids treated with TGFβ1 (200 pM) and 4-OHT (schematic), stained for Paneth (Lyz1), Enteroendocrine (ChgA) and Goblet cells (Muc2). Red arrows indicate cells of interest. Scale bars, 10 µm (Crypt) and 50 µm (whole organoid). N = 3 replicates. b) Schematic (top) and representative IF images (bottom) of control and 5-FU-treated (20 µM; 24 h) Lgr5-GFP-expressing organoids. Scale bar, 10 µm (Crypts), 50 µm (whole organoids). N = 3 replicates. c) Schematic (top) and brightfield images (bottom) showing organoid morphology following a 24-hour pulse of 5-FU (20 µM) and after 2 successive passages. Scale bar, 10 µm. N = 3 replicates. d) Expression of indicated ISC signature genes in organoids treated with 5-FU (20 µM; 48 h) was quantified by RT-qPCR and normalized to Ywhaz expression. Data are plotted as mean ± SEM. P= Lgr5, 0.011; Olfm4, 0.0038; Ascl2 = 0.055; Lrig1, 0.088. N = 3 replicates. e) Expression of indicated revSC signature genes in organoids treated with 5-FU (20 µM; 48 h), quantified by RT-qPCR, and normalized to Ywhaz expression. Data are plotted as mean ± SEM. P= Clu, 0.05; Anxa1, 0.003; F3, 9.6E-3. N = 3 experiments performed in triplicate (d-e). f) Clu-CreERT2;lsl-tdTom;Lgr5-GFP organoids treated with 5-FU (20 µM) and 4-OHT (top) as indicated (Schematic), and lineage-traced offspring visualized after damage, as indicated. Scale bars, 10 µm. Unpaired, one-tailed t-test; *p ≤ 0.05; ** p < 0.01; ***p < 0.001 (d and e). P, passage; d, day; h, hour; 4-Hydroxytamoxifen, 4-OHT; Clu-CreERT2;lsl-tdTom;Lgr5-GFP-DTR, ClutdTom;Lgr5-GFP. N = 3 replicates. Source numerical data available in source data.
Extended Data Fig. 8 Analysis of human colon organoids.
a) Schematic of experimental design (top) and representative bright-field images of human colon organoid (HCO) cultures (bottom). P, passage number; d, day. +/- denotes presence or absence of TGFβ/inhibitor. Scale bar, 500 µm. b) qRT-PCR analysis indicating changes in revSC marker expression (CLU, F3, ANXA1) and SMAD7. Expression normalized to GAPDH, ‘+A-83-01/-TGFβ1’ represents the control. The mean±range of N = 2 replicates, performed in triplicate, is plotted. Source numerical data available in source data.
Extended Data Fig. 9 Combinator Identifies a differentiation path for Goblet cells through secretory progenitors and differentiated organoid production.
a) Unsupervised clustering of single cell RNA-Seq profiles of cells from unirradiated 0dpi cells from this study and Ayyaz et al. (2019) that were integrated and aligned on a UMAP using scVI1. Different clusters are identified by different colors and labelled according to their cell type identity. Clusters identified at higher cluster resolutions were collapsed into groups corresponding to their gross cell type. b) Combinator rankings of cells in uninjured intestinal epithelium based on a goblet cell signature (Rab27a, Rab27b, Sytl2, Atoh1, Foxa3, Hepacam2, Qsox1) were overlaid on the UMAP in a. For combinator pseudotime analysis, only cells from the intestinal epithelium were retained. c) Smoothened pseudotime heatmap created using the individual rankings of cells according to their expression of all permutations of a goblet cell signature. 100 positions along a pseudotime were simulated and percentage of cells from all indicated clusters relative to each other at each position was calculated and smoothened to create the pseudotime map. d) Intestinal organoids from ClutdTom;Lgr5:GFP mice were plated in matrigel and treated with DMSO or the indicated small molecule signalling modulators to drive differentiation of Lgr5+ intestinal stem cells into enterocytes (IV), Paneth cells (CD) or Goblet cells (ID). Fixed organoids were stained with the indicated antibodies to visualize the corresponding differentiated cell. Scale bar=100 μm.
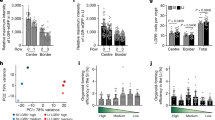
Extended Data Fig. 10 The absence of Yap signalling reduces revSC induction and delays regeneration.
a) Representative IHC staining of Yap in the proximal and distal regions of the small intestine at 0 and 2 dpi (12 Gy) of wild-type mice. Scale bar, 50 μm. Dotted boxes denote enlarged regions, scale bar, 20 μm. Yellow and white arrows depict Yap+ and Yap−nuclear staining, respectively (left). Quantification of mean nuclear intensity of DAB staining per crypt expressed as mean ± SD (right). N = 3 mice per condition. 0 dpi P = 0.68, 2 dpi P = 0.43. b) Representative images of RNAscope indicating Clu expression in Vil-CreERT2; Yapfl/fl; Tazfl/fl (-TAM) and Vil-CreERT2; Yap-/-; Taz-/- ( + TAM) knockout mice at 3 or 5 dpi (left). Scale bar, 100 µm. Quantification of Clu+ cells in different regions of the intestine are presented as a box and whisker plot (right). Whiskers show maximum and minimum values, box extends from the 25thto 75th percentiles, with median represented by the centre line. N = 2 mice per condition. 3 dpi P = 0.005, 5 dpi P = 0.2. c) Immunostaining of Olfm4 in Vil-CreERT2;Yapfl/fl;Tazfl/fl and Vil-CreERT2;Yap-/-;Taz-/- mice at 0-, 3-, and 5 dpi (12 Gy). Scale bar, 50 μm. Arrows denote intestinal crypts with re-emerging Olfm4 signal (left). Quantification of Olfm4+ crypts per 1000 μm is presented as relative percentage (right). Dashed line indicates median. N = 3 mice per condition. 0 dpi P = 0.67, 3 dpi P = 0.0015, 5 dpi P = 1.62E-6. d) Kaplan–Meier curves of control (-TAM) and YTT-/- ( + TAM) mice following 12 Gy IR. All YTT-/- mice required euthanasia by 5 dpi. Control and triple KO mice: Vil-CreERT2; Yapfl/fl; Tazfl/fl;Tgfbr2fl/fl. Control, N = 5 (2 M, 3 F); YTT-/-, N = 9 (3 M, 6 F). P = 0.0002. Statistical analysis via unpaired, two-tailed t-test (a-c), or log-rank test (d). Numerical source data available in source data.
Supplementary information
Supplementary Information
Supplementary Fig. 1.
Supplementary Table 1
Supplementary Tables 1–13, lists of resources, and supplementary statistics from figures.
Source data
Source Data Fig. 1
Graph and statistical source data.
Source Data Fig. 3
Graph and statistical source data.
Source Data Fig. 4
Graph and data.
Source Data Fig. 5
Graph and statistical source data.
Source Data Fig. 6
Graph and statistical source data.
Source Data Fig. 7
Graph and statistical source data.
Source Data Fig. 7
Unprocessed gel image.
Source Data Fig. 8
Graph and statistical source data.
Source Data Extended Data Fig. 1
Graph and statistical source data.
Source Data Extended Data Fig. 5
Graph and statistical source data.
Source Data Extended Data Fig. 7
Graph and statistical source data.
Source Data Extended Data Fig. 8
Graph and statistical source data.
Source Data Extended Data Fig. 10
Graph and statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fink, M., Njah, K., Patel, S.J. et al. Chromatin remodelling in damaged intestinal crypts orchestrates redundant TGFβ and Hippo signalling to drive regeneration. Nat Cell Biol 26, 2084–2098 (2024). https://doi.org/10.1038/s41556-024-01550-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41556-024-01550-4
This article is cited by
-
Gastrointestinal acute radiation syndrome: current knowledge and perspectives
Cell Death Discovery (2025)