Abstract
Video capsule endoscopy (VCE) is an important technology with many advantages (non-invasive, representation of small bowel), but faces many limitations as well (time-consuming analysis, short battery lifetime, and poor image quality). Artificial intelligence (AI) holds potential to address every one of these challenges, however the progression of machine learning methods is limited by the avaibility of extensive data. We propose Galar, the most comprehensive dataset of VCE to date. Galar consists of 80 videos, culminating in 3,513,539 annotated frames covering functional, anatomical, and pathological aspects and introducing a selection of 29 distinct labels. The multisystem and multicenter VCE data from two centers in Saxony (Germany), was annotated framewise and cross-validated by five annotators. The vast scope of annotation and size of Galar make the dataset a valuable resource for the use of AI models in VCE, thereby facilitating research in diagnostic methods, patient care workflow, and the development of predictive analytics in the field.
Similar content being viewed by others
Background & Summary
Video Capsule Endoscopy (VCE) is a minimally invasive gastroenterological imaging procedure used to capture video footage of a patient’s digestive tract. This is especially relevant for the small intestine, which is not readily accessible through conventional endoscopic procedures like colonoscopy and esophagogastroduodenoscopy. However, this comes with limitations such as a time-consuming manual analysis1, technical restrictions (e.g., battery runtime2 or a lack of active locomotion), and heterogeneous image quality. In 16.5% of cases, the capsule does not pass through the ileocecal valve, resulting in incomplete small intestine examinations3.
VCE is currently primarily employed for the detection of internal bleeding4,5. However, the potential use cases for VCE are far more expansive. The indications for capsule endoscopy are evolving alongside technological advancements, such as the introduction of colon capsule endoscopy6, thereby expanding its use e.g. in pediatric populations and for inflammatory bowel disease7.
The major drawback of VCE is the large amount of video footage generated, as medical staff are required to watch hours of recorded video. In these recordings, the section of interest is a tiny subset of the total video, and fluctuating image quality renders large parts unusable for diagnostic purposes. The use of Artificial Intelligence (AI) in VCE is already reducing the diagnostic evaluation time needed to interpret the large amount of VCE footage. With the rise of AI in VCE, the procedure has the potential to become more widely used and thereby more cost-efficient, as observed in other modalities, such as AI-assisted X-Ray evaluation8. Lately, the integration of Edge AI emphasizes the growing need for efficient, miniaturized algorithms for low-power devices9,10, which opens up new possibilities for real-time analysis within VCE systems.
The successful development of AI models requires substantial quantities of high-quality data, as well as precise and rigorous annotations11. However, the availability of large datasets is scarce; the VCE-datasets thus far publicly available are either relatively small12,13,14, or are limited to specific questions (e.g., quality, ulcers, bleeding, polyps, anatomy)12,15– 18. To further drive the progress of AI in VCE, the creation of large, preprocessed, and annotated datasets is necessary16,19. Most academic research projects process their own data, which is tailored to their specific tasks20,21 and do not make their datasets publicly available. The drawback of such an individualistic approach is that it necessitates a disproportionate amount of resources, limiting the progress of research. The existence of large, high-quality datasets could reduce the cost and effort involved in developing research for VCE and other medical technologies22.
In this publication, we introduce a dataset that marks a considerable advancement in the field of capsule endoscopic research. In the ___domain of VCE there are a few openly accessible datasets, an overview of these is given in Table 6. Galar positions itself to be one of the largest datasets in the field. With 29 distinct labels, incorporating a broad range of functional, anatomical, and pathological annotations across 3,513,539 frames, Galar is primed for application in the field of machine learning.
Furthermore, the Galar dataset consists of VCE data from two endoscopy centers in Germany, with two different capsule systems (OlympusTM Endocapsule 10 System, PillCamTM SB2, SB3, and Colon2 Capsule Endoscopy Systems23,24). As multidisciplinary and multicenter VCE research is needed for the clinical use of AI in patient diagnosis16,25, this further elevates Galar in its usefulness.
In summary, we provide a multicentric, multisystem dataset with high frame count and the most diverse and detailed annotations to date. These characteristics establish Galar as a robust resource for training machine learning models in video capsule endoscopy.
Methods
Videos were collected from the University Hospital Carl Gustav Carus (Dresden, Germany) and from an outpatient practice for gastroenterology (Dippoldiswalde, Germany). VCE recordings were obtained from August 2011 to March 2023 using the OlympusTM Endocapsule 10 System (Hamburg, Germany) as well as the PillCamTM SB2, SB3, and Colon Capsule Endoscopy Systems (Meerbusch, Germany)23,24. The videos were initially generated in proprietary data formats and were converted to the Moving Picture Experts Group (MPEG) format. The video resolution ranged from 336 × 336 pixels (OlympusTM) to 576 × 576 pixels (PillCamTM). Out of the 449 recordings, 80 videos were pre-selected for annotation based on the related findings by selecting only pathological videos for annotation. To de-identify VCE recordings, randomly generated study IDs were assigned, and the videos were cut. Afterwards, videos were transferred to university servers. There each video in the dataset was labeled framewise, resulting in 3,513,539 labeled frames.
This study was approved by the Ethics Committee of the University Hospital Carl Gustav Carus at the Technical University of Dresden on December 16, 2022 (Ethics ID: BO-EK-534122022), confirming adherence to the ethical principles of the Declaration of Helsinki. Due to the retrospective anonymization of the data and their collection during clinically indicated routine interventions, explicit consent was not required. This is additionally supported by the Ethics Committee’s approval, a consultation with the data privacy officer, and local law. Section 34, Paragraph 1 of the Saxon Hospital Act (SächsKHG) explicitly allows the collection and analysis of this type of data.
Data preparation
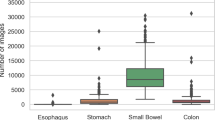
CVAT26 is a web-based, open-source image- and video annotation tool. Using CVAT, five annotators (a team of experienced gastroenterologists and trained medical students) labelled the data. The labels were categorized into three main groups: The technical group consists of labels concerning the image quality, where good view indicates a reduction of the view by less than 50%, reduced view indicates a reduction of the view by over 50%, and no view indicates a reduction of the view by over 95%. Furthermore, a distinction is made between bubbles and dirt as factors contributing to the degradation of image quality. The anatomical group consists of typical landmarks: z-line, pylorus, papilla of Vater, ileocecal valve and the different sections of the gastrointestinal tract: mouth, esophagus, stomach, small intestine, colon. The final group is the pathological group, which consists of the most frequent pathologies found in VCE and some less frequent findings: ulcer, polyp, active bleeding, blood, erythema, erosion, angiectasia, inflammatory bowel disease, foreign body, esophagitis, varices, hematin, celiac, cancer, lymphangiectasis. The pathologies esophagitis, varices and celiac did not occur in any of the videos. Figure 1 gives an overview of example images of the 26 labels in the dataset, Fig. 2 displays the number of annotated frames per label.
Annotation Process
An early decision was made to label each frame in the dataset individually, with the annotation process occurring in multiple stages. From each of the videos, every unique frame was extracted using Python (v3.9.8)27 and FFMPEG (v4.0.6)28. Frames originating from the PillCamTM capsule system were cropped to remove black borders. A timestamp, visible in the top right corner, was also removed. No further pre-processing was done for the videos from the OlympusTM capsule system. Subsequently, the frames were uploaded to CVAT, where frames were annotated by our team. Frames containing unrecognizable features were given the label unknown. Then, all frames labeled with a pathology were cross-validated with the confirmation of a secondary annotator. Any frames still possessing the unknown label were reviewed by a gastroenterologist with 10 years of experience in endoscopy and were relabeled accordingly.
Data Records
The Galar VCE dataset can be found in the open access repository figshare29. It consists of 3,513,539 frames, each labeled with 29 labels, and has a total size of ~ 580GB. A detailed overview of the structure of the dataset is shown in Fig. 3. Each video in the dataset was labeled framewise. The dataset contains the folders Frames and Labels. The Labels folder contains CSV files, where each file has a header starting with the index column, followed by the columns of the 29 possible labels described in Data preparation and ending with the frame column, which refers to the corresponding frame the labels belong to. The Frames folder consists of 80 sub-folders, each containing the frames associated with a study. Table 1 shows the number of videos, resolution, and distribution of frames per capsule system. Additionally, a metadata file is provided, containing patient age, gender, and capsule system used.
Six videos contain technical annotations: 5, 8, 9, 13, 14, 22. A total of 35,733 frames were annotated with this label category. The creation of technical annotations was found to be more resource intensive compared to the other categories, as the visibility is more volatile and prone to sudden change. As the category is highly relevant for machine learning (ML) applications in VCE, the labels were included for completeness.
Technical Validation
The dataset was used to train multiple ResNet-50 models30. The data was split into a set for training and validation consisting of 60 videos, and a test set comprised of the remaining 20 videos. K-fold cross-validation was performed on the data in the training and validation set. The videos from the test set all originate from the Dippoldiswalde practice and there is no overlap of these videos with those from the train set.
The labels dirt and bubbles form a multi-label classification problem, while the labels good view, reduced view, and no view as well as the section labels mouth, esophagus, stomach, small intestine, and colon require multi-class classification. Some of the other more frequently occurring pathological (e.g., blood or polyp) labels are trained on separately.
For the classification of the multi-label and the multi-class models, 5-fold cross-validation was employed. The binary classification of the pathologic labels was done using 2-fold cross-validation, as some labels were not contained in a sufficient number of videos. To ensure that the frames of one patient are not spread over the training and test set and to get the best possible distribution of the labels over the folds, sklearn’s StratifiedGroupKFold31 method was applied.
The ResNet-50 model pre-trained on ImageNet32 was fine-tuned for 10 epochs using PyTorch (v2.0.1)33. Following this, fine-tuning was done for each of the target tasks. These models were trained over 100 epochs (with early stopping), with a 128 Batch size and a 0.001 learning rate. For each image, a Resize transform was applied, to scale the image down to 224 × 224. Additionally, the transforms ShiftScaleRotate, RGBShift, GaussNoise and RandomBrightnessContrast were each applied with a 30% likelihood to each image. The small subset of images which contain the technical annotation were fine-tuned similarly, excepting the epochs, which were capped at 50 with early stopping.
As measurements for the classification performance, the F-1 score, the Area under the Receiver Operating Characteristic Curve (AUROC) as well as the accuracy were calculated using the TorchMetrics (v1.0.3)34 Python library.
Tables 2, 3, 4, and 5 show results for the classification models. The model fine-tuned for dirt and bubbles, along with the two multi-class models, performed decently with accuracy value up to 93% for the labels stomach and 92% for small intestine.
The binary models for pathological labels encountered challenges to accurately identify positive samples. To improve performance, weighted sampling as well as weighted loss was explored. For weighted sampling, the probability for an image to be sampled was based on the occurrence of its class, as a fraction of the total dataset. For the more complex multi-label problem, each unique combination of labels was assigned a weight, again as a portion of the total dataset. This made the weights dynamic, based on the target the model is trained on.
Although these strategies helped to improve performance on some labels, other required heavy parameter optimization. This underscores the difficulty and necessity of improving and developing AI methods to address the challenges of imbalanced label distrubution and multi-source data. Consequently, it highlights the importance of a multicentric, multisystem dataset with extensive annotations of pathologies.
Usage Notes
With Galar we provide the largest public VCE dataset, both in terms of the number of features labeled per image and the total number of annotated images. The large number of ground truth labeled images allows for supervised training of machine learning models and is a significant contribution to the landscape of publicly available VCE datasets.
If the dataset is to be employed for machine learning applications, it is essential to carefully partition the data into training and validation sets. The comparative rarity of select labels, especially over others in the same class, must be respected. Additionally, the data originates from two different VCE systems and was collected at two different study sites. Patients of varying age and gender are also present in the dataset. This information must be considered when generating splits. The metadata file, found in the figshare repository, provides information regarding the capsule system and patient age and gender, per individual study.
The dataset is provided compressed, in the 7-Zip (.7z) format. The data must be uncompressed before it may be viewed and modified; common operating systems (Windows, Linux, MacOs) by default provide archive utility which enables this.
By licensing the dataset under a Creative Commons Attribution 4.0 International (CC BY 4.0) License which allows sharing, copying, and redistribution, as well as adaptation and transformation, we hope to advance research in the field. For more details about Creative Commons licensing, please refer to https://creativecommons.org.
Code availability
The code employed for the technical validation can be accessed via our public GitHub Repository: https://github.com/EKFZ-AI-Endoscopy/GalarCapsuleML. The repository contains a full guide on running the code, tuning hyperparameters, and generating statistics.
References
Ahmed, M. et al. Video Capsule Endoscopy in Gastroenterology. Gastroenterology Research 15, 47–55, https://doi.org/10.14740/gr1487 (2022).
Kwack, W. G. et al. Current Status and Research into Overcoming Limitations of Capsule Endoscopy. Clinical endoscopy 49, 8–15, https://doi.org/10.5946/ce.2016.49.1.8 (2016).
Liao, Z. et al. Indications and detection, completion, and retention rates of small-bowel capsule endoscopy: a systematic review. Gastrointestinal Endoscopy 71, 280–286, https://doi.org/10.1016/j.gie.2009.09.031 (2010).
Goenka, M. K. et al. Capsule endoscopy: Present status and future expectation. World Journal of Gastroenterology 20, 10024–10037, https://doi.org/10.3748/wjg.v20.i29.10024 (2014).
Iddan, G. et al. Wireless capsule endoscopy. Nature 405, 417–417, https://doi.org/10.1038/35013140 (2000).
Spada, C. et al. Colon capsule endoscopy: What we know and what we would like to know. World Journal of Gastroenterology : WJG 20, 16948–16955, https://doi.org/10.3748/wjg.v20.i45.16948 (2014).
Nemeth, A. et al. Video capsule endoscopy in pediatric patients with Crohn’s disease: a single-center experience of 180 procedures. Therapeutic Advances in Gastroenterology 11, 1756284818758929, https://doi.org/10.1177/1756284818758929 (2018).
Mun, S. K. et al. Artificial Intelligence for the Future Radiology Diagnostic Service. Frontiers in Molecular Biosciences, 7, https://www.frontiersin.org/articles/10.3389/fmolb.2020.614258 (2021).
Werner, J. et al. Precise Localization Within the GI Tract by Combining Classification of CNNs and Time-Series Analysis of HMMs., 174–183, https://doi.org/10.1007/978-3-031-45676-3_18 (2024).
Wang, Y. et al. A locally-processed light-weight deep neural network for detecting colorectal polyps in wireless capsule endoscopes. J. Real-Time Image Process. 18(4), 1183–1194, https://doi.org/10.1007/s11554-021-01126-7 (2021).
Wang, S. et al. Annotation-efficient deep learning for automatic medical image segmentation. Nature Communications 12, 5915, https://doi.org/10.1038/s41467-021-26216-9 (2021).
Smedsrud, P. H. et al. Kvasir-Capsule, a video capsule endoscopy dataset. Scientific Data 8, 142, https://doi.org/10.1038/s41597-021-00920-z (2021).
Akihito, Y. et al. The SEE-AI Project Dataset, https://doi.org/10.34740/KAGGLE/DS/1516536 (2022).
Cychnerski, J. et al. ERS: a novel comprehensive endoscopy image dataset for machine learning, compliant with the MST 3.0 specification. https://arxiv.org/abs/2201.08746 (2022).
Charoen, A. et al. Rhode Island gastroenterology video capsule endoscopy data set. Scientific Data 9, 602, https://doi.org/10.1038/s41597-022-01726-3 (2022).
Iakovidis, D. K. et al. Software for enhanced video capsule endoscopy: challenges for essential progress. Nature Reviews Gastroenterology & Hepatology 12, 172–186, https://doi.org/10.1038/nrgastro.2015.13 (2015).
Palak, H. et al. Deepak Gunjan, Prof. Nidhi Goel, Prof. S. Indu. AI-KODA Dataset: An AI-Image Dataset for Automatic Assessment of Cleanliness in Video Capsule Endoscopy as per Korea-Canada Scores., https://doi.org/10.6084/m9.figshare.25807915.v1 (2024).
Thakur, A. et al. VCE-AnomalyNet: A New Dataset Fueling AI Precision in Anomaly Detection for Video Capsule Endoscopy, https://doi.org/10.22541/au.171387106.63353485/v1 (2024).
Park, J. et al. Recent Development of Computer Vision Technology to Improve Capsule Endoscopy. Clinical Endoscopy 52, 328–333, https://doi.org/10.5946/ce.2018.172 (2019).
Hwang, Y. et al. Improved classification and localization approach to small bowel capsule endoscopy using convolutional neural network. Digestive Endoscopy 33, 598–607, https://doi.org/10.1111/den.13787 (2021).
Mascarenhas, S. et al. Artificial Intelligence and Capsule Endoscopy: Automatic Detection of Small Bowel Blood Content Using a Convolutional Neural Network. GE - Portuguese Journal of Gastroenterology 29, 331–338, https://doi.org/10.1159/000518901 (2021).
Zhang, A. et al. Shifting machine learning for healthcare from development to deployment and from models to data. Nature Biomedical Engineering 6, 1330–1345, https://doi.org/10.1038/s41551-022-00898-y (2022).
PillCam SB 3 Capsule ∣ Medtronic (UK). https://www.medtronic.com/covidien/en-gb/products/capsule-endoscopy/pillcam-capsules/pillcam-sb-3-capsule.html.
Kapselendoskopie - Gastroenterologie - Olympus Medizintechnik. https://www.olympus.de/medical/de/Produkte-und-L.
Yang, Y. J. et al. The Future of Capsule Endoscopy: The Role of Artificial Intelligence and Other Technical Advancements. Clinical Endoscopy 53, 387–394, https://doi.org/10.5946/ce.2020.133 (2020).
CVAT. https://www.cvat.ai/.
Python. https://www.python.org/.
FFMPEG, et al. https://ffmpeg.org/.
Le Floch, M. et al. Galar - a large multi-label video capsule endoscopy dataset. Figshare+. https://doi.org/10.25452/figshare.plus.25304616.v1 (2025) .
Xu, W. et al. ResNet and its application to medical image processing: Research progress and challenges. Computer Methods and Programs in Biomedicine 240, 107660, https://doi.org/10.1016/j.cmpb.2023.107660 (2023).
Sklearn’s StratifiedGroupKFold. scikit-learn, sklearn.model_selection.StratifiedGroupKFold.html.
ImageNet. https://www.image-net.org/.
PyTorch. https://pytorch.org/.
Detlefsen. TorchMetrics - Measuring Reproducibility in PyTorch. Journal of Open Source Software 7, 4101, https://doi.org/10.21105/joss.04101 (2022).
Acknowledgements
This research was funded by the BMBF (German Federal Ministry of Education and Research) as part of the SEMECO cluster4future FKZ 03ZU1210GA and 03ZU121HB.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
M.L.F.: methodology, investigation, data curation, writing original draft, visualization. F.W.: methodology, software, technical validation, writing original draft, visualization. L.M.: methodology, software, data curation, writing original draft, visualization. C.W.: methodology, data annotation. A.P.: methodology, data annotation. K.V.: methodology, data annotation. P.H.: data curation, investigation. S.H.K.: methodology, investigation, clinical supervision. J.L.S.: methodology, investigation, review and editing. C.S.: data curation. M.E.G.: methodology, data curation, data annotation. M.H.: review and editing. S.S.: data curation, technical and clinical supervision. A.M.: methodology, review and editing. A.H.: methodology, review and editing. J.N.K.: review and editing, supervision. S.P.: conceptualization, investigation, data curation. J.H.: conceptualization, review and editing, supervision, N.H.: conceptualization, investigation, data curation, writing original draft, review, and editing, visualization, supervision, project administration, funding acquisition. F.B.: conceptualization, investigation, data curation, review, and editing, visualization, supervision, project administration, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Le Floch, M., Wolf, F., McIntyre, L. et al. Galar - a large multi-label video capsule endoscopy dataset. Sci Data 12, 828 (2025). https://doi.org/10.1038/s41597-025-05112-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-025-05112-7