Abstract
Evidence from previous studies have demonstrated that gut microbiota are closely associated with occurrence of interstitial cystitis/bladder pain syndrome (IC/BPS), yet the causal link between the two is not well known. In this study, we performed a two-sample Mendelian randomization (MR) analysis to determine the possible causal association between gut microbiota with IC/BPS. Gut microbiota summary level data were derived from the genome-wide association study (GWAS) conducted by MiBioGen and the IC/BPS GWAS summary level data were obtained from the GWAS Catalog. Next, we performed an MR study to investigate the causal link between gut microbiota and IC/BPS. The primary method for causal analysis was the inverse variance weighted (IVW), and the MR results were validated through multiple sensitivity analyses. A positive association was found between IC/BPS and eight gut microbial taxa, including genus Bacteroides, genus Haemophilus, genus Veillonella, genus Coprococcus1, genus Butyricimonas, family Bacteroidaceae, family Christensenellaceae, and order Lactobacillales. Sensitivity analysis revealed lack of significant pleiotropy or heterogeneity in the obtained results. This MR analysis reveals that a causal association exists between some gut microbiota with IC/BPS. This finding may is expected to guide future research and development of IC/BPS preventions and treatments based on the bladder-gut axis. However, given the clinical complexity and diagnostic challenges of IC/BPS, along with the limitations of using large-scale GWAS summary data for analysis, our MR results require further validation through additional research.
Similar content being viewed by others
Introduction
Interstitial cystitis (IC), also known as bladder pain syndrome (BPS), manifests as chronic pelvic pain that is primarily associated with bladder (pain, pressure, discomfort) and is accompanied by persistent lower urinary tract symptoms lasting over six weeks, in the absence of an infection or specific etiological factors1. Currently, the etiology of IC/BPS is unknown, but the disease causes severe debilitation and affects the quality of life in affected individuals2. The incidence of IC/BPS has been arising, with an estimated rate ranging from approximately 0.01–2.3%. Moreover, females have a five times higher prevalence compared with that of males3. The precise etiology and pathogenesis of IC/BPS are not fully understood. Depending on the presence or absence of Hunner lesions, IC/BPS can be divided into Hunner-type interstitial cystitis (HIC) and BPS, which can be differentiated through cystoscopy3. However, diagnosing BPS remains highly challenging due to the significant variability in clinical presentations among individuals. Although several therapeutic approaches have been performed, the effectiveness of current treatments for IC/BPS is poor, with the recurrence rate reported to be high during long-term follow-up4. Some patients even require destructive surgery such as cystectomy5. Therefore, to alleviate the burden of IC/BPS, it is imperative to investigated the etiology and identify more effective therapeutic targets.
The pathophysiological mechanisms of IC/BPS are multifactorial, yet our understanding of them remains limited6. Implying intercommunication between the bladder and the gut, the conceptualization of the bladder-gut-brain axis suggests that gut functionality may influence functional urological disorders, including IC/BPS7. Gut microbiota have been reported to modulate gut functionality8, and dysbiosis leads to the occurrence of inflammatory, metabolic, mental, and immune-related diseases in humans9. Current studies have indicated that the composition of the human gut microbiota is influenced by various factors such as diet10, aging11, and antibiotic use12. Although the phenotype of the gut microbiota is shaped by postnatal environments, it has been reported that the host’s genetic state also affects the composition of gut microbiota13. Given the overlapping microbes in the diverse symptoms of gut and bladder diseases implies that the microbial community may modulate the communication between the bladder and the brain-gut axis14,15. Both the urine microbiota and the gut microbiota are essential components of the human microbiome, and some studies have suggested that the gut may be a potential origin of the urinary microbiota15. Numerous studies have demonstrated that dysbiosis of the urine microbiota is associated with various urinary diseases, including IC/BPS16. However, research on the relationship between the gut microbiota and IC/BPS remains relatively limited. In recent years, high-throughput sequencing has helped to uncover the relationship between gut microbiota and IC/BPS. Studies employing 16S rRNA sequencing methods have demonstrated dysbiosis in the gut microbiota of individuals with IC/BPS compared to controls17. This observation was further supported by an animal study, where administration of anaerobes cultured from the stool of IC/BPS patients exacerbated pelvic allodynia when orally gavaged to experimental mice18.
Although above studies have linked gut microbiota to IC/BPS, they are primarily observational in nature, often based on small sample sizes and are susceptible to confounding factors. Consequently, research has not been unable to uncover the specific causal connection between the two. Therefore, conducting a thorough genetic-level assessment of the causal association between gut microbiota and IC/BPS is imperative.
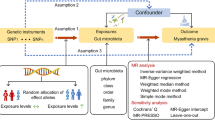
The Mendelian randomization (MR) method can elucidate the causal association between exposure and outcome by utilizing single nucleotide polymorphisms (SNPs) as instrumental variables (IVs)19. Because genetic variations are randomly allocated at conception, MR studies are less prone to the influence of reverse causation or confounding factors which is often observed in traditional observational methods20. In recent years, the hypothesis of a causal association between gut microbiota and various diseases has been assessed and confirmed by multiple MR studies21,22,23.
In this investigation, we obtained summarized GWAS data on gut microbiota and IC/BPS from publicly available extensive genome-wide association studies (GWAS). We utilized a two-sample MR approach to analyze these data, unveiling the causal effect of specific gut microbiota on the risk of IC/BPS. The aim of our study was to reveal a new biological marker for the diagnosis and treatment of patients with challenging IC/BPS, as well as to offer partial evidence for bladder-gut communication within the bladder-gut-brain axis.
Methods
Data sources
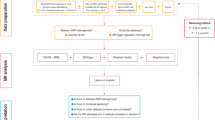
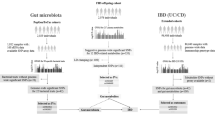
The complete design of this study is shown in Fig. 1. Given the chronic nature of IC/BPS, GWAS data were derived using the diagnostic term “Interstitial cystitis (chronic)” in line with the ICD-10 classification. The GWAS data for (chronic) interstitial cystitis (ID: GCST90044234), involving 240 cases and 456,108 controls of European ancestry, were obtained from the GWAS Catalog (https://www.ebi.ac.uk/gwas/)24. In the GWAS data we obtained, chronic interstitial cystitis is defined as “a condition with recurring discomfort or pain in the urinary bladder and the surrounding pelvic region without an identifiable disease. Severity of pain in interstitial cystitis varies greatly and often is accompanied by increased urination frequency and urgency”. This definition corresponds to the ICD-10 diagnosis code N30.1 [Interstitial cystitis (chronic)], and the GWAS data we used included patients meeting this diagnosis code. While there are some variations in the definitions of IC/BPS across different guidelines2, our comparison revealed that the ICD-10 classification’s definition of IC/BPS is largely consistent with those in these guidelines. Therefore, we believe that using the ICD-10 classification’s definition for diagnosing and selecting patients with IC/BPS is appropriate and reliable.
The summary-level data related to gut microbiota (https://mibiogen.gcc.rug.nl/) were derived from the extensive GWAS MiBioGen consortium. In this GWAS meta-analysis, 211 bacterial taxa units were analyzed, which included 9 phyla, 16 classes, 20 orders, 35 families, and 131 genera, with 18,340 participants from 24 cohorts across 11 countries25. To ensure accuracy, 196 taxa were retained after exclusion of 15 bacterial taxa with unknown family and genus in this study.
Selection of instrumental variables
The following criteria were used to select significant instrumental variables: (1) We set a threshold of p < 1 × 10−5 (locus-wide significance) to identify SNPs that were significantly associated with the exposure data instead of p < 1 × 10−8 (genome-wide significance) because the number of SNPs obtained using the latter threshold was limited26. (2) The “TwoSampleMR” package was used to obtain instrumental variables from independent genetic loci in the 1000 Genomes EUR data. The clumping distance was set at 10,000 kb, along with a linkage disequilibrium (LD) threshold of R2 < 0.001. The SNPs with the most significant p-values linked to the 196 bacterial taxa were subjected to clumping. (3) To ensure the causal relationship outcome between gut microbiota and IC/BPS was not affected by alleles, palindromic SNPs were excluded during GWAS data harmonization. (4) The F-statistic was utilized to test the strength of instrumental variables to evaluate the impact of weak instrument bias on causal effect estimates. If the associated F-statistic exceeded 10, weak instrumental bias was deemed non-existent27.
Statistical analysis
To estimate the potential causality between gut microbiota and IC/BPS, several methods including random-effects inverse variance weighted (IVW), weighted median, and MR-Egger were employed. In the MR analysis, the IVW method was applied as the primary approach28. The IVW method, which includes the Wald estimator and Delta method, was used to initially calculate the ratio estimates for individual SNPs. Subsequently, these individual estimates were aggregated to obtain the primary causal estimate29. To obtain more robust estimates across various scenarios, MR-Egger and weighted median methods were utilized to enhance IVW esimates, although they yielded lower efficiency (resulting in wider confidence intervals)30.
To explore the existence of potential heterogeneity and pleiotropy in the MR results, we performed sensitivity analysis. Heterogeneity was assessed using Cochrane’s Q test, and it was considered present when the instrumental variable had a p-value < 0.05. Potential horizontal pleiotropy was explored by the MR-Egger regression method and was considered present if intercept’s p-value < 0.0531. Pleiotropy outliers were identified by the MR-PRESSO method, and causal effect estimates were obtained using the IVW method on the plausible SNPs32. In addition, to determine whether a single SNP was driving the MR estimates, we performed leave-one-out analysis33. All statistics were performed using The “TwoSampleMR” package (version 0.5.8) in R (version 4.3.2), which can be accessed at https://www.r-project.org/.
Ethics declarations
Additional review or approval by an ethics committee was not needed for this study because the GWAS summary statistics data were already publicly available. All participating studies had obtained informed consent in accordance with protocols approved by their institute’s ethics committee.
Results
Table 1 shows that identified 106 SNPs associated with IC/BPS from eight bacterial taxa using the IVW method after quality control. In addition, the IVW method revealed a positive relationship between these eight bacterial taxa and the risk of IC/BPS (Table 1 and Fig. 2): genus Bacteroides (OR = 4.273, 95% CI:1.361–13.413, p = 0.013), genus Haemophilus (OR = 2.175, 95% CI:1.172–4.036, p = 0.014), genus Veillonella (OR = 2.379, 95% CI:1.037–5.458, p = 0.041), genus Coprococcus1 (OR = 2.704, 95% CI:1.118–6.542, p = 0.027), genus Butyricimonas (OR = 2.265, 95% CI:1.151–4.455, p = 0.018), family Bacteroidaceae (OR = 4.273, 95% CI:1.361–13.413, p = 0.013), family Christensenellaceae (OR = 2.623, 95% CI:1.045–6.605, p = 0.040), and order Lactobacillales (OR = 2.142, 95% CI:1.024–4.480, p = 0.043). These findings suggest that these microbiota might increase the incidence of IC/BPS. Among the eight causal associations, the F-statistic was larger than 10 for all IVs, suggesting low risk of weak instrument bias in the estimates, as detailed in Table 1.
Scatter plots illustrating the causal effect of gut microbiota on IC/BPS. (A) Genus Bacteroides (B) Genus Haemophilus (C) Genus Veillonella (D) Genus Coprococcus1 (E) Genus Butyricimonas (F) Family Bacteroidaceae (G) Family Christensenellaceae (H) Order Lactobacillales. Each black dot indicates a SNP, with the x-axis denoting the SNP effects on gut microbiota and the y-axis representing SNP effects on IC/BPS. The slope of the line corresponds to causality estimates calculated using three MR methods. SNP single-nucleotide polymorphism, MR Mendelian randomization, IC/BPS interstitial cystitis/bladder pain syndrome.
The sensitivity analysis did not reveal any significant heterogeneity or pleiotropy (Table 1). No significant heterogeneity was detected among these IVs based on the results of the Cochran’s Q test. Furthermore, the MR-Egger regression analysis results confirmed that there was no significant horizontal pleiotropy. In the MR-PRESSO analysis, no outliers were detected, further supporting the MR-Egger regression findings. Lastly, we confirmed that the MR estimates were not driven or biased by any single SNP because no outlier SNPs were identified by the Leave-one-out test (Fig. 3).
Leave-one-out plots for the causal effect of gut microbiota on IC/BPS. (A) Genus Bacteroides (B) Genus Haemophilus (C) Genus Veillonella (D) Genus Coprococcus1 (E) Genus Butyricimonas (F) Family Bacteroidaceae (G) Family Christensenellaceae (H) Order Lactobacillales. The line depicts the 95% confidence interval derived from the inverse variance weighted method.
Discussion
To our knowledge, this study is the first extensive MR investigation into the causal effect of gut microbiota on IC/BPS. While the gut microbiota have been identified to be correlated with IC/BPS in previous research17, establishing a causal link has been challenging due to the potential impact of confounding factors. Our results, however, indicate that specific gut microbiota (1 order taxon, 2 family taxa, and 5 genus taxa) are causally associated with the risk of IC/BPS. This insight offers a new angle regarding the diagnosis and treatment of IC/BPS.
In the human body, the gut-brain axis has been documented34, with evidence suggesting that microbes are involved in this axis, known as the microbiota-gut-brain axis35. Previous research has indicated that the gut-brain axis and the microbiota-gut-brain axis contribute to various diseases36,37,38,39,40, and there is inflammatory signal transmission between the gut and the brain41. Given the frequent coexistence of functional urological disorders, including IC/BPS, with specific gastrointestinal dysfunctions (such as irritable bowel syndrome) and mental disorders (such as depression), we speculate that there are interactions among the bladder, intestines, and brain. Based on this, a hypothesis is proposed regarding the bladder-gut-brain axis in the human body7,14. The role of microbiota in mediating communication between the bladder and the gut-brain axis has been postulated based on their essential functions in the gut-brain axis. Clinical studies have identified correlations between gut microbiota alterations and the occurrence of functional urological disorders, such as IC/BPS, chronic prostatitis/chronic pelvic pain syndrome (CP/CPPS), and overactive bladder (OAB)17,42,43. However, causation remains unproven. In this study, we demonstrate that gut microbiota have a causal effect on IC/BPS, providing new perspectives regarding the potential interactions between the bladder and gut. Moreover, we provide a partial preliminary evidence for further investigation into the bladder-gut-brain axis.
Our study identified causal associations between eight gut microbiota and IC/BPS. These microbiota taxa were all positively correlated with the risk of IC/BPS. However, due to the limited research on this relationship, no studies have reported changes in these microbiota taxa in patients with IC/BPS. While the clinical observational study mentioned earlier reported statistically significant decreases in the abundance of specific microbiota in fecal specimens from IC/BPS patients, trends of increased abundance were also observed in some gut microbiota, although not statistically significant17. However, observational studies are often affected by numerous confounding factors, especially given that IC/BPS treatments involve dietary management and antimicrobial drug use3. This complexity makes it difficult to interpret the findings obtained in studies on gut microbiota in IC/BPS patients. Therefore, we believe our MR study, less susceptible to confounding factors, provides a valuable foundation for further research.
In this research, we found that genus Bacteroides and family Bacteroidaceae were both causally related to IC/BPS, with positive results observed in both the IVW and weighted median analyses for genus Bacteroides. Moreover, the genus Bacteroides, belonging to the family Bacteroidaceae, is one of the most prevalent species in the human gut microbiota, involved in many biological processes. This species can be transmitted from mother to child during vaginal delivery, becoming part of the human microbiota in the earliest stages of life44. Bacteroides are typically probiotics in the gut, with the potential to become opportunistic pathogens when they translocate from the gut to other parts of the body, contributing to or exacerbating infections. This translocation may be triggered by factors such as compromised immune systems, intestinal barrier disruption, surgical injury, excessive antibiotic use, and aging45. Current research suggests that one potential origin of the urinary microbiota may be the gut15. Additionally, other studies have indicated a correlation between dysbiosis of the urinary microbiota and IC/BPS16. Therefore, it is worth investigating whether the translocation of Bacteroides from the gut could be one of the reasons for the positive correlation between Bacteroides and the risk of IC/BPS.
Genus Haemophilus is a common pathogen that can colonize the oral, respiratory, and digestive tracts of humans, causing various infectious diseases. While urinary and genital infections caused by Haemophilus are relatively less common in humans compared to other sites, there are some reports of potential occurrence46,47,48,49. Genus Veillonella is an opportunistic pathogen found in the human respiratory, digestive, and urogenital tracts, with the potential to cause urinary and genital infections50. Our MR results indicated a positive correlation between the increased abundance of genera Haemophilus and Veillonella in the gut and the risk of IC/BPS. However, the underlying mechanisms remain unclear, and additional studies are necessary to validate this finding.
Genus Coprococcus1 and genus Butyricimonas are major producers of butyrate, which is considered beneficial for the gut, with anti-inflammatory properties and capacity to improve intestinal barrier function and mucosal immunity51. Family Christensenellaceae is considered to be one of the most heritable taxonomic groups13. Although research has demonstrated that the abundance of family Christensenellaceae is increased in patients with childhood asthma52, it is generally considered a probiotic, and its abundance is negatively correlated with inflammation53. However, contrary to previous research findings, this MR study suggests that the higher abundance of family Christensenellaceae, genera Coprococcus1 and Butyricimonas in the gut may elevate the risk of IC/BPS. Further research is advocated to validate our findings.
In this study, we found that Lactobacillales was the only microbial group on the order level that was causally associated with IC/BPS. Typically, the order Lactobacillales have several benefits to the human body. However, our results indicated that a higher abundance of the order Lactobacillales in the gut may contribute to the risk of IC/BPS. Although there is no research on the connection between gut Lactobacillales and IC/BPS, in a study on CP/CPPS, another functional urological disorder, researchers found that the gut Lactobacillales abundance was significant increased in mice with experimental autoimmune prostatitis (EAP)54. The potential biological mechanisms linking the two need to be further investigated.
This MR study offers several important findings. Firstly, it is the first two-sample MR analysis to investigate the potential causal relationship between gut microbiota and IC/BPS. Traditional observational studies are susceptible to bias due to factors such as reverse causality and confounding variables. Our approach introduces a novel method for screening candidate gut microbiota, thereby offering a valuable tool for identifying new biomarkers. Secondly, compared to small-scale randomized controlled trials, the summary-level genetic data from large-scale GWAS are derived from larger population samples, enhancing the study’s credibility. Lastly, our study conducted sensitivity analysis, which confirmed the absence of heterogeneity or pleiotropy, underscoring the statistical robustness of our findings.
However, this study has some limitations. Firstly, during the selection of instrumental variables based on the threshold of p < 1 × 10−8 (genome-wide significance), few SNPs were obtained, which restricted further analysis. Therefore, we selected a more lenient threshold of p < 1 × 10−5 (locus-wide significance) to obtain for SNPs, which may potentially influence the results. Secondly, we analyzed gut microbiota only at the taxonomic level of the genus and above, which may not have included more specific levels such as species or strains. Employing advanced metagenomic sequencing would enhance specificity and accuracy in gut microbiota GWAS outcomes. Thirdly, the large-scale GWAS summary data used in MR studies may be influenced by selection bias, sociodemographic factors, cultural differences, and other factors during the statistical process. This impact may be particularly pronounced given the inherent clinical complexity and diagnostic challenges of IC/BPS. Fourthly, the use of GWAS summary statistics of IC/BPS patients rather than raw data in the study resulted in the inability to perform subgroup analysis. In addition, obtaining more detailed IC/BPS patient inclusion/exclusion criteria or other individual patient information from GWAS summary statistics was difficult. Fifthly, since the study cohort primarily consisted of individuals of European ancestry, any extrapolation of our findings to other ethnic groups should be conducted with caution. Sixthly, although the instrumental variables chosen for the analysis were closely linked to gut microbiota taxa and met the core MR assumptions, we cannot rule out the potential of weak instrument bias. Finally, during the collection of gut microbiota samples, the microbial abundance may vary depending on the participants diet, medication, and other factors.
Conclusions
In summary, we employed a two-sample MR analysis with publicly available summary-level data from GWAS to explore the causal relationship between gut microbiota and IC/BPS. Through this approach, we identified potential pathogenic flora associated with the development of IC/BPS. While extrapolating the MR result to guide clinical intervention might be inappropriate at this stage, these microbial communities may serve as innovative biomarkers in IC/BPS-related research, offering new opportunities for the treatment and prevention of IC/BPS. This study also offers partial preliminary evidence for future investigations into the bladder-gut communication and its potential implications in clinical practice.
However, our study has several limitations. Similar to other MR studies, the large-scale GWAS summary data we used may be influenced by selection bias, sociodemographic factors, cultural differences, and other factors during the statistical process. Additionally, GWAS summary data, unlike raw data, do not provide more detailed information such as individual patient subtypes or specific inclusion/exclusion criteria. Given the clinical complexity and diagnostic challenges of IC/BPS, these limitations suggest that our MR results require further validation through additional research. In the future, we will conduct animal experiments or large and prospective cohort studies to validate our findings and explore the associated mechanisms.
Data availability
All the statistics can be found in GWAS Catalog (https://www.ebi.ac.uk/gwas/) and GWAS MiBioGen consortium (https://mibiogen.gcc.rug.nl/). Further inquiries can be directed to the corresponding author.
References
Hanno, P. M., Erickson, D., Moldwin, R. & Faraday, M. M. Diagnosis and treatment of interstitial cystitis/bladder pain syndrome: AUA guideline amendment. J. Urol. 193, 1545–1553 (2015).
Malde, S., Palmisani, S., Al-Kaisy, A. & Sahai, A. Guideline of guidelines: Bladder pain syndrome. BJU Int. 122, 729–743 (2018).
Homma, Y. et al. Clinical guidelines for interstitial cystitis/bladder pain syndrome. Int. J. Urol. 27, 578–589 (2020).
Dellis, A. E. & Papatsoris, A. G. Bridging pharmacotherapy and minimally invasive surgery in interstitial cystitis/bladder pain syndrome treatment. Exp. Opin. Pharmacother. 19, 1369–1373 (2018).
Mateu Arrom, L. et al. Long-term follow-up after cystectomy for bladder pain syndrome: pain status, sexual function and quality of life. World J. Urol. 37, 1597–1603 (2019).
Nickel, J. C. & Doiron, R. C. Hunner lesion interstitial cystitis: The bad, the good, and the unknown. Eur. Urol. 78, e122–e124 (2020).
Leue, C. et al. Functional urological disorders: a sensitized defence response in the bladder–gut–brain axis. Nat. Rev. Urol. 14, 153–163 (2017).
Jandhyala, S. M. et al. Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803 (2015).
Tlaskalová-Hogenová, H. et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol. Immunol. 8, 110–120 (2011).
Perler, B. K., Friedman, E. S. & Wu, G. D. The role of the gut microbiota in the relationship between diet and human health. Annu. Rev. Physiol. 85, 449–468 (2023).
Ling, Z., Liu, X., Cheng, Y., Yan, X. & Wu, S. Gut microbiota and aging. Crit. Rev. Food Sci. Nutr. 62, 3509–3534 (2022).
Pennycook, J. H. & Scanlan, P. D. Ecological and evolutionary responses to antibiotic treatment in the human gut microbiota. FEMS Microbiol. Rev. 45, fuab018 (2021).
Goodrich, J. K. et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014).
Jung, J., Kim, A. & Yang, S.-H. The innovative approach in functional bladder disorders: The communication between bladder and brain-gut axis. Int. Neurourol. J. 27, 15–22 (2023).
Perez-Carrasco, V., Soriano-Lerma, A., Soriano, M., Gutiérrez-Fernández, J. & Garcia-Salcedo, J. A. Urinary microbiome: Yin and Yang of the urinary tract. Front. Cell. Infect. Microbiol. 11, 617002 (2021).
Fu, C. et al. The microbiota in patients with interstitial cystitis/bladder pain syndrome: A systematic review. BJU Int. https://doi.org/10.1111/bju.16439 (2024).
Braundmeier-Fleming, A. et al. Stool-based biomarkers of interstitial cystitis/bladder pain syndrome. Sci. Rep. 6, 26083 (2016).
Rahman-Enyart, A. et al. Acyloxyacyl hydrolase is a host determinant of gut microbiome-mediated pelvic pain. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 321, R396–R412 (2021).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian randomization. JAMA 318, 1925–1926 (2017).
Smith, G. D. & Ebrahim, S. ‘Mendelian randomization’: Can genetic epidemiology contribute to understanding environmental determinants of disease?. Int. J. Epidemiol. 32, 1–22 (2003).
Luo, Q. et al. Effects of gut microbiota and metabolites on heart failure and its risk factors: A two-sample mendelian randomization study. Front. Nutr. 9, 899746 (2022).
Ni, J.-J. et al. Gut microbiota and psychiatric disorders: A two-sample Mendelian randomization study. Front. Microbiol. 12, 737197 (2021).
Ren, F., Jin, Q., Liu, T., Ren, X. & Zhan, Y. Causal effects between gut microbiota and IgA nephropathy: A bidirectional Mendelian randomization study. Front. Cell. Infect. Microbiol. 13, 1171517 (2023).
Jiang, L., Zheng, Z., Fang, H. & Yang, J. A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 53, 1616–1621 (2021).
Kurilshikov, A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165 (2021).
Sanna, S. et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605 (2019).
Li, P. et al. Association between gut microbiota and preeclampsia-eclampsia: A two-sample Mendelian randomization study. BMC Med. 20, 443 (2022).
Bowden, J. et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 36, 1783–1802 (2017).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Chen, X. et al. Kidney damage causally affects the brain cortical structure: A Mendelian randomization study. eBioMedicine 72, 103592 (2021).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525 (2015).
Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018).
Long, Y., Tang, L., Zhou, Y., Zhao, S. & Zhu, H. Causal relationship between gut microbiota and cancers: A two-sample Mendelian randomisation study. BMC Med. 21, 66 (2023).
Mayer, E. A., Nance, K. & Chen, S. The gut-brain axis. Annu. Rev. Med. 73, 439–453 (2022).
Cryan, J. F. et al. The microbiota-gut-brain axis. Physiol. Rev. 99, 1877–2013 (2019).
Hartstra, A. V. et al. Infusion of donor feces affects the gut–brain axis in humans with metabolic syndrome. Mol. Metab. 42, 101076 (2020).
Jacobs, J. P. et al. Cognitive behavioral therapy for irritable bowel syndrome induces bidirectional alterations in the brain-gut-microbiome axis associated with gastrointestinal symptom improvement. Microbiome 9, 236 (2021).
Ahluwalia, V. et al. Enhancement of functional connectivity, working memory and inhibitory control on multi-modal brain MR imaging with Rifaximin in Cirrhosis: Implications for the gut-liver-brain axis. Metab. Brain Dis. 29, 1017–1025 (2014).
Nikrad, N. et al. The effect of calorie-restriction along with thylakoid membranes of spinach on the gut-brain Axis pathway and oxidative stress biomarkers in obese women with polycystic ovary syndrome: A randomized, double-blind, placebo-controlled clinical trial. J. Ovarian. Res. 16, 216 (2023).
Wang, Y. et al. Probiotics and fructo-oligosaccharide intervention modulate the microbiota-gut brain axis to improve autism spectrum reducing also the hyper-serotonergic state and the dopamine metabolism disorder. Pharmacol. Res. 157, 104784 (2020).
Agirman, G., Yu, K. B. & Hsiao, E. Y. Signaling inflammation across the gut-brain axis. Science 374, 1087–1092 (2021).
Okamoto, T. et al. Altered gut microbiome associated with overactive bladder and daily urinary urgency. World J. Urol. 39, 847–853 (2021).
Shoskes, D. A. et al. Analysis of gut microbiome reveals significant differences between men with chronic prostatitis/chronic pelvic pain syndrome and controls. J. Urol. 196, 435–441 (2016).
Wexler, H. M. Bacteroides : The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007).
Zafar, H. & Saier, M. H. Gut bacteroides species in health and disease. Gut Microbes 13, 1–20 (2021).
Nørskov-Lauritsen, N. Classification, identification, and clinical significance of haemophilus and Aggregatibacter species with host specificity for humans. Clin. Microbiol. Rev. 27, 214–240 (2014).
Franco-Acosta, A., Espadafor-López, B., Rosales-Castillo, A., Navarro-Marí, J. M. & Gutiérrez-Fernández, J. Emergence of genital infections due to Haemophilus pittmaniae and Haemophilus sputorum. Infect. Dis. Now 52, 227–229 (2022).
Hansson, S., Svedhem, A., Wennerström, M. & Jodal, U. Urinary tract infection caused by Haemophilus influenzae and Haemophilus parainfluenzae in children. Pediatr. Nephrol. 22, 1321–1325 (2007).
Mathiasen, A. S. F. et al. Haemophilus influenzae septicaemia and urinary tract infection associated with nefrocalcinosis: Case report. Diagn. Microbiol. Infect. Dis. 107, 116001 (2023).
Ding, Z. et al. Detecting and quantifying Veillonella by real-time quantitative PCR and droplet digital PCR. Appl. Microbiol. Biot. 108, 45 (2024).
Liu, H. et al. Butyrate: A double-edged sword for health?. Adv. Nutr. 9, 21–29 (2018).
Lee-Sarwar, K. A. et al. Integrative analysis of the intestinal metabolome of childhood asthma. J. Allergy Clin. Immunol. 144, 442–454 (2019).
Waters, J. L. & Ley, R. E. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biol. 17, 83 (2019).
Du, H.-X. et al. Gut microflora modulates Th17/Treg cell differentiation in experimental autoimmune prostatitis via the short-chain fatty acid propionate. Front. Immunol. 13, 915218 (2022).
Acknowledgements
We would like to express our gratitude to the participants and investigators involved in the GWAS utilized in the present study.
Funding
This study was supported by the National Natural Science Foundation of China (Grant no: 81860142) .
Author information
Authors and Affiliations
Contributions
P.J. contributed to the writing of the manuscript and the visualization. C.L., Z.S., and H.L. contributed to data collection and analysis. D.C. and J.C. supervised the study. H.M. and C.L. contributed to the study design and manuscript revision. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, P., Li, C., Su, Z. et al. Mendelian randomization study reveals causal effects of specific gut microbiota on the risk of interstitial cystitis/bladder pain syndrome (IC/BPS). Sci Rep 14, 18405 (2024). https://doi.org/10.1038/s41598-024-69543-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-69543-9
Keywords
This article is cited by
-
Causal effect of gut microbiota on venous thromboembolism: a two-sample mendelian randomization study
Thrombosis Journal (2024)