Abstract
Natural Killer (NK) cells are pivotal in immune responses to viral infections, malignancies, autoimmune diseases, and transplantation. Assessment of NK cell adhesion, migration, and cytotoxicity is fundamental for in vitro studies. We propose a novel live-cell tracking method that addresses these three major aspects of NK cell function using human NK cells and primary porcine aortic endothelial cells (PAECs) in two-dimensional (2D) static assays and an in-house cylindrical 3D microfluidic system. The results showed a significant increase of NK cytotoxicity against pTNF-activated PAECs, with apoptotic cell death observed in the majority of dead cells, while no difference was observed in the conventional Delfia assay. Computed analysis of NK cell trajectories revealed distinct migratory behaviors, including trajectory length, diameter, average speed, and arrest coefficient. In 3D microfluidic experiments, NK cell attachment to pTNF-activated PAECs substantially increased, accompanied by more dead PAECs compared to control conditions. NK cell trajectories showed versatile migration in various directions and interactions with PAECs. This study uniquely demonstrates NK attachment and killing in a 3D system that mimics blood vessel conditions. Our microscope method offers sensitive single-cell level results, addressing diverse aspects of NK functions. It is adaptable for studying other immune and target cells, providing insights into various biological questions.
Similar content being viewed by others
Introduction
Natural killer (NK) cells are part of the innate immune system, responsible for clearing virally infected and transformed cells. They also play an essential role in allo- and xeno-graft rejections1,2,3,4. They uniquely detect and kill target cells without prior sensitization5. NK cytotoxicity is regulated by the expression of activating and inhibitory receptor signaling at the immune synapses with the target cells6. Cytokines, including interleukin (IL)-2, IL-12, IL-15, type-I interferons, and tumor necrosis factor (TNF), activate NK cells7,8. The primary mechanisms for NK cytotoxicity are granule exocytosis (perforin, granzymes) and death ligands (FasL, TRAIL)9. Various cytotoxicity assays measuring target cell death have been reported10,11,12,13. Nowadays, the classical 51Chromium (51[Cr])-release assay is used less since it depends on a radioactive substance. Non-radioactive compounds like Delfia and flow cytometry-based assays require the detachment of adherent target cells before labeling or direct analysis11,12, which can lead to the disruption of surface markers and adhesion molecules. Calcein AM is more adaptable for adherent cells; however, incomplete release or entrapped calcein AM within apoptotic bodies can result in the underestimation of specific lysis10. Overall, assays using target cells in suspension are unsuited for monitoring continuous dynamics of cellular interactions leading to NK cytotoxicity. Since assessment of NK interaction with endothelial cells is critical in the field of cancer treatment and transplantation medicine, it is essential to develop in vitro methods that overcome the limitations of existing assays.
Apart from cytotoxicity, NK cell motility and tissue infiltration are fundamental to NK cell function. The process of trans-endothelial migration and local recruitment requires a series of adhesion steps to the endothelial layer of blood or lymphatic vessels, including chemotaxis activation, rolling and firm adhesion, and transmigration14,15. Cell migration is tightly regulated by surface receptors, cytoskeleton structure, cytokine-chemokine attractants, and target cell interactions14. Transwell assays with and without endothelial layers are used to measure NK cell migration in vitro. Moreover, microscope-based experiments on flat surfaces of endothelial cells have provided informative results on NK cell adhesion16,17. NK cells exhibit distinct migration and contact dynamics depending on IL-2-activation18, different NK phenotypes, and cell differentiation stages19,20,21. 3D approaches using collagen gels in microwells yield more relevant data, resembling the extracellular microenvironment22. Lee et al. showed marked differences where a subpopulation of NK cells moved slowly and in a constrained manner within the matrix, while others moved more freely21. The development of automated cell tracking using different computational algorithms has also helped generate robust results and reducing bias for large datasets22,23. It is, however, still difficult to correlate NK cytotoxicity with migration on adherent target cells, as no accurate assay currently exists that can assess both simultaneously.
NK cells can acquire unique functions and migratory properties in specific tissues24,25. In transplantation and cardiovascular contexts, the endothelium in blood vessels of transplanted organs is the first cell layer in contact with the recipient’s NK cells. Studying their interactions in a vessel-like system can deepen our understanding of the cellular interactions and immune responses under flow conditions. To this end, 3D microfluidic technologies and commercial chips have been developed for culturing endothelial cells (ECs) under the luminal flow26,27. However, in these systems ECs grow in a rectangular-shaped microchannel, which does not accurately represent the microvasculature’s geometry, particularly in terms of shear stress26,28. Chrobak et al. developed a 3D cylindrical microchannel using needles as molds29, which was further improved by Sfriso et al. with the use of a peristaltic pump and reservoir tubes to obtain a closed microfluidic circuit30. The latter system allows adjusting laminar shear stress levels, replicating the conditions found in various vessel tissues, such as arteries, veins, and capillaries. Recently, the same group demonstrated that the formation of a glycocalyx layer, a significant component of the blood vessel luminal surface, depends on laminar shear stress31. With adjustable shear stress and glycocalyx layer assessment advantage, this 3D cylindrical microchannel provides a physiologically relevant structure that we applied in the present study to investigate circulating NK cell interactions with the endothelial layer.
Overall, we present a complete workflow to study NK cell functions in 2D and 3D systems using an enhanced microscope-based method. Combined assessment of NK motility and cytotoxicity provides valuable insights into how NK cells interact with and efficiently eliminate their target cells under both static and flow conditions. We describe segmentation and tracking strategies for high-throughput analysis of NK cell trajectories on endothelial cell layers, overcoming limitations of current cytotoxicity and migration assays. Furthermore, the live-cell imaging approach in a 3D flow system offers a more realistic view and closely resembles in vivo conditions.
Materials and methods
Chemicals, culture media, and supplements
(3-Aminopropyl) tiethoxysilane (APTES), bovine serum albumin (BSA), dextran from Leuconostoc spp., ethylenediaminetetraacetic acid (EDTA), fetal bovine serum (FBS), glutaraldehyde, human plasma, fibronectin, were from Sigma-Aldrich (St. Louis, MO, USA). The following buffers, culture media, and supplements were all from Gibco (Grand Island, NY, USA): Dulbecco’s Modified Eagle Medium (DMEM), Dulbecco’s Phosphate-Buffered Saline (PBS 1×), AIM-V medium, FluoroBrite DMEM, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), minimum essential medium amino acids solution (EAA), and non-essential amino acids solution (NEAA); penicillin/streptomycin, sodium pyruvate, Collagen-I Bovine 5 mg/ml. L-Alanyl-L-Glutamine (L-Ala/L-Glu) was obtained from Bioswisstec AG (Schaffhausen, Switzerland). Ficoll-Paque PLUS density gradient media was from GE Healthcare (Uppsala, Sweden). The human NK cell isolation kit and LS columns were purchased from Miltenyi Biotec (Bergisch Gladbach, Germany); human recombinant interleukin 2 (IL-2) was from Novartis Pharma (Proleukin, Switzerland), 7-Aminoactinomycin D (7-AAD) from BD Biosciences (La Jolla, CA, USA); DELFIA EuTDA Cytotoxicity Reagents from PerkinElmer (Waltham, MA, USA). Hoechst 33342 solution (20 mM) and CellTrace Yellow Cell Proliferation Kit (Invitrogen), 16% formaldehyde solution (FA), methanol-free, were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Draq7 (0.3mM) was from BioStatus (Shepshed, UK). Recombinant porcine TNF (pTNF) and human TNF (hTNF) was from R&D Systems (Minneapolis, MN, USA), and SYLGARD 184 Silicone Elastomer Kit from Dow Inc. (Midland, MI, USA). Details in Supplementary Table S1.
Endothelial cells culture
Primary porcine aortic endothelial cells (PAECs), wild-type, kindly provided by Eckhard Wolf (Ludwig-Maximilians-University of Munich). Cells were isolated mechanically from porcine thoracic aorta obtained from female and male German landrace outbred pigs32 upon euthanasia after terminal experiments performed by different groups at LMU Munich, complying with the 3R principles. Cells were cultured in DMEM medium supplemented with 10% heat-inactivated FBS, 2 mM L-Ala/L-Glu, 1 mM sodium-pyruvate, 20 mM HEPES, 1× EAA and 1× NEAA, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37ºC, 5% CO2. For live cell imaging, 16,000 PAECs per well were seeded in 96-well µ-Plates with black walls and optically clear flat bottom (Ibidi, Fitchburg, WI, USA) in phenol red-free FluoroBrite DMEM supplemented as described above. As indicated PAECs were treated with 100 ng/ml pTNF overnight.
The human endothelial cell line EA.hy926 was kindly provided by Prof. Michelangelo Foti (University of Geneva) and cultured in the same media as PAECs. For live cell imaging, 16,000 EA.hy926 cells per well were seeded in 96-well micro-plates with black walls and optically clear flat bottom (Ibidi, Fitchburg, WI, USA) in phenol red-free FluoroBrite DMEM supplemented as described above. As indicated EA.hy926 cells were treated with 100 ng/ml hTNF overnight.
Isolation and stimulation of human NK cells
Human NK cells were isolated from healthy donors’ buffy coats obtained from the Blood Transfusion Centre of the University Hospitals Geneva, with approval from the ethical committee of our institution (CER18-00552). Blood samples were collected upon receiving an informed consent and immediately anonymized. The experimental protocols were conducted according to Swiss Federal regulations and the guidelines of the University of Geneva. Peripheral blood mononuclear cells (PBMC) were separated by density gradient centrifugation in Ficoll-Paque PLUS at 900×g, 20 min at room temperature, without brake. Once collected, cells were washed twice in PBS-EDTA by centrifugation at 200×g for 10 min. Thereafter, PBMC were used for NK cell purification by negative selection using NK isolation kits according to the manufacturer’s instructions. Flow cytometry analysis demonstrated 90–95% purity of CD3–CD56+CD16+ NK cells. NK cells were cultured at 1 × 106 cells/ml for 5–9 days in AIM-V medium supplemented with heat-inactivated plasma, 20 mM HEPES, 1 mM sodium-pyruvate, 2 mM L-Glu/L-Ala, 1× NEAA and 1× EAA, 100 U/ml penicillin and 100 µg/ml streptomycin at 37ºC, 5% CO2. On day one, 200 IU/ml IL-2 was added, thereafter, 50 IU/ml IL-2 every two days of culture.
Characterization of NK cells by flow cytometry
Cell surface staining was performed on freshly isolated, and IL-2 activated NK cells at 4°C in the dark for 25 min with fluorophore-conjugated monoclonal antibodies directed against CD16, CD25, CD56, CD57, CD69, CD94, NKp30, NKp44, NKp46, CD158e, NKG2A, NKG2D (Supplementary Table S2). After washing, cell pellets were resuspended in 300 µl of staining buffer and labeled with 7-AAD for dead cell exclusion. Attune NxT was used for the sample acquisition and FlowJo (version 10) for data analysis.
Cytotoxicity assay using standard release assay
Non-radioactive DELFIA EuTDA Cytotoxicity Reagents (PerkinElmer, Waltham, MA, USA) were used for NK cell-mediated cytotoxicity assays according to the manufacturer’s instructions. Briefly, target cells (PAECs) were loaded for 20 min with fluorescence-enhancing ligand BATDA, washed three times with DMEM at 200×g, 4 min. NK and target cells were then plated in round-bottom 96-well plates at different effector-to-target (E:T) ratios (10:1, 5:1, and 1:1). Triplicates were prepared for each condition. Maximum release was obtained using labeled target cells that were completely lysed with Lysis Buffer (PerkinElmer), while spontaneous release was measured from labeled target cells in the absence of effector cells. After 2 h of incubation at 5% CO2 and 37 °C, the supernatants were mixed with 200 µl Europium salt solution to form a chelate and measured in a time-resolved fluorometer (EnVision 2014 Multilabel reader, PerkinElmer). Cytotoxicity was calculated with formulas provided by the manufacturer as a percentage (%) of specific lysis.
3D microfluidic system for flow cytotoxicity assay
The microfluidic channels were made as described previously30, using polydimethylsiloxane (PDMS, from Silicone Elastomer kit), and mold needles (0.55 mm diameter) to create a cylindrical channel simulating small vessels. Inlet and outlet holes were created with a 2 mm biopsy punch. After oxygen plasma activation, the channels were treated sequentially with APTES, glutaraldehyde, fibronectin (50 µg/ml), and collagen-I (100 µg/ml). Ultimately, PAECs were seeded twice in the microchannels at a concentration of 1–2 × 106 cells/ml. First, 50 µl of cell suspension was added and incubated without flow for 1 h. The chip was then inverted, and another 50 µl of PAEC suspension was added, followed by overnight incubation at 37 °C to allow cell adhesion and monolayer formation. The endothelial cells reached confluence one day after cell seeding. The chip was subsequently connected to the peristaltic pump (Ismatec, Glattburg, Switzerland) and a reservoir using silicon tubing (MaagTechnic, Dübendorf, Switzerland). The culture medium was replaced with 4 ml of flow medium composed of DMEM-10 with 4% dextran and 1% BSA to reach a viscosity corresponding to 2.1 dyn/m2×seq at 37 °C. Laminar sheer stress was fixed at 2.0 dyn/cm2 using the peristaltic pump and kept for an additional 48 h at 37ºC, 5% CO2 before proceeding with 3D cytotoxicity assay. These flow conditions resemble those of arterial branches with a diameters ranging from 600 to 3000 μm33 and blood-like viscosity; however, the shear stress was approximately five times lower in order to avoid detachment of the endothelial cells from the channels.
Live-cell imaging cytotoxicity assays in static 2D and 3D microfluidic system
PAECs’ nuclei were stained with 2 µg/ml Hoechst for 1 h in the incubator and washed with PBS once using an automatic dispenser BioTek EL406 (Agilent, Santa Clara, CA, USA). NK cells were labeled with cell trace yellow (CTY), 0.2 µl per million cells for 20 min in PBS 1× at 37ºC. Once washed, NK cells were placed in phenol-red free FluoroBrite DMEM-10 containing 1 µl/ml of Draq7, to evaluate cell viability, and added to target cells at a 1:1 E:T ratio.
In the static 2D assays, NK cells seeded in the 96-well plates were centrifuged at 300×g for 1 min in order to reach the same microscope focal plane as PAECs. An automated spinning disc microscope (ImageXpress Micro Confocal High-Content Imaging System, 10× objective, Supplementary Table S3) was used to capture video time-lapse images of the interactions of NK cells with PAEC monolayers during 120 min at 5 min intervals. Three technical replicates with four regions per replicate were acquired. Time-lapse videos were created by MetaXpress. Analysis of dead and apoptotic cells was done using Fiji, ImageJ2 2.9.0 software (Supplementary Table S4). Microscopic image domains were split into segments to detect individual cells (cell segmentation) based on Hoechst staining signals and nuclei morphology. The percentage of total dead cells was determined according to the equation \(\:Total\:dead\:cells\:\left(\%\right)=\left(\frac{{{Live\:cells}_{\left(t0\right)}-Live\:cells}_{\left(t120\right)}}{{Live\:cells}_{\left(t0\right)}}\right)\times\:100\), where t0 and t120 correspond to the counting at time zero and at the endpoint (120 min), respectively. The counts of apoptotic cells included apoptotic bodies with intact membranes (intensified blue signals due to DNA condensation), and late apoptotic cells with Draq7+ signal. Thus, cell death through apoptosis was calculated by the equation \(\:Apoptotic\:cells\:\left(\%\right)=\left(\frac{{Apoptotic\:cells}_{{t}_{120}}}{{Live\:cells}_{{t}_{0}}}\right)\times\:100\). Lastly, necrotic cells were counted manually based on their distinct features consisting of normal nuclei shape with Draq7+ signals, but without intensified blue signals, and percentages were calculated as for apoptotic cells.
In the 3D microfluidic system, after 48 h under flow conditions, the channels were checked under the microscope before adding NK cells for quality control. If the endothelial layer was not 100% confluent, this channel was not used for NK cell perfusion experiments. For both wild-type PAECs and human endothelial EA.hy926 cells, we did not observe any cell detachment before or after the perfusion for 48 h at 2 dyn/cm2 shear stress. Image acquisition was performed with a Leica DMi8 wide-field microscope at 10× magnification to capture the entire chip (1 cm) connected to the reservoir (Supplementary Table S3). 4 ml of flow medium FluoroBrite DMEM − 10 containing 2 × 106 CTY-labeled NK cells with 1 µl/ml Draq7 added were used per channel and injected into the 15 ml reservoir tubes. The peristaltic pump perfused the NK cell solution through the system for 2 h. Images were acquired at one focal plane - no z-stacks with the thickness of 130 μm from the 10× objective lens. The recording one area was of 0.1 cm length per channel continuously for 40 min. Images captured the adhesion of NK cells to the endothelial cell monolayer under flow, as well as all dead PAECs of the whole channel at t0 and t120 min for endpoint analysis. The number of attached NK cells and Draq7+ PAECs were quantified using Fiji, ImageJ2 2.9.0 software.
Automatic detection and tracking of cell bright centers
A Python program was developed for the automatic detection of NK cell bright centers and their trajectories. A series of 16-bit monochrome images were first loaded. A median filter with kernel size of one pixel was applied to remove single-pixel high-frequency noise. Instead of using traditional segmentation and centroid-finding methods, each individual image frame was processed to locate gaussian-like intensity distributions, which effectively mapped the ___location of each cell bright center. The “locate” module from the trackpy Python library was used to accomplish this task, which is an implementation of the Crocker-Grier centroid finding algorithm34. A set of parameters were experimentally defined to reach the optimum detection results. The initial central detection diameter (initial approximation) was defined as 31 pixels, the minimum integrated intensity under a single distribution was defined as 9,000, the minimum separation between bright centers was 15 pixels, and the gaussian blurring filter kernel size was three pixels. The coordinates (x and y position in the unit of pixels) from each detected cell bright center were stored for each image frame as tabular data. Coordinates were transformed to µm by using the calibration constant of 0.684 μm/pixel. Once the positions were determined for each frame, trajectories were built by using the “link” module from the trackpy Python library, which is an implementation of the Crocker-Grier linking algorithm34. Due to the high number of detected bright centers and the large range of motion, the linking algorithm was run on an adaptive search mode, which could adaptively change the size of the linking trajectory if needed, with the goal of accelerating convergence. A maximum search radius of 80 pixels and an adaptive stop value of 30 pixels were defined. In case of complex situations, the search radius could be automatically limited to the stop value, reducing the search possibilities and computing time. The linking process grouped each detected bright center and assigned an identifier in the form of a sequential number. The trajectory information was stored as tabular data, in such a way that spatial and temporal information could be extracted during the trajectory analysis stage.
Analysis of NK cell trajectories
To assure a density of NK cells in the optical field suitable for analysis of cell trajectories in the 2D cytotoxicity assays, the E:T ratio was reduced to 1:20. The trajectories of single NK cells were tracked and classified as large, intermediate, and small based on the total effective displacement for each cell, also referred to as the trajectory diameter (µm). Trajectory diameter was defined as the distance between the position of the cell detected in the first and last frame (µm). Three classes were categorized based on the trajectory diameter (small < 10, intermediate 10–100 and large > 100 μm) for further analysis of trajectory length-to-diameter ratio, average speed (µm/min) and arrest coefficient. Arrest coefficient was defined as the percentage of time that the cell remained in arrest based on the threshold on instantaneous speed (0.2 μm/min)21. Tracking of NK cells was performed by analyzing every frame (frame rate: 4 min) of 120 min recording with a program built in Python. Data were analyzed from a heterogeneous population of at least 1,500 NK cells in nine different regions of three technical replicates.
Immunofluorescence staining and imaging of 2D and 3D systems
For immunofluorescence staining of the 2D system after the live-cell imaging session, cells were fixed in 4% buffered formaldehyde for 20 min at room temperature, followed by eight washes with DPBS, and blocked overnight at 4ºC using 3% BSA in DPBS. Next, the samples were incubated overnight at 4ºC with properly titrated antibodies (Supplementary Table S2), followed by secondary antibodies for 25 min at room temperature in the dark. An ImageXpress Confocal microscope was used for acquiring images at 10× and 20× (Supplementary Table S3).
For the 3D microfluidics system, the channels were disconnected from the perfusion system, washed three times with DPBS and fixed with freshly prepared 4% formaldehyde, washed, and blocked. Antibody staining was performed following the same protocol as described above for the 2D system. Finally, the chip was washed and incubated for 15–30 min with Hoechst (1 µg/ml). Images were acquired using a confocal Leica SP8 microscope at 20×; 3D images were created by Imaris software (Supplementary Table S3, S4).
Statistical analysis
Means of two groups were compared using paired and unpaired t-test (indicated in the figure legends). Error bars show ± SD. Differences were considered significant when p ≤ 0.05. The analysis was performed using Prism GraphPad.
Results
2D live-cell tracking is more sensitive to study NK cytotoxicity than conventional release assays
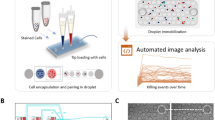
In the conventional Delfia release assay, PAEC targets are detached from the culture plates, labeled with BATDA, and seeded into U-bottom plates to interact with NK cells. After centrifugation and 2 h of co-culture, the TDA released from dead PAECs allows the determination of the percentage of lysed target cells (Fig. 1A). Unlike the Delfia assay, in the current 2D live-cell tracking method, the PAECs targets are cultured to 100% confluence in 96-well flat-bottom plates (Fig. 1B). NK cells isolated from human PBMCs were activated with IL-2 for 5–9 days. PAEC nuclei were stained with Hoechst, and NK cells were labeled with CTY for downstream analysis. Live/Dead staining with Draq7 in the culture media identified cells with disrupted membranes. An automated time-lapse microscope captured thousands of NK‒PAECs interactions over a 2 h period.
Comparison of Delfia and microscope-based cytotoxicity assays. (A) Graphical images describe the steps of Delfia assay and (B) the sample preparation for live cell-imaging microscopy. BATDA (Bis(acetoxymethyl)-2,2′:6′,2″-terpyridine-6,6’’-dicarboxylate) is a non-fluorescent compound that permeates cell membranes. TDA (Terbium-based fluorescence) is used as a label in the assay as it chelates produce long-lived fluorescence signals. (C) Endothelial cell monolayer used in the microscope-based assay. Porcine endothelial cells (PAECs) were stained with the endothelial cell marker CD31 (red). The nuclei were stained with Hoechst (cyan) before the live imaging session. Scale bar = 50 μm.
To set up our assays and ensure quality control, we confirmed the activation status of the NK cells after IL-2 stimulation by analyzing the upregulation of activating markers such as NKp44, NKp46, NKp30, CD69, and CD25 via flow cytometry (Supplementary Fig. S1). In parallel, staining with the endothelial marker CD31 (also known as platelet and endothelial cell adhesion molecule-1, PECAM-1) was used to demonstrate a preserved monolayer of PAECs (Fig. 1C). To assess the potential phototoxicity of the fluorescent dyes and microscope lasers, PAEC viability tests were conducted. The PAECs were stained with Hoechst in media containing Draq7 but without NK cell co-culture. The cells were exposed to the same set of lasers for 2 h and 4 h, and no significant effects on PAECs were observed. The percentage of dead cells after 2 h time-lapse analysis for control and pTNF-stimulated PAECs was 0.76 ± 0.5% and 0.55 ± 0.3% (P = 0.3671), respectively (Supplementary Fig. S2). Furthermore, we observed PAEC division even after extended imaging times up to 4 h, indicating the cells remained in a healthy condition (Supplementary Video 1).
TNF activates endothelial cells, leading to increased NK cell adhesion and cytotoxicity35,36. Here, we compared the sensitivity of the standard Delfia release assay and the 2D live-cell tracking method in detecting cytotoxicity against PAECs, with and without pTNF overnight stimulation. As shown in Fig. 2A, Delfia release assays showed a maximum killing of 32.8 ± 17.1% and 23.7 ± 25.6% specific lysis at the highest E:T ratio of 10:1, and 13.2 ± 9.8% and 10.8 ± 14.1% at an E:T ratio of 1:1, for control and pTNF-stimulated PAECs, respectively. Thus, no significant difference between killing of pTNF-activated and inactivated PAECs were revealed, (P = 0.1046, parametric paired t-test) (Fig. 2A). In 2D live-cell tracking assays, the images captured at the end-point clearly demonstrate PAEC lysis with disappearance of the stained nuclei (Fig. 2B). In contrast to the conventional assay, the 2D live-cell tracking method accounted for significant differences at an E:T ratio of 1:1, 44.3 ± 18.6% and 62.4 ± 17.7% of cell death by NK cells in control and pTNF-activated PAECs, respectively (P < 0.0001). Pooled data after automated quantification of dead cells showed that the NK cytotoxicity against pTNF-treated and control PAECs was donor-dependent. However, increases of NK cytotoxicity following TNF activation of PAECs were consistently observed, ranging from 8.7 to 32.5% (mean 20%) (Fig. 2C). In conclusion, the utilization of the live-cell tracking method enables the examination of PAECs’ initial interaction with NK cells up to the 2 h endpoint (Supplementary Video 2). The analysis demonstrated significantly greater killing of PAECs at an E:T ratio of 1:1, with an increased response to pTNF stimulation, compared to standard cytotoxicity assays. (Fig. 2C). The same donors were also tested at an E:T ratio of 1:20 during a 2 h assay, which more closely represents physiological conditions. Across three different healthy donors, the assay was sensitive enough to detect differences in NK cytotoxicity between untreated and pTNF-treated cells (Fig. 2D). The effect was donor-dependent and consistent with our observations at an E:T ratio of 1:1. As proof of concept that our system can assess NK allo-cytotoxicity against human endothelial cells, we co-cultured the cell line EA.hy926 with human NK cells. As shown in Fig. 2E, we detected statistically significant differences in the killing of NK cells between control and hTNF-activated EA.hy926 cells.
Human NK cytotoxicity against PAECs. (A) Percentages of specific lysis at different effector to target (E:T) ratios obtained by a Delfia release assay. Porcine endothelial target cells (PAEC) were treated with 100 ng/ml of pTNF overnight (pink) or left without TNF (Φ, black), human NK cells were pre-activated by IL-2. Data from three healthy donors were pooled. (B) Microscopy images of NK cells (green) co-cultured with PAECs (blue nuclei), at an E:T ratio of 1:1, in the presence of Draq7 staining dead cells (pink). Images were captured at time 0 and 120 min. Scale bar = 100 μm. (C) Pooled data showing the percentage of total dead PAECs lysed by NK cells at an E:T ratio of 1:1 using the 2D live-cell tracking method. Data represent four healthy donors, each shown in a different color, along with their technical replicates. Data were analyzed by Fiji. P < 0.0001 parametric paired t-test. (D) Pooled data showing the percentage of total dead PAECs and (E) total dead human endothelial cells EA.hy926 after 2 h co-incubation with human primary NK cells at an E:T ratio of 1:20. Data represent three healthy donors, each shown in a different color, along with their technical replicates.
Apoptosis is the predominant pathway of NK cytotoxicity against porcine endothelial cells under 2D static conditions
During the 2 h time-lapse microscopy, we examined cell morphology and fluorescent signals to differentiate between the two primary lytic pathways induced by NK cells, apoptotic and necrotic-like cell death. Hoechst dye enters living cell nuclei, while Draq7 enters nuclei only upon cell membrane disruption. Necrotic-like cell death is detected through double staining (Hoechst and Draq7). Apoptosis, characterized by apoptotic bodies with intact membranes and enhanced Hoechst staining signals. In vivo, phagocytes engulf apoptotic cells leading to their elimination37. However, in their absence, as in our experimental setting, the membranes of apoptotic bodies break down at later stages, resulting in staining with Draq7. This process is known as secondary necrosis or late apoptosis38,39 (Fig. 3A). During the two-hour assay, about 20% of the dead PAECs detached from the plate surface and moved out of the focal plane of the microscope. When the percentages of apoptotic and necrotic cells were analyzed to the attached PAECs at the endpoint, a trend toward increased apoptosis was observed in PAECs treated with pTNF (Fig. 3B, left). The predominant death mechanism observed in PAEC was apoptosis, accounting for 96.2 ± 3.8% of the dead cells in control PAECs, and 97.7 ± 1.5% in pTNF-stimulated PAECs, respectively (Fig. 3B, right). Future studies are necessary to determine whether granule release or death receptor signaling is responsible.
2D live-cell tracking allows analysis at a single-cell level and reveals cell death pathways. Two conditions were tested; PAECs overnight activated with porcine tumor necrosis factor (pTNF) or not (Φ). Colors represent PAECs’ nuclei (blue), cells with disrupted membranes (pink), NK cells (yellow). (A) Pictures showing apoptotic and necrotic cell death of pTNF-activated PAECs lysed by NK cells were captured during a 20 min time-lapse video using an effector-to-target (E:T) ratio of 1:1. Scale bar = 50 μm. (B) Quantification of PAEC death occurring by apoptosis and necrosis after 120 min interaction with human NK cells at E:T ratio = 1:1. Nested-plot for the percentage of apoptotic and necrotic cells in total counted adherent PAECs, in control and pTNF-treated targets (left), and bar-plot comparing differences in apoptosis within the dead PAEC population between control and pTNF-treated PAECs (right). Data were from three different healthy donors, analyzed by paired t-test.
2D live-cell tracking and automated cell segmentation reveal distinct NK cell migration behavior on pTNF-activated and control PAECs
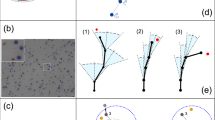
In-house program development enabled the segmentation and analysis of NK cell migration trajectories from a heterogeneous population of over 3,500 individual NK cells. Our analysis revealed three distinct types of movement based on trajectory diameter: small (less than 10 μm), intermediate (ranging from 10 to 100 μm), and large (over 100 μm) (Fig. 4A). The diameter of the trajectory was determined by measuring the distance a single cell traveled from one point to another, and the length of the trajectory refers to the total distance a cell travels within the same diameter (Fig. 4B). As shown in Fig. 4C, NK cells encountering pTNF-activated PAECs exhibited nearly triple the number of small trajectories (< 10 μm) compared to the control group. Figure 4D (left) provides an overview of migratory behavior of more than 1,500 NK cells and highlights a significant difference between the two groups tested, with a P-value of < 0.0001. Figure 4D (right) shows a plot zoomed in on the trajectory diameter from 0 to 20 μm, confirming the increased number of small trajectories in pTNF-treated samples. Furthermore, in small and intermediate trajectories, NK cells on pTNF-activated PAECs exhibited significantly higher ratios of trajectory length to diameter than controls (Fig. 4E). Within the same diameter range, it is apparent that NK cells on pTNF PAECs took longer routes to explore the region and to interact with surrounding cells. There were no discernable differences (P = 0.7739) in the average speed of NK cells in trajectories smaller than 10 μm (Fig. 4F, left). However, a slight increase in speed was observed on pTNF-treated cells in the intermediate range (0.55 ± 0.03 versus 0.43 ± 0.05 μm/min for control, P = 0.0150, Fig. 4F, middle). In large trajectories, NK cells showed a trend migrated faster at 1.94 ± 0.2 μm/min on pTNF-treated PAECs versus 1.5 ± 0.23 μm/min on control PAECs, P = 0.1184 (Fig. 4F, right). These findings were consistent with the arrest coefficient data shown in Fig. 4G. The arrest coefficient is defined as the percentage of time a cell remains below a speed threshold. When a cell’s instantaneous speed is lower than the threshold speed (0.2 μm/min), it is considered to be in an arrest phase. For large trajectories, NK cells on control PAECs spent more time in the arrest phase, indicating that they required more time to complete their trajectories (P = 0.073). Altogether, these data support the notion that NK cells display increased dynamics when interacting with pTNF-activated endothelial cells as opposed to untreated cells.
Automated cell tracking of NK cell trajectories using the 2D live-cell imaging method provides valuable information about cellular behavior during target cell elimination. (A) The movement of each NK cell in the field of view was tracked and classified into three different types: small, intermediate, and large. Each color assigns a single NK’s trajectory during 120 min of the time-lapse video, overlayed on the representative frame. Scale bar = 100 μm. (B) Definition of trajectory diameter (dotted yellow) and trajectory length (dashed purple) using an example of the displacement of one NK cell (blue) recorded over 120 min. (C) Upper panels display the NK trajectories in control and pTNF-treated samples in one field of view of a well where the coordinates of x- and y-axis are given in pixels [px]. Lower panels display the percentage of different NK movements in control (Φ) and pTNF-activated PAECs. (D) Correlation between individual NK cells’ trajectory length and diameter (both in µm) is shown for all detected trajectories (left), with a zoomed-in view of those with diameters smaller than 20 μm (right). Comparison in NK behavior in response to control (Φ) and pTNF-stimulated PAECs is illustrated using violin-plots for (E) the trajectory length-to-diameter ratio, (F) NK cell average speed, and (G) the NK arrest coefficient for each of the three trajectory diameters. Data were analyzed from the average of NK cells per field of view and three technical replicates, with the total of at least 1,500 NK cells per group. Paired t-test was used for statistical analysis.
3D microfluidic system simulating vessel-like conditions to evaluate NK cell functions under laminar shear stress
The 3D microfluidic system developed by Chrobak et al. and Sfriso et al.29,30 was modified to study the cellular interactions between circulating NK cells and PAECs that adhere to the walls of the channels. The study focused specifically on adhesion, migratory behavior, and NK cell-mediated target cell destruction. Sfriso et al. had reported that PAECs maintain typical endothelial markers when cultured in microfluidic channels, including CD31 and VE-Cadherin30. After 48 h of culturing under 2 dyn/cm2 (equivalent to 0.2 Pa) shear stress and using media with blood-like viscosity, we successfully established a PAEC monolayer throughout the entire 3D channel (Fig. 5A). As illustrated in Fig. 5A, NK cell-containing medium was injected into the 15 ml reservoir. The cells passed through the inlet, travelled through a 0.55 mm-diameter tube that was covered by an endothelial monolayer, and exited through the outlet. Additionally, without TNF treatment, we observed a significant and dense layer of heparan sulfate, which is a major component of the glycocalyx layer present in all blood vessels31 (Fig. 5B). Recirculation is an important aspect of this system because it enables cell-cell communication through soluble mediator molecules such as cytokines and chemokines that are released from the initial interactions of NK and PAECs. We characterized the expression of adhesion molecules in 3D channels under both static and flow conditions. Flow conditions consisted of flow media (DMEM containing 4% Dextran and 1% BSA) at a shear stress of 2 dyn/cm² and 48 h of culture. There was no significant difference in the expression of pVCAM-1 and pICAM-1 in 3D static and flow conditions, without additional stimulation. However, when PAECs were stimulated with pTNF, the expression of pICAM-1 and pVCAM-1 significantly increased, reaching up to 100% of total cells per field (Fig. 5C).
3D-microfluidic system and the NK functionality against PAECs under the flow. (A) Illustration of the cylindrical 3D chip depicting the three spatial coordinates (magenta, yellow, and green) (left). Channel’s length: 1 cm, dimeter: 550 μm. Confocal imaging shows a monolayer of pig endothelial cells (PAECs) covering the inner wall of the channel. Cells were stained with anti-alphaGal antibody (red), and nuclei were stained with Hoechst (cyan). The image was acquired using a Leica confocal SP8 microscope at 20× magnification. Scale bar = 100 μm. (B) The presence of a glycocalyx layer on PAECs in the 3D system under flow conditions without TNF treatment. Cells were stained with anti-heparan sulfate (red), and nuclei were stained with Hoechst (cyan). Images were acquired using a Leica confocal SP8 microscope at 20× magnification. Scale bar = 25 μm. (C) Adhesion molecules on porcine endothelium in 3D static and flow conditions, with and without TNF treatment. At the endpoint, the experimental channels were washed, and the cells were fixed before staining for porcine Vascular adhesion molecule-1 (pVCAM-1) and intercellular adhesion molecule-1 (pICAM-1). Nuclei were stained with Hoechst (cyan). The figure shows the maximum projection of 20 z-stacks with 0.5 μm per stack. Percentage of pICAM-1 and pVCAM-1 positive cells was quantified by dividing the number of positive cells by the total number of nuclei per field. Data represent two independent experiments. Scale bar = 60 μm.
To assess the possible effects of plastic tubing and hard channel surfaces on NK cell function, freshly isolated NK cells underwent a 4 h flow test and were compared to NK cells kept in a flask in an incubator. The flow cytometry findings demonstrate that the NK phenotypes were identical in both conditions (Supplementary Fig. S3). Therefore, it can be concluded that the tubing system did not have any noticeable impact on the NK cells during the 4 h flow test.
Using live cell imaging, we observed the interactions between NK cells and the endothelium under the shear stress and blood-like viscosity generated from a peristaltic pump and a 3D round-section channel. Within 20 min, variations in NK cell attachment on control and pTNF-stimulated PAECs became apparent. Figure 6A shows the tendency of NK cells to accumulate in certain regions of the channels in pTNF-treated PAECs. Despite the fast flow, under microvessel-like shear stress and viscosity, NK cells came to a complete stop without deceleration. The NK cells strongly adhered to the PAECs and remained attached throughout the 40-minute observation period by time-lapse microscopy. No detachment of the NK cells from the examined areas was observed (Supplementary Video 3A). Flow medium supplemented with Draq7 dye displayed live killing by NK cells, as evidenced by red dead PAECs during the process (Supplementary Video 3B). The entire channel was examined after 2 h of NK perfusion to evaluate the attachment and lysis of PAECs by NK cells. As shown in Fig. 6B, the data revealed a notable rise in the count of NK cells adhered to pTNF-stimulated PAECs in comparison to control cells at E:T ratios of 1:1 and 1:20. In correlation with the NK cell adhesion, the number of PAEC lysis events in pTNF-treated channels was found to be statistically higher at an E:T ratio of 1:1. The reason we did not detect a comparable number of attached NK cells and dead PAECs in pTNF-treated cells was due to the detachment of some Draq7+ PAECs and unlabeled apoptotic bodies following the flow medium during the 2 h time-lapse imaging (Supplementary Video 3C). The number of dead PAECs per attached NK cell showed no significant difference between control and pTNF-treated groups across three healthy donors. This suggests that one NK cell may kill the same number of target cells under both control and pTNF conditions when attached to the channel. Activating PAECs with pTNF appears to only enhance the stickiness of the endothelium, attracting more NK cells (Supplementary Fig. S6). Testing the 3D microfluidic system with human endothelial cells, namely EA.hy926 cells, showed good adhesion to the channels before and after 48 h of perfusion. However, we could detect no significant differences in terms of adhesion and killing by NK cells between treated and non-treated hTNF samples, most likely because of the large variability observed among the three different donors (Fig. 6B, right).
NK cell adhesion and lysis of PAECs under the flow. (A) Live-cell wide-field microscopy of NK cells on control (Φ, left) and pTNF-stimulated (right) PAECs at given time points. The channels showing NK cells (green) and dead cells (Draq7+, red) were overlaid on the bright field image to correlate with the PAECs monolayer. For easier visualization and quantification, only the fluorescent channels are shown on the right. Arrows show the three spatial coordinates (magenta, yellow and green), with magenta indicating the flow direction. Scale bar = 100 μm. (B) Analysis throughout the entire 3D channels seeded with control (Φ) or pTNF-stimulated PAECs, at the endpoint (120 min), E:T ratio 1:1. Bottom left plot shows the number of attached NK and dead PAECs at an E:T ratio of 1:1; middle plot shows the number of attached NK and dead PAECs at an E:T ratio of 1:20; right plot shows the number of attached NK cells and dead EA.hy926 cells at an E:T ratio of 1:20. Data were analyzed from three independent experiments with three different healthy donors, multiple paired t-test. Scale bar = 200 μm. (C) The bar-plot shows number of NK cells attached to one region of the channel (0.1 mm) under flow, as well as and the number of cells that migrated on top of control (Φ) and pTNF-stimulated (right) PAECs. Data were analyzed from three different experiments consisting of different healthy donors, one specific region per channel. (D) The left panel displays NK cell trajectories in control (Φ) and pTNF-treated PAECs in one field of view (0.1 mm) of a 3D channel during a 20 min time-lapse, with x- and y-axis coordinates given in pixels (px). The right panel displays the percentage of different NK cell movements, categorized by small and intermediate diameter trajectories, in control (Φ) and pTNF-activated PAECs. The magenta arrow indicates the flow direction. (E) Average speed of NK cell movements on control (Φ) and pTNF-treated target cells. Data were analyzed from a heterogeneous population of NK cells in one region of a 3D channel, from one healthy donor, using an in-house developed Python program. Statistics were obtained using a multiple paired t-test.
Interestingly, a detailed analysis of PAEC and NK interactions revealed NK cell trajectories moving in different directions on top of the PAEC monolayer (Fig. 6D). In certain instances, specific cells even exhibited migration against the flow direction (indicated by white arrows) (Supplementary Video 4). This phenomenon has recently been reported for neutrophils46. Variations in the movement of NK cells were observed within different regions of a single 3D channel. Some regions depicted stationary adherence of NK cells to the PAEC layer, while other areas showed a higher tendency to migrate. This observation requires further investigation. Among the total NK cells that adhered to the endothelium, there was a noticeable tendency of more NK cells exhibiting crawling behavior in pTNF-stimulated PAECs, as compared to the control (Fig. 6C). While a trend was observed, this difference did not reach statistical significance (P = 0.5007). This result was due to the analysis being conducted within a single examined area across three independent experiments, rather than encompassing the entire channel as shown in Fig. 6B. Due to technical limitations, we were unable to acquire timely time-lapse videos for all channel scans. Thus, we compared NK trajectories in one specific region from a single healthy donor, which demonstrated a high number of crawling NK cells (Fig. 6D-E, S4). As shown in Fig. 6D (left panel), under flow conditions, NK cells displayed diverse migration patterns in multiple directions. Interestingly, we found more small trajectories (< 10 μm) in the control group of PAECs (Fig. 6D, right panel), while no large trajectories (> 100 μm) were detected, unlike in the 2D setting. NK cells moved marginally faster with an average speed of 1.66 ± 0.57 μm/min in pTNF-stimulated PAECs, compared to a rate of 1.18 ± 0.28 μm/min in the control group (P = 0.048) (Fig. 6E). The average speed of NK cells in the 3D microfluidic system was twice as high as the speed observed in intermediate trajectories in the 2D settings (Fig. 4F, middle). However, there were no differences in trajectory length to diameter ratio and arrest coefficient between control and pTNF-activated targets (Supplementary Fig. S4). This suggests that NK cells behave similarly in terms of trajectory distance and area exploration under the influence of flow and shear stress. Altogether, we showed NK cell adhesion and migratory patterns in the presence of flow shear stress. Although increasing the sample size is necessary, our in-house developed program applies equally well to microfluidic systems, revealing various aspects of NK cell migratory behavior.
Discussion
The standard cytotoxicity release assays (Delfia, 51[Cr], and Calcein assays) demonstrate considerable variability among cell lines, largely due to the varying maximum and spontaneous release measurements10,13. Our microscope-based assay proves to be a practical method for assessing NK cytotoxicity in adherent cells, yielding highly sensitive results. This method, from a single experiment, provides a comprehensive understanding of NK cell killing dynamics and cell death pathways. Several studies have shown that NK cells can initiate different death pathways in target cells including apoptosis, necrosis, and programmed necrotic death such as necroptosis, and pyroptosis9,47,48. Typical cytotoxicity assays employ high E:T ratios (3:1, 5:1, 10:1, or higher). However, in many clinical situations, target cells may outnumber cytotoxic lymphocytes49,50. Therefore, such high E:T ratios may not accurately represent how NK cells eliminate target cells in vivo. Cytotoxicity release Delfia assays solely measure overall lysis at the endpoint, so it is not feasible to trace the events during the 2 h interaction time between NK cells and PAECs.
Remarkably, at a relatively low E:T ratio (1:1), and in the absence of phagocytic cells, we discovered that about 97% of PAEC death during interactions with NK cells occurred through apoptosis. Apoptosis is characterized by cell shrinkage, membrane blebbing, and the formation of apoptotic bodies with intact membranes51. Our findings align with a recent study by Pan et al.., which highlights the significance of apoptosis in NK-mediated killing at a more physiological E:T ratio (1:2)52. Another study on cytotoxic lymphocytes demonstrated that perforin played a major role in over 90% of NK killing events, resulting in apoptosis of every damaged target cell through rapid sequential delivery53. Necroptosis exhibits a similar morphology to that of necrotic cell death, characterized by cell body swelling and eventual explosion. By contrast, pyroptotic cells show less swelling and generate multiple bubble-like protrusions that resemble apoptotic bodies54. We cannot rule out the possibility of pyroptotic cell death in our study, since there is insufficient research that distinctly identifies morphological differences between pyroptosis and apoptosis. Future use of live-cell imaging with specific pathway inhibitors such as zVAD-fmk, necrostatin-, ferrostatin-1, and dimethyl fumarate can further examine apoptosis, necroptosis, ferroptosis, and pyroptosis.
Another advantage of our 2D system is its capacity to track individual cell interactions simultaneously with cytotoxicity in high throughput, an aspect that has yet to receive systematic and thorough investigation. A previous report by Önfelt’s group assessed NK cytotoxicity and migration in parallel using cell tracers and ultrasound to facilitate cell-cell interactions55. This technique represents a valuable tool for studying NK cell function against target in suspension. However, in the context of adherent endothelial cell layers, it is necessary to modify the cell culture conditions, labeling system, and analysis strategies to mimic the physiological interactions between NK cells and the endothelium in blood vessels. Our segmentation and tracking strategies allowed us to visualize thousands of NK cells moving on top of the endothelial cell monolayer. The categorization of small trajectories (< 10 μm) resembles confined movement, while large trajectories (> 100 μm) are similar to directed movement, as previously described in the literature based on mean square displacement (MSD) data55,56. In the time frame of 2 h, the large trajectories detected in our experiments were shorter in comparison to findings from previous studies, which usually last more than 8 h19,55. We did not prolong the time-lapse microscope session, as IL-2-activated NK cells can efficiently kill target cells in less than 2 h. Therefore, in our study, the trajectory categorization is based on diameter ranges rather than MSD modulation. We found a heterogeneous migratory behavior of NK cells on both control and pTNF-activated PAECs. The significant increase in confined-like movement (< 10 μm) of NK cells on pTNF-activated targets, along with the high trajectory length-to-diameter ratio, indicates that NK cells explored more extensively within the same region. Moreover, NK cells displayed more dynamic behavior, as evidenced by the increased average speed in intermediate and larger trajectories. The increase in confined-like movement on pTNF-treated PAECs can be explained by the increased number of cells expressing pVCAM-1 in 2D static conditions, and both pVCAM-1 and pICAM-1 in 3D flow conditions, which led to an increase in NK cell adhesion (Supplementary Fig. S5 and Fig. 5C). Previous studies have shown a major role of pVCAM-1 in facilitating leukocyte rolling, firm adhesion to the endothelium, and leukocyte transmigration45. Our novel 2D system will now be utilized to examine cell surface markers and intracellular proteins after cytotoxicity and migration experiments, and to study the molecular mechanisms occurring during the cellular interactions using specific blocking reagents. Our cell trajectory studies conducted in the 2D system can provide valuable information on NK cell migratory behavior in tissues and capillaries where shear stress is minimal or absent.
In the 3D microfluidic system, NK cells displayed migratory behavior akin to that seen in 2D static conditions. Some regions of the artificial vessel exhibited restricted NK cell movement under flow and vessel shear stress, while in other areas, NK cells migrated in various directions on the endothelium, irrespective of the flow orientation, similar to recent findings with neutrophils in the Ibidi system57. Due to technical constraints and data storage limitations, we were unable to draw a solid conclusion regarding the distinctions in NK crawling behavior between control and pTNF-stimulated PAECs in 3D microfluidic channels. However, in the 3D system, NK cells were unable to execute large diameter trajectories. Instead, they displayed a range of trajectories spanning from small to intermediate, with the longest recorded distance being 35.12 μm in pTNF-treated PAECs during the 20 min time-lapse. This might be explained by the fast flow and shear stress limiting the ability of NK cells to migrate long distances. As z-stacks were not included in the acquisition, there is a possibility of miscounting the trajectory length in the third dimension of the 3D channels. Certain NK cells may detach from the observed endothelial layer and shift laterally or towards the upper section of the tube. To further explore this notion, it is necessary to extend the acquisition time and incorporating z-stacks in 3D imaging. A key advantage of the 3D system is its cylindrical shape, which allows for the generation of homogenous shear stress, cell distribution, and velocity along the endothelial walls30. Opposite, geometrically symmetric distribution and a height-to-width ratio of 0.53 are not present in commercially available rectangular duct chambers. This results in non-uniform, asymmetric spreading of particles/cells along the flow direction, leading to uneven E:T ratios where the EC are located58. Shear stress significantly impacts the EC alignment and the composition and arrangement of the glycocalyx. Moreover, endothelial dysfunction is strongly associated with the shedding of this important layer59,60. The glycocalyx is hypothesized to shield EC surface, preventing excessive leukocyte attachment, transmigration, and curbs tissue inflammation61.
Although our microscope-based assays provide valuable insights, we acknowledge that artefacts introduced by the labelling process, such as photobleaching, can affect data accuracy. Future research could incorporate quantitative techniques, as flow cytometry, to assess CD107a expression on the NK cells and the release of lytic granules (granzyme, perforin) following co-culture with PAEC monolayers in the 3D system. Additionally, quantitative PCR or Western blot targeting specific marker genes after perfusion could be developed to further validate and strengthen the findings.
In summary, we present a novel live-cell imaging system adaptable to both 2D and 3D experiments for exploring various aspects of NK cell functions. We demonstrated the killing dynamics and heterogenous migratory behavior of NK cells interacting with primary ECs under both activated and inactivated conditions. Additionally, our data highlighted distinct features of NK cell movement in the presence of flow and shear stress compared to static 2D conditions. The integrated 2D and 3D workflow offers a more comprehensive view of NK cell functions. Substantial opportunities exist to enhance and advance this live imaging 3D system to more precisely replicate diverse physiological conditions observed in vivo. Lastly, our system is versatile in addressing various biological inquiries regarding the interactions between immune cells and endothelial cells, including T cells, B cells, macrophages, and neutrophils.
Data availability
Data is provided within the manuscript or supplementary information files. Additional data are available from the authors upon request.
References
Biron, C. A., Nguyen, K. B., Pien, G. C., Cousens, L. P. & Salazar-Mather, T. P. Natural killer cells in antiviral defense: Function and regulation by innate cytokines. Annu. Rev. Immunol. 17, 189–220. https://doi.org/10.1146/annurev.immunol.17.1.189 (1999).
Itescu, S. et al. Role of natural killer cells, macrophages, and accessory molecule interactions in the rejection of pig-to-primate xenografts beyond the hyperacute period. Hum. Immunol. 59, 275–286. https://doi.org/10.1016/s0198-8859(98)00026-3 (1998).
Wolf, N. K., Kissiov, D. U. & Raulet, D. H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. 23, 90–105. https://doi.org/10.1038/s41577-022-00732-1 (2023).
Gill, S., Olson, J. A. & Negrin, R. S. Natural killer cells in allogeneic transplantation: Effect on engraftment, graft- versus-tumor, and graft-versus-host responses. Biol. Blood Marrow Transpl. 15, 765–776. https://doi.org/10.1016/j.bbmt.2009.01.019 (2009).
Zamora, A. E., Grossenbacher, S. K., Aguilar, E. G. & Murphy, W. J. Models to study NK cell biology and possible clinical application. Curr. Protoc. Immunol. 110, 143711–143714. https://doi.org/10.1002/0471142735.im1437s110 (2015).
Davis, D. M. et al. The human natural killer cell immune synapse. Proc. Natl. Acad. Sci. U S A. 96, 15062–15067. https://doi.org/10.1073/pnas.96.26.15062 (1999).
Caligiuri, M. A. Human natural killer cells. Blood. 112, 461–469. https://doi.org/10.1182/blood-2007-09-077438 (2008).
Zwirner, N. W. & Domaica, C. I. Cytokine regulation of natural killer cell effector functions. Biofactors. 36, 274–288. https://doi.org/10.1002/biof.107 (2010).
Ramirez-Labrada, A. et al. All about (NK Cell-Mediated) death in two acts and an unexpected encore: Initiation, execution and activation of adaptive immunity. Front. Immunol. 13, 896228. https://doi.org/10.3389/fimmu.2022.896228 (2022).
Somanchi, S. S., McCulley, K. J., Somanchi, A., Chan, L. L. & Lee, D. A. A Novel Method for Assessment of Natural Killer cell cytotoxicity using image cytometry. PLoS One. 10, e0141074. https://doi.org/10.1371/journal.pone.0141074 (2015).
Buller, C. W. & Mathew, S. O. NK Cell isolation and cytotoxicity by Radioactive Chromium Release assay and DELFIA-EuTDA cytotoxicity assay. Methods Mol. Biol. 2463, 221–233. https://doi.org/10.1007/978-1-0716-2160-8_16 (2022).
Kim, J. et al. A Flow Cytometry-based whole blood natural killer cell cytotoxicity assay using overnight cytokine activation. Front. Immunol. 11, 1851. https://doi.org/10.3389/fimmu.2020.01851 (2020).
Neri, S., Mariani, E., Meneghetti, A., Cattini, L. & Facchini, A. Calcein-acetyoxymethyl cytotoxicity assay: Standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin. Diagn. Lab. Immunol. 8, 1131–1135. https://doi.org/10.1128/CDLI.8.6.1131-1135.2001 (2001).
Muller, W. A. Leukocyte-endothelial-cell interactions in leukocyte transmigration and the inflammatory response. Trends Immunol. 24, 327–334. https://doi.org/10.1016/s1471-4906(03)00117-0 (2003).
Ley, K., Laudanna, C., Cybulsky, M. I. & Nourshargh, S. Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689. https://doi.org/10.1038/nri2156 (2007).
Tomin, K., Goldfarb, R. H. & Albertsson, P. In vitro assessment of human natural killer cell migration and invasion. Methods Mol. Biol. 1441, 65–74. https://doi.org/10.1007/978-1-4939-3684-7_6 (2016).
Starnes, T. et al. The chemokine CXCL14 (BRAK) stimulates activated NK cell migration: Implications for the downregulation of CXCL14 in malignancy. Exp. Hematol. 34, 1101–1105. https://doi.org/10.1016/j.exphem.2006.05.015 (2006).
Olofsson, P. E. et al. Distinct Migration and Contact dynamics of resting and IL-2-Activated human natural killer cells. Front. Immunol. 5, 80. https://doi.org/10.3389/fimmu.2014.00080 (2014).
Mace, E. M., Gunesch, J. T., Dixon, A. & Orange, J. S. Human NK cell development requires CD56-mediated motility and formation of the developmental synapse. Nat. Commun. 7, 12171. https://doi.org/10.1038/ncomms12171 (2016).
Vanherberghen, B. et al. Classification of human natural killer cells based on migration behavior and cytotoxic response. Blood. 121, 1326–1334. https://doi.org/10.1182/blood-2012-06-439851 (2013).
Lee, B. J. & Mace, E. M. Acquisition of cell migration defines NK cell differentiation from hematopoietic stem cell precursors. Mol. Biol. Cell. 28, 3573–3581. https://doi.org/10.1091/mbc.E17-08-0508 (2017).
Olofsson, P. E. et al. A collagen-based microwell migration assay to study NK-target cell interactions. Sci. Rep. 9, 10672. https://doi.org/10.1038/s41598-019-46958-3 (2019).
Martinez, A. L. et al. Quantifying human natural killer cell migration by imaging and image analysis. Methods Mol. Biol. 2463, 129–151. https://doi.org/10.1007/978-1-0716-2160-8_10 (2022).
Gregoire, C. et al. The trafficking of natural killer cells. Immunol. Rev. 220, 169–182. https://doi.org/10.1111/j.1600-065X.2007.00563.x (2007).
Melsen, J. E., Lugthart, G., Lankester, A. C. & Schilham, M. W. Human circulating and tissue-resident CD56(bright) natural killer cell populations. Front. Immunol. 7, 262. https://doi.org/10.3389/fimmu.2016.00262 (2016).
Hattori, K. et al. Microfluidic perfusion culture chip providing different strengths of shear stress for analysis of vascular endothelial function. J. Biosci. Bioeng. 118, 327–332. https://doi.org/10.1016/j.jbiosc.2014.02.006 (2014).
Wilson, D. & Hube, B. Hgc1 mediates dynamic Candida albicans-endothelium adhesion events during circulation. Eukaryot. Cell. 9, 278–287. https://doi.org/10.1128/EC.00307-09 (2010).
Esch, M. B., Post, D. J., Shuler, M. L. & Stokol, T. Characterization of in vitro endothelial linings grown within microfluidic channels. Tissue Eng. Part. A. 17, 2965–2971. https://doi.org/10.1089/ten.tea.2010.0371 (2011).
Chrobak, K. M., Potter, D. R. & Tien, J. Formation of perfused, functional microvascular tubes in vitro. Microvasc Res. 71, 185–196. https://doi.org/10.1016/j.mvr.2006.02.005 (2006).
Sfriso, R. et al. 3D artificial round section micro-vessels to investigate endothelial cells under physiological flow conditions. Sci. Rep. 8, 5898. https://doi.org/10.1038/s41598-018-24273-7 (2018).
Milusev, A., Rieben, R. & Sorvillo, N. The endothelial glycocalyx: A possible therapeutic target in Cardiovascular disorders. Front. Cardiovasc. Med. 9, 897087. https://doi.org/10.3389/fcvm.2022.897087 (2022).
Milusev, A. et al. Glycocalyx dynamics and the inflammatory response of genetically modified porcine endothelial cells. Xenotransplantation. 30 ARTN e1282010.1111/xen.12820 (2023).
Stefan Silbernagl, A. D. Color Atlas of Physiology. 6th edn, (2011).
Crocker, J. Methods of digital video microscopy for colloidal studies. J. Colloid Interface Sci. 179, 12 (1996).
Puga Yung, G., Schneider, M. K. J. & Seebach, J. D. The role of NK cells in Pig-to-human xenotransplantation. J. Immunol. Res. 2017 (4627384). https://doi.org/10.1155/2017/4627384 (2017).
Goodman, D. J., Von Albertini, M., Willson, A., Millan, M. T. & Bach, F. H. Direct activation of porcine endothelial cells by human natural killer cells. Transplantation. 61, 763–771. https://doi.org/10.1097/00007890-199603150-00016 (1996).
Arandjelovic, S. & Ravichandran, K. S. Phagocytosis of apoptotic cells in homeostasis. Nat. Immunol. 16, 907–917. https://doi.org/10.1038/ni.3253 (2015).
Krysko, D. V., Berghe, V., D’Herde, T., Vandenabeele, P. & K. & Apoptosis and necrosis: Detection, discrimination and phagocytosis. Methods. 44, 205–221. https://doi.org/10.1016/j.ymeth.2007.12.001 (2008).
Bertheloot, D., Latz, E. & Franklin, B. S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell. Mol. Immunol. 18, 1106–1121. https://doi.org/10.1038/s41423-020-00630-3 (2021).
Rao, R. M., Yang, L., Garcia-Cardena, G. & Luscinskas, F. W. Endothelial-dependent mechanisms of leukocyte recruitment to the vascular wall. Circ. Res. 101, 234–247. https://doi.org/10.1161/CIRCRESAHA.107.151860b (2007).
Schneider, M. K. & Seebach, P. F. J. D. Adhesive interactions between human natural killer cells and porcine endothelial cells. Scand. J. Immunol. 54, 70–75 (2001).
Lechleitner, S., Gille, J., Johnson, D. R. & Petzelbauer, P. Interferon enhances tumor necrosis factor-induced vascular cell adhesion molecule 1 (CD106) expression in human endothelial cells by an interferon-related factor 1-dependent pathway. J. Exp. Med. 187, 2023–2030. https://doi.org/10.1084/jem.187.12.2023 (1998).
Burke-Gaffney, A. & Hellewell, P. G. Tumour necrosis factor-alpha-induced ICAM-1 expression in human vascular endothelial and lung epithelial cells: modulation by tyrosine kinase inhibitors. Br. J. Pharmacol. 119, 1149–1158. https://doi.org/10.1111/j.1476-5381.1996.tb16017.x (1996).
Stocker, C. J. et al. Cloning of porcine intercellular adhesion molecule-1 and characterization of its induction on endothelial cells by cytokines. Transplantation. 70, 579–586. https://doi.org/10.1097/00007890-200008270-00008 (2000).
Schneider, M. K. et al. Rolling adhesion of human NK cells to porcine endothelial cells mainly relies on CD49d-CD106 interactions. Transplantation. 73, 789–796. https://doi.org/10.1097/00007890-200203150-00023 (2002).
Grönloh, M. L. B., Tebbens, M. E., Kotsi, M., Arts, J. J. G. & van Buul, J. D. ICAM-2 regulates diapedesis hotspots by allowing neutrophil crawling against the direction of flow. LID. https://doi.org/10.1530/VB-23-0005.
Prager, I. & Watzl, C. Mechanisms of natural killer cell-mediated cellular cytotoxicity. J. Leukoc. Biol. 105, 1319–1329. https://doi.org/10.1002/JLB.MR0718-269R (2019).
Backes, C. S. et al. Natural killer cells induce distinct modes of cancer cell death: Discrimination, quantification, and modulation of apoptosis, necrosis, and mixed forms. J. Biol. Chem. 293, 16348–16363. https://doi.org/10.1074/jbc.RA118.004549 (2018).
Galon, J. & Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 18, 197–218. https://doi.org/10.1038/s41573-018-0007-y (2019).
Shimasaki, N., Jain, A. & Campana, D. NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19, 200–218. https://doi.org/10.1038/s41573-019-0052-1 (2020).
Wyllie, A. H., Kerr, J. F. & Currie, A. R. Cell death: the significance of apoptosis. Int. Rev. Cytol. 68, 251–306. https://doi.org/10.1016/s0074-7696(08)62312-8 (1980).
Pan, R., Ryan, J., Pan, D., Wucherpfennig, K. W. & Letai, A. Augmenting NK cell-based immunotherapy by targeting mitochondrial apoptosis. Cell 185, 1521–1538 e1518 (2022). https://doi.org/10.1016/j.cell.2022.03.030
Lopez, J. A. et al. Rapid and unidirectional perforin pore delivery at the cytotoxic immune synapse. J. Immunol. 191, 2328–2334. https://doi.org/10.4049/jimmunol.1301205 (2013).
Chen, X. et al. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell. Res. 26, 1007–1020. https://doi.org/10.1038/cr.2016.100 (2016).
Forslund, E. et al. Novel microchip-based tools facilitating live cell imaging and assessment of functional heterogeneity within NK cell populations. Front. Immunol. 3, 300. https://doi.org/10.3389/fimmu.2012.00300 (2012).
Khorshidi, M. A. et al. Analysis of transient migration behavior of natural killer cells imaged in situ and in vitro. Integr. Biol. (Camb). 3, 770–778. https://doi.org/10.1039/c1ib00007a (2011).
Gronloh, M. L. B. et al. Endothelial transmigration hotspots limit vascular leakage through heterogeneous expression of ICAM-1. EMBO Rep. https://doi.org/10.15252/embr.202255483 (2023).
Aminian, M., Bernardi, F., Camassa, R. & McLaughlin, R. M. Squaring the Circle: Geometric skewness and symmetry breaking for Passive Scalar Transport in ducts and pipes. Phys. Rev. Lett. 115, 154503. https://doi.org/10.1103/PhysRevLett.115.154503 (2015).
Kroon, J. et al. Flow-induced endothelial cell alignment requires the RhoGEF Trio as a scaffold protein to polarize active Rac1 distribution. Mol. Biol. Cell. 28, 1745–1753. https://doi.org/10.1091/mbc.E16-06-0389 (2017).
Moore, K. H., Murphy, H. A. & George, E. M. The glycocalyx: A central regulator of vascular function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 320, R508–R518. https://doi.org/10.1152/ajpregu.00340.2020 (2021).
Constantinescu, A. A., Vink, H. & Spaan, J. A. Endothelial cell glycocalyx modulates immobilization of leukocytes at the endothelial surface. Arterioscler. Thromb. Vasc Biol. 23, 1541–1547. https://doi.org/10.1161/01.ATV.0000085630.24353.3D (2003).
Acknowledgements
This work was supported by the Swiss National Science Foundation (SNSF # CRSII5_198577). The authors extend their gratitude to Dr. Anastasia Milusev for sharing her knowledge and techniques involved in developing the 3D microfluidic system. Additionally, we appreciate the group of Prof. Eckhard Wolf from Ludwig-Maximilian-University of Munich for providing porcine endothelial cells. The authors would also like to thank Dr. Dimitri Moreau, Stefania Vossio, Dr. Vincent Mercier, and Dr. Jérome Bosset for their invaluable microscopy technical support and analysis.
Author information
Authors and Affiliations
Contributions
TAT conducted most of the experimental work, data analysis, and interpretation, and wrote and edited the manuscript. VG conducted part of the experimental work, and interpretation, and wrote and edited part of the manuscript. OU developed the in-house built Python program for the analysis of trajectories and cell segmentation. DRG conducted part of the experimental work and flow cytometry analysis. RR conducted several revisions of the manuscript. TBMA supervised the development of the in-house Python program for analysis of trajectories and cell segmentation. GPY supervised the project, contributed to data interpretation and critical discussions, and conducted several revisions of the manuscript. JDS supervised the project, contributed to data interpretation and critical discussions, and conducted several revisions of the manuscript. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the work.
Corresponding authors
Ethics declarations
Competing interests
CSL Behring partially funded GPY’s salary while all other authors attest that the research was conducted without any commercial or financial relationships that could create a potential conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary Material 2
Supplementary Material 3
Supplementary Material 4
Supplementary Material 5
Supplementary Material 7
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tran, T., Galdina, V., Urquidi, O. et al. Assessment of NK cytotoxicity and interactions with porcine endothelial cells by live-cell imaging in 2D static and 3D microfluidic systems. Sci Rep 14, 24199 (2024). https://doi.org/10.1038/s41598-024-75217-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-75217-3