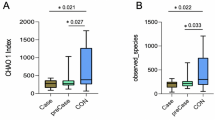
Abstract
The local arrangement of microbes can profoundly impact community assembly, function and stability. However, our understanding of the spatial organization of the human gut microbiome at the micron scale is limited. Here we describe a high-throughput and streamlined method called Split-And-pool Metagenomic Plot-sampling sequencing (SAMPL-seq) to capture spatial co-localization in a complex microbial consortium. The method obtains microbial composition of micron-scale subcommunities through split-and-pool barcoding. SAMPL-seq analysis of the healthy human gut microbiome identified bacterial taxa pairs that consistently co-occurred both over time and across multiple individuals. These co-localized microbes organize into spatially distinct groups or ‘spatial hubs’ dominated by Bacteroidaceae, Ruminococcaceae and Lachnospiraceae families. Using inulin as a dietary perturbation, we observed reversible spatial rearrangement of the gut microbiome where specific taxa form new local partnerships. Spatial metagenomics using SAMPL-seq can unlock insights into microbiomes at the micron scale.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Raw sequencing reads are available from PRJNA1196417. The Refseq 16S database (https://www.ncbi.nlm.nih.gov/refseq/targetedloci/16S_process/) was used for species-level taxonomic identification, and SILVA 138.1 (https://www.arb-silva.de/documentation/release-1381/) was used for taxonomic assignment of all ASVs. Source data are provided with this paper.
Code availability
Scripts for read processing are implemented in BASH and R. They are available from GitHub at https://github.com/wanglabcumc/SAMPL-seq (ref. 61).
Change history
11 February 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41564-025-01951-7
References
Donaldson, G. P., Lee, S. M. & Mazmanian, S. K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14, 20–32 (2016).
Gilbert, J. A. et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018).
Tropini, C., Earle, K. A., Huang, K. C. & Sonnenburg, J. L. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21, 433–442 (2017).
Ho, P.-Y., Good, B. H. & Huang, K. C. Competition for fluctuating resources reproduces statistics of species abundance over time across wide-ranging microbiotas. eLife 11, e75168 (2022).
Wu, F. et al. Modulation of microbial community dynamics by spatial partitioning. Nat. Chem. Biol. 18, 394–402 (2022).
Reichenbach, T., Mobilia, M. & Frey, E. Mobility promotes and jeopardizes biodiversity in rock–paper–scissors games. Nature 448, 1046–1049 (2007).
Rakoff-Nahoum, S., Coyne, M. J. & Comstock, L. E. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr. Biol. 24, 40–49 (2014).
Sheth, R. U. et al. Spatial metagenomic characterization of microbial biogeography in the gut. Nat. Biotechnol. 37, 877–883 (2019).
Riva, A. et al. A fiber-deprived diet disturbs the fine-scale spatial architecture of the murine colon microbiome. Nat. Commun. 10, 4366 (2019).
Koh, A., Vadder, F. D., Kovatcheva-Datchary, P. & Bäckhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011).
Earle, K. A. et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015).
Shi, H. et al. Highly multiplexed spatial mapping of microbial communities. Nature 588, 676–681 (2020).
Mondragón-Palomino, O. et al. Three-dimensional imaging for the quantification of spatial patterns in microbiota of the intestinal mucosa. Proc. Natl Acad. Sci. USA 119, e2118483119 (2022).
Cao, Z. et al. Spatial profiling of microbial communities by sequential FISH with error-robust encoding. Nat. Commun. 14, 1477 (2023).
Lötstedt, B., Stražar, M., Xavier, R., Regev, A. & Vickovic, S. Spatial host–microbiome sequencing reveals niches in the mouse gut. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01988-1 (2024).
Welch, J. L. M., Hasegawa, Y., McNulty, N. P., Gordon, J. I. & Borisy, G. G. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017).
Grodner, B. et al. Spatial mapping of mobile genetic elements and their bacterial hosts in complex microbiomes. Nat. Microbiol. https://doi.org/10.1038/s41564-024-01735-5 (2024).
Zilionis, R. et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12, 44–73 (2017).
Blattman, S. B., Jiang, W., Oikonomou, P. & Tavazoie, S. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat. Microbiol. 5, 1192–1201 (2020).
Kuchina, A. et al. Microbial single-cell RNA sequencing by split-pool barcoding. Science 371, eaba5257 (2021).
Bloom, J. D. Estimating the frequency of multiplets in single-cell RNA sequencing from cell-mixing experiments. PeerJ 6, e5578 (2018).
Yamawaki, T. M. et al. Systematic comparison of high-throughput single-cell RNA-seq methods for immune cell profiling. BMC Genomics 22, 66 (2021).
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020).
Knight, R. et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16, 410–422 (2018).
Gotelli, N. J. Null model analysis of species co-occurrence patterns. Ecology 81, 2606–2621 (2000).
Ryan, M. J. et al. Development of microbiome biobanks – challenges and opportunities. Trends Microbiol. 29, 89–92 (2021).
Blanchet, F. G., Cazelles, K. & Gravel, D. Co-occurrence is not evidence of ecological interactions. Ecol. Lett. 23, 1050–1063 (2020).
Frioux, C. et al. Enterosignatures define common bacterial guilds in the human gut microbiome. Cell Host Microbe 31, 1111–1125.e6 (2023).
Le Bastard, Q. et al. The effects of inulin on gut microbial composition: a systematic review of evidence from human studies. Eur. J. Clin. Microbiol. Infect. Dis. 39, 403–413 (2020).
Cornick, N. A., Jensen, N. S., Stahl, D. A., Hartman, P. A. & Allison, M. J. Lachnospira pectinoschiza sp. nov., an anaerobic pectinophile from the pig intestine. Int. J. Syst. Evol. Microbiol. 44, 87–93 (1994).
Riva, A. et al. Identification of inulin-responsive bacteria in the gut microbiota via multi-modal activity-based sorting. Nat. Commun. 14, 8210 (2023).
Lozupone, C. A., Stombaugh, J. I., Gordon, J. I., Jansson, J. K. & Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 489, 220–230 (2012).
Wu, G., Zhao, N., Zhang, C., Lam, Y. Y. & Zhao, L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 13, 22 (2021).
Miquel, S. et al. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261 (2013).
Kim, H., Jeong, Y., Kang, S., You, H. J. & Ji, G. E. Co-culture with Bifidobacterium catenulatum improves the growth, gut colonization, and butyrate production of Faecalibacterium prausnitzii: in vitro and in vivo studies. Microorganisms 8, 788 (2020).
Huang, Y. et al. High-throughput microbial culturomics using automation and machine learning. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01674-2 (2023).
Hoskins, L. C., Boulding, E. T., Gerken, T. A., Harouny, V. R. & Kriaris, M. S. Mucin glycoprotein degradation by mucin oligosaccharide-degrading strains of human faecal bacteria. Characterisation of saccharide cleavage products and their potential role in nutritional support of larger faecal bacterial populations. Microb. Ecol. Health Dis. 5, 193–207 (1992).
Van den Abbeele, P. et al. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J. 7, 949–961 (2013).
Ge, X. et al. SRS-FISH: a high-throughput platform linking microbiome metabolism to identity at the single-cell level. Proc. Natl Acad. Sci. USA 119, e2203519119 (2022).
Mahowald, M. A. et al. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc. Natl Acad. Sci. USA 106, 5859–5864 (2009).
Wolfe, B. E. & Dutton, R. J. Fermented foods as experimentally tractable microbial ecosystems. Cell 161, 49–55 (2015).
Levy, R. & Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl Acad. Sci. USA 110, 12804–12809 (2013).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Wilkins, O. G., Capitanchik, C., Luscombe, N. M. & Ule, J. Ultraplex: a rapid, flexible, all-in-one fastq demultiplexer. Wellcome Open Res. 6, 141 (2021).
Shen, W., Le, S., Li, Y. & Hu, F. SeqKit: a cross-platform and ultrafast toolkit for FASTA/Q file manipulation. PLoS ONE 11, e0163962 (2016).
Rognes, T., Flouri, T., Nichols, B., Quince, C. & Mahé, F. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584 (2016).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Buschmann, T. DNABarcodes: an R package for the systematic construction of DNA sample tags. Bioinformatics 33, 920–922 (2017).
Pruesse, E., Peplies, J. & Glöckner, F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829 (2012).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Yilmaz, P. et al. The SILVA and ‘All-species Living Tree Project (LTP)’ taxonomic frameworks. Nucleic Acids Res. 42, D643–D648 (2014).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst. 1695, 9 (2006).
Kembel, S. W. et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010).
Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011).
Kassambara, A. ggpubr: ‘ggplot2’ based publication ready plots. R package version 0.6.0 https://rpkgs.datanovia.com/ggpubr/ (2018).
Pedersen, T. L. ggraph: an implementation of grammar of graphics for graphs and networks. R package version 2.2.1.9000 https://github.com/thomasp85/ggraph (2022).
Ji, B. W. et al. Quantifying spatiotemporal variability and noise in absolute microbiota abundances using replicate sampling. Nat. Methods 16, 731–736 (2019).
Gohl, D. M. et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 34, 942–949 (2016).
Caporaso, J. G. et al. EMP 16S Illumina Amplicon Protocol. https://doi.org/10.17504/protocols.io.nuudeww (2018).
Richardson, M. SAMPL-Seq, V1.0 (2025); https://doi.org/10.5281/zenodo.14619156
Acknowledgements
We thank P. Sims, D. Vitkup, L. Dietrich, K. Tchourine for immensely helpful insights and discussions; G. Gerber and T. Gibson for strengthening the computational rigour of the method; and members of the Wang Laboratory for providing a supportive environment. H.H.W. acknowledges funding support from the NSF (MCB-2025515), NIH (2R01AI132403, 1R01DK118044, 1R01EB031935, 1R21AI146817), ONR (N00014-18-1-2237), Burroughs Wellcome Fund (1016691), ARO (W911NF-22-2-0210), DARPA (HR0011-23-2-0001), Irma T. Hirschl Trust, and the Schaefer Research Award. M.R., R.U.S. and F.V.-C. were supported by the NSF Graduate Research Fellowship Program (DGE-1644869). R.U.S. was supported by the Fannie and John Hertz Foundation Fellowship. This study was supported in part by the Columbia University Digestive and Liver Disease Research Center (funded by NIH grant 5P30DK132710) through use of its bioinformatic and single-cell analysis core.
Author information
Authors and Affiliations
Contributions
M.R., R.U.S. and H.H.W. conceived the project. R.U.S., M.R. and T.M. developed and validated the protocol. M.R., R.U.S., D.R., Y.H., L.L. J.L. and G.U. performed experiments. M.R., S.Z., Y.Q. and F.V.-C. analysed the data. M.R. and H.H.W. generated and edited figures. H.H.W. supervised the overall project. M.R., H.H.W. and S.Z. wrote the manuscript with input from co-authors. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
H.H.W. is a scientific advisor of SNIPR Biome, Kingdom Supercultures, Fitbiomics, VecX Biomedicines, Genus PLC, and a scientific co-founder of Aclid and Foli Bio, all of which are not involved in the study. R.U.S. is a co-founder of Kingdom Supercultures. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Microbiology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Characterization of SAMPL-seq steps and comparison with other methods.
(a) Histograms showing the effect of the number of in situ PCR cycles on both the ASVs per particle and reads per particle based on different number of PCR cycles. (b) Plot summarizing the overall ease of use and performance of various microbial spatial analysis methods. (c) Table comparing the performance of different spatial analysis methods.
Extended Data Fig. 2 Homogenized fecal mixing experiment (M1).
(a) Histograms of particle sizes for the replicates. (b,c) Correlation of ASV prevalence by SAMPLE-seq and ASV relative abundance by bulk 16S sequencing for Core 1 at particle size 15-30 μm (b) and Core 2 (c). SAMPL-seq abundances are averaged between replicates (excluding Spike-in). (d) Histogram of the ASV per particle distribution by size (excluding Spike-in). (e) Barplot of the singlet rate of each replicate, grouped by particle size.
Extended Data Fig. 3 Defined community mixing experiment (M2).
(a) An outline of the process for producing the fecal mixing library. Zymo Gut Microbial Standard and Zymo High Concentration Spike-in are separately embedded at equal cell ratios at 1x or 3x concentration replicates. They are then combined during the cryofracturing step, and are then size sorted, amplified and sequenced in aliquots of 10,000 particles, of average size ~50μm, which were used for further analysis (b) Histograms of particle sizes for the replicates. (c) Scatterplots of technical (amplification) and biological (concentration) replicates, using both the relative abundance based on summed reads, and ASV prevalence among particles, which is the percentage of particles an ASV is found (excluding the Spike-in). (d) Scatterplot of ASV relative abundance and prevalence compared to absolute reference provided by the manufacturer. SAMPL-seq abundances are averaged between replicates (excluding Spike-in). (e) Histograms of the ASV per particle distribution by concentration (excluding Spike-in). (f) Barplot of the multiplet rate of each replicate, grouped by concentration. (g) Plot showing mixing rates of two defined communities (M1A and M1B), with each colored dot corresponding to a classified particle. (h) Heatmap of particles clustered by Bray-Curtis similarity and the Ward’s method.
Extended Data Fig. 4 Correlation between ASVs from different murine gut compartments.
(a) Clustered heatmap of prevalent mouse ASVs grouped by gut compartments, with summed abundances at the end of the row. ASVs are clustered by Jaccard overlap across the dataset. (b, c) Correlation between ASV relative abundance (b) and prevalence among particles (c) between mouse gut compartments. Colon and feces samples showed the highest correlation among samples. (d) Principal Coordinate Analysis (PCoA) plot of particles derived from different mouse gut compartments using Simpson distance, colored by gut compartments. Dashed circles correspond to the 95% confidence interval for each compartment using the multivariate t-distribution.
Extended Data Fig. 5 Particle-level data of the human gut microbiome from stool profiling.
(a, b) Using filters for 20-40 μm, distributions of particle sizes (a) and ASVs per particle (b) for longitudinal human stool samples from H11 are shown. (c, d) Using filters for 20-40 μm, distributions of particle size (c) and ASVs per particle (d) for interpersonal samples are shown. (e,f) Particles from longitudinal (e) or interpersonal (f) are clustered within each day using the Simpson overlap, and ASVs are clustered using their Jaccard overlap across all days. (g) Rarefaction plot for longitudinal samples of unique ASV-particle pairs for prevalent ASVs (>1% prevalence in particles). (h, i) Rarefaction plots for interpersonal samples of unique ASV-particle pairs (h) for prevalent ASVs (>1% particle prevalence) or unique ASV-ASV co-presence (i) in a particle (observed >3 times).
Extended Data Fig. 6 Technical validations of H11 Day 4 SAMPL-seq libraries.
(a) Scatterplots of amplification (technical) replicates showed high correlation. Correlation between spatial (biological) replicates also showed high correlation. Homogenized sample showed high correlation, but increased particle prevalence relative to intact libraries. (b, c) Correlation of bulk ASV relative abundance (b) or ASV prevalence (c) between longitudinal SAMPL-seq libraries. (d, e) Correlation of bulk ASV relative abundance (d) or ASV prevalence (e) between interpersonal SAMPL-seq libraries. (f, g) Correlation of ASV relative abundance between interpersonal (f) or H11 longitudinal (g) samples with their corresponding bulk measurements.
Extended Data Fig. 7 Large scale ASV compositional patterns.
(a) Heatmap of overall ASV abundance in the dataset of prevalent ASVs (>1%), clustered by Jaccard overlap. (b) Heatmap of overall ASV abundance of prevalent ASVs (>1%) across 5 humans (H1, H10, H11, H18, H19), clustered by Bray-Curtis distance. (c) Heatmap of family-level relative abundance of human fecal samples. Families are clustered using the Jaccard overlap, and families conserved across all individuals are indicated in green.
Extended Data Fig. 8 Pairwise ASV colocalization analysis.
(a) Barplot of the number of significant ASV pairs found in each sample. (b) Scatterplot between ASV-pair Z-Scores between intact and disrupted samples. (c) Scatterplots of ASV-Pair Z-scores between fresh and frozen samples. (d) Pairwise ASV spatial associations in five people. Each heatmap shows all statistically significant spatial associations between pairs of ASVs for each individual (H1, H10, H11, H18, H19). Colors in the heatmap correspond to Z-scores and stars correspond to statistical significance (p < 0.05 BH FDR Corrected). ASVs are labeled in 3 possible colors (red, green, blue) if they belong to a conserved spatial cluster group (P1, P2, P3) found across 3 or more individuals. Common taxonomic families are labeled next to each ASV label on the y-axis. (e) Heatmap of significant co-associations found on 2 or more days in H11. (f) Heatmap of significant co-associations found in 3 or more people.
Extended Data Fig. 9 Phylogenetic Distance Distributions and Relationships Between Clusters.
Histograms of simulated MPD distributions for L1-L4 (a) or P1-P3 (b) spatial hubs. The red line indicates the observed MPD in the cluster, while blue dashed lines indicate the 95% confidence interval around the mean of simulations. P-value was calculated using the two-sided Z-score. (c) Scatterplot of Z-score for associations found in both for longitudinal and interpersonal samples with the corresponding correlation. (d) Contingency table of the sign of longitudinal versus interpersonal associations. (e) Contingency table of ASV presence across the clusters. Chi-squared test of independence (χ2 = 22.4, df = 6, p = 0.001).
Extended Data Fig. 10 Inulin supplementation.
Heatmaps of bulk relative abundance, Z-score sum, and change in total Z-score for ASVs that had a total Z-score change >10 over the course of inulin supplementation.
Supplementary information
Supplementary Information
Supplementary Tables 1–11.
Supplementary Data 1
Raw gel data for the gel in Fig. 1.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Richardson, M., Zhao, S., Lin, L. et al. SAMPL-seq reveals micron-scale spatial hubs in the human gut microbiome. Nat Microbiol 10, 527–540 (2025). https://doi.org/10.1038/s41564-024-01914-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41564-024-01914-4