Abstract
Melissa officinalis is a perennial medicinal plant traditionally used for its diverse biological activities, including antiviral properties. This study investigates the antiviral efficacy of various extracts, including water, acetone, alkaloid, non-alkaloid, ethanol, and methanol extracts, against influenza A (H1N1), SARS-CoV-2, and MERS-CoV. The water extract demonstrated significant inhibitory effects on SARS-CoV-2 (IC50 = 421.9 µg/mL) and MERS-CoV (IC50 = 222.1 µg/mL) in Vero E6 cells (an African green monkey kidney cell line), with a CC50 of 4221 µg/mL, indicating a favorable selectivity index. Additionally, it exhibited strong activity against H1N1 in Madin-Darby canine kidney cell line (MDCK cells) (IC50 = 57.30 µg/mL, CC50 = 3073 µg/mL). Among all the extracts, the methanol extract showed the highest antiviral activity. It has IC50 = 2.549 µg/ml and selectivity index (SI) = 230 against H1N1.While it showed IC50 = 10.83 µg/ml against SARS-CoV-2 and 9.82 µg/ml against MERS-CoV with SI values of 54.2 and 59.77, respectively. Molecular docking studies revealed that 5-Methyl-5 H-naphtho[2,3-c]carbazole,7 H-Dibenzo[b, g]carbazole, 7-methyl, hesperidin, luteolin-7-glucoside-3′-glucuronide, Melitric acid A, and other compounds exhibited high binding affinities to the receptor-binding domains (RBDs) of SARS-CoV-2 and MERS-CoV spike glycoproteins, suggesting their potential to interfere with viral entry. Furthermore, GC-MS-identified bioactive compounds, including docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), paromomycin, and phenolic acids, demonstrated additional antiviral potential. These results underscore the potential of M. officinalis extracts as natural antiviral agents, offering a foundation for further in vitro and in vivo validation and potential therapeutic applications against respiratory viral infections, including coronaviruses and influenza viruses.
Similar content being viewed by others
Introduction
Respiratory infections are a significant cause of morbidity and mortality worldwide. Among these, coronaviruses such as SARS-CoV-2 and MERS-CoV, along with influenza viruses, particularly H1N1, have demonstrated their potential to cause global pandemics1. The outbreak of severe acute respiratory syndrome coronavirus (SARS-CoV) in late 2002, originating in Foshan, Guangdong Province, China, rapidly spread to 29 countries, infecting 8098 individuals with a case fatality rate of 10%2,3. Similarly, Middle east respiratory syndrome coronavirus (MERS-CoV), first identified in Jeddah, Saudi Arabia, in 2012, led to over 2,600 laboratory-confirmed cases and 943 deaths, reflecting a high mortality rate of around 34–36%4,5. Subsequent outbreaks, such as the 2015 MERS outbreak in South Korea, which resulted in 186 cases and 38 fatalities, reinforced the pandemic potential of coronaviruses6.
In late 2019, SARS-CoV-2 emerged in Wuhan, China, leading to the COVID-19 pandemic. Symptoms of SARS-CoV-2 infections range from mild respiratory illness to severe pneumonia, with vulnerable populations exhibiting higher mortality rates7. The virus rapidly spread worldwide, causing COVID-19, a disease characterized by fever, fatigue, and dry cough. In more severe cases, patients experienced shortness of breath, chills, muscle pain, loss of taste or smell, vomiting, and diarrhea. Older individuals and those with pre-existing conditions were at higher risk of severe complications8,9.
Influenza virus infections are also a common public health concern, particularly in winter months. Influenza can present throughout the year with symptoms varying from mild upper respiratory tract infections—characterized by sore throat, fever, runny nose, muscle aches, and headache—to severe pneumonia10. Seasonal influenza viruses, specifically types A and B, typically spread through respiratory droplets and small aerosols emitted by infected individuals during coughing or sneezing. The viruses can remain infectious in airborne particles and on surfaces for several hours, increasing the risk of widespread transmission11.
Natural plant-derived compounds provide an alternative for antiviral drug development, offering bioavailability, lower cost, and fewer side effects. Approximately 40% of currently available pharmaceuticals originate from plant sources, demonstrating the significance of exploring medicinal plants for novel antiviral agents12. Melissa officinalis (lemon balm), a member of the Lamiaceae family, has a long history of medicinal use. Native to the Mediterranean and Western Asia, it is widely cultivated across North America and Europe13.
Several studies have reported the pharmacological benefits of M. officinalis, including antimicrobial, anti-inflammatory, antioxidant, and antiviral properties14,15. The plant has demonstrated efficacy against various pathogens, including herpes simplex virus (HSV-1), human immunodeficiency virus (HIV-1), and influenza viruses16. However, its specific antiviral mechanisms against coronaviruses, particularly SARS-CoV-2 and MERS-CoV, remain largely unexplored.
This study aims to address this gap by evaluating the antiviral efficacy of M. officinalis extracts against SARS-CoV-2, MERS-CoV, and H1N1. Through cytotoxicity assays, viral inhibition studies, and molecular docking approaches, we aim to identify key bioactive compounds responsible for the observed antiviral effects. Furthermore, we assess the drug-likeness and toxicity profiles of these compounds to determine their therapeutic potential. Understanding the antiviral mechanisms of M. officinalis can contribute to the search for plant-derived antiviral therapies. By investigating how natural extracts inhibit viral replication and transmission, this research provides valuable insights into alternative treatment options for emerging respiratory viruses. The results of this study hold significant implications for antiviral drug discovery, particularly in the context of future pandemics.
Materials and methods
Viruses
The study utilized MERS-CoV-NRCE-HKU270/2013 (camel coronavirus), hCoV-19/Egypt/NRC-03/2020 (SARS-CoV-2), and Influenza A virus [A/Puerto Rico/8/34 (H1N1)]. These viruses were used to determine the antiviral activity of water and alcoholic extracts of Melissa officinalis.
Cell line
African green monkey kidney (Vero E6) cells and Madin-Darby Canine Kidney (MDCK) cells were obtained from Dr. Richard Webby at St. Jude Children’s Research Hospital, Department of Virology and Molecular Biology, USA. The cells were cultured in 75 cm2 tissue culture flasks using DMEM medium supplemented with 1% Penicillin/Streptomycin and 10% fetal bovine serum (FBS). The cells were propagated to confluence over multiple passages, aliquoted, and stored in liquid nitrogen for later use.
Plant material
M. officinalis was procured from a herbal store in Jeddah, Saudi Arabia. The plants were authenticated and characterized by Prof. Dr. Abdo Marei Hamed, Professor of Plant Ecology, Botany Department, Al-Azhar University. The plant classification was based on the United States Department of Agriculture (USDA) Natural Resources Conservation Service guidelines.
Preparation of water extract
A total of 3 g of dried plant material was mixed with 40 mL of deionized water and boiled for 3 min. The mixture was covered and left to cool overnight at room temperature under sterile conditions. The solution was clarified by centrifugation at 3000 rpm for 3 min. The supernatant was lyophilized in a petri dish in an oven set at 50 °C, and the resulting extract was weighed (276 mg/g). The extract was dissolved in an equal volume (w/v) of deionized water at a concentration of 1 g/mL.
Alcoholic extract preparation
A total of 100 g of M. officinalis herbal powder was subjected to extraction with 400 mL of absolute ethanol, methanol, and acetone separately. Ultrasonication in a water bath at 40 °C was performed for three hours to enhance the extraction process. The samples were further processed in a shaking water bath at 70 °C for 24 h to facilitate solvent evaporation. The resulting solutions were filtered using Whatman filter paper and prepared for GC-MS analysis. The extraction yields for ethanol, methanol, and acetone were 265, 303, 161 mg/g, respectively.
Preparation of alkaloid and non-alkaloid extracts
The alkaloid and non-alkaloid extracts were prepared following Elkady’s method17. A dried sample (100 g) was soaked in 80% ethanol for three days, filtered, and evaporated using a Buchi rotary evaporator. The extract was mixed with 80 mL of distilled water and 20 mL of glacial acetic acid, vigorously shaken, and filtered. An equal volume of chloroform was added, and the solution was shaken for two hours, forming two distinct layers. The upper aqueous layer was discarded, and the pH of the lower layer was neutralized using 0.1 N HCl. Further extraction was performed using chloroform for three hours, yielding an alkaloid-rich fraction (96 mg/g) and a non-alkaloid fraction (188 mg/g), both of which were freeze-dried for further analysis.
Gas chromatography-mass spectrometry (GC-MS) analysis
GC-MS analysis was performed using an Agilent Technologies 7000 GC/MS Triple Quad system at the King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia. The separation was conducted using an HP-5MS capillary column (30 m x 0.25 mm ID x 0.25 μm film thickness). The temperature was programmed from 40 to 220 °C at a rate of 4 °C/min. The injector temperature was set at 250 °C with a helium carrier gas flow rate of 20 mL/min. The mass spectrometer operated at 70 eV ionization potential with an ion source temperature of 180 °C, scanning a range of 50–350 Da. The mass spectra and retention times of compounds were compared with known standards in the GC-MS library.
Cytotoxicity and antiviral activity assays
MTT assay
The cytotoxic effects of the extracts were evaluated in Vero E6 cells for coronaviruses and MDCK cells for influenza virus using the MTT assay according to Mosmann (1983)18. Plant extracts were serially diluted two-fold in Dulbecco’s Modified Eagle Medium (DMEM). Briefly, Vero E6 cells were seeded into 96-well plates at a density of 3 × 10⁵ cells/mL (100 µL per well) and incubated at 37 °C in a 5% CO₂ atmosphere for 24 h. The cells were then treated with various concentrations of both aqueous and alcoholic extracts from the tested plants, in triplicate. After 24 h of incubation, the culture medium containing the extracts was discarded, and the cell monolayers were washed three times with sterile phosphate-buffered saline (PBS). MTT solution (20 µL of 5 mg/mL stock) was added to each well, followed by incubation at 37 °C for 4 h. After incubation, the medium was aspirated, and the resulting formazan crystals were dissolved in 200 µL of acidified isopropanol (0.04 M HCl in absolute isopropanol, equivalent to 0.073 mL HCl/50 mL isopropanol). The 24-hour incubation period was selected because cells require approximately one day to adhere, integrate, and form a confluent monolayer when cultured at appropriate densities. Previous studies have demonstrated that 24 h is optimal for ensuring full cell attachment and coverage of the culture surface before exposure to viruses or test agents19. Additionally, this timeframe aligns with standard protocols for cytotoxicity and antiviral assays, maintaining cell viability while minimizing metabolic stress that may interfere with experimental outcomes20.
The absorbance of the formazan solutions was measured at 540 nm with 620 nm as the reference wavelength, using a FLUOstar multi-well plate reader (USA). The percentage of cytotoxicity was calculated using:
The plot of % cytotoxicity versus sample concentration was used to calculate the concentration which exhibited 50% cytotoxicity (IC50).
Virus titration
Vero E6 and MDCK cells Vero E6 cells were seeded into 96-well plates in growth medium supplemented with 10% fetal bovine serum (FBS) and 2% antibiotic-antimycotic solution. The plates were incubated overnight at 37 °C in a 5% CO2 atmosphere to allow for cell attachment and growth. Once the monolayer reached confluence, a 1-log serial dilution of the virus stock was prepared. Fifty microliters (50 µL) of each dilution was added to the wells containing cells monolayers. After 1 h of incubation to facilitate viral adsorption, the virus inoculum was removed, and 160 µL of infection medium was added to each well. The plates were then incubated for 72 h at 37 °C in 5% CO2. The TCID₅₀/50 µL value was calculated using the Reed and Muench equation by determining the percentage of cytopathic effect (CPE) in each well. Many studies recommend a 1-hour incubation period as optimal for viral adsorption, including Finkel et al. (2021), who demonstrated that prolonged adsorption can interfere with host protein synthesis and viral replication kinetics21. Based on these considerations, we adopted a 1-hour incubation period to maintain experimental accuracy and reproducibility. The optimal incubation time for viral adsorption depends on multiple factors, including the type of virus, host cell type, viral concentration, and cell density. Additionally, the interaction between the virus and cell surface receptors plays a crucial role in determining the adsorption efficiency. For instance, coronaviruses such as SARS-CoV-2 and MERS-CoV bind to ACE2 and DPP4 receptors, respectively, requiring sufficient time for stable attachment before entry22. In contrast, influenza A virus (H1N1) utilizes sialic acid receptors and may exhibit faster adsorption kinetics23. Furthermore, the mechanism of viral entry—whether through direct membrane fusion or receptor-mediated endocytosis—affects the required incubation time. These factors collectively influence the selection of optimal adsorption periods in our study, ensuring efficient viral attachment while preventing premature internalization or degradation.
Antiviral activity assay
The antiviral assay was performed according to the method of Feoktistova et al.24, with modifications. A 96-well U-shaped plate was prepared one day after seeding Vero E6 cells (for coronaviruses) and MDCK cells (for the influenza virus), ensuring a 90% confluent monolayer. Infection medium (100 µL) was added to all wells in rows 2 to 12, while 180 µL was added to row 1. Twenty microliters (20 µL) of the antiviral test sample stock solution was added to wells in row 1 (A1 to H1), resulting in a 1:10 dilution. A 2-fold serial dilution was performed across columns 1 to 10 by transferring 100 µL from one column to the next. For cell controls, wells in rows 11 and 12 received 200 µL of infection medium. Additionally, 100 µL of TCID50 virus dilution was added to the appropriate wells, ensuring a final volume of 300 µL per well. Virus dilutions were added to the designated wells (rows 1 to 12, columns E to H), and the plate was incubated at 37 °C for 1 h to allow for virus adsorption. After incubation, confluent monolayers of Vero E6 and MDCK cells were treated with 100 µL of the prepared mixture and incubated for 72 h at 37 °C in 5% CO2. Following the incubation period, the cells were: Fixed with 10% formaldehyde for at least 2 h at room temperature. Washed and stained with 50 µL of 0.5% crystal violet stain (prepared by dissolving 0.5 g crystal violet in 80 mL distilled water and 20 mL methanol) for 10 min. Air-dried, followed by the addition of 200 µL of methanol to each well to dissolve the stain. The optical density (OD) of the stained cells was measured using an ELISA reader. The OD values were normalized, with 100% viability assigned to non-treated control cells. The percentage of cytotoxicity and viral replication reduction was calculated by comparing the average OD of treated wells (with or without virus) to the control wells. Mean values and the standard error of the mean (SEM) were calculated from at least three independent experiments. A schematic diagram of the experimental setup is provided in Fig. 1S for enhanced clarity.
Plaque titration assay
The plaque titration assay was performed to determine the plaque-forming units (PFUs), following the method described by Tobita (1975)25. Six-well tissue culture plates were seeded with Vero E6 and MDCK cells cells at a density of 1 × 105 cells per well and incubated until the cells reached 90–100% confluence (one-day post-seeding). The monolayers were washed twice with phosphate-buffered saline (PBS), and the virus stock was serially diluted 10-fold in Dulbecco’s Modified Eagle Medium (DMEM). A total of 100 µL of the undiluted virus and each serial dilution were mixed with 400 µL of DMEM and inoculated onto the Vero E6 cell monolayers, followed by incubation at 37 °C for 1 h to allow for viral adsorption. After incubation, the residual inoculum was aspirated, and each well was immediately overlaid with 2 mL of DMEM-based medium, containing 1% agarose type 1 (Lonza, Basel, Switzerland), 1% antibiotic-antimycotic solution, and 1 µg/mL Tosyl-L-phenylalanine chloromethyl ketone (TPCK)-treated trypsin. The plates were incubated at 37 °C in a 5% CO2 atmosphere for 3 days, and plaque formation was monitored microscopically daily. Once clear plaques were visible, 1 mL of 10% formaldehyde was added to each well and incubated at room temperature for 1 h to fix the cells and inactivate the virus. The formaldehyde was then discarded, the plates were rinsed with water, and the wells were air-dried. To visualize the plaques, 1 mL of staining solution (containing 1% crystal violet and 20% methanol in distilled water) was added to each well and incubated at room temperature for 5 min. The stain was then discarded, the wells were rinsed with water, and the plates were dried before plaque counting. The virus titer (PFU/mL) was calculated using the formula:
PFU/mL = Number of plaques×Reciprocal of virus dilution×Multiplication factor to adjust inoculum to 1 mL.
Plaque reduction assay for evaluating the antiviral activity of water extracts against MERS-CoV and SARS-CoV-2
The plaque reduction assay was performed following the method described by Hayden et al. (1980)26 in a six-well plate. Vero E6 cells were seeded at a density of 1 × 103 cells/mL and cultivated for 24 h at 37 °C to achieve confluence. The MERS-CoV or SARS-CoV-2 virus was diluted to a concentration yielding countable plaques at a 103 dilution and then mixed with the safe concentration of the tested extract, as determined by the cytotoxicity assay. The virus-extract mixture was incubated at 37 °C for 1 h before being added to the Vero E6 cell monolayers. The growth medium was removed from the cell culture plates, and 100 µL of the virus-extract mixture was inoculated into each well. After a 1-hour adsorption period, 3 mL of DMEM supplemented with 2% agarose was added onto the cell monolayer. The plates were left undisturbed to allow the overlay medium to solidify, and then incubated at 37 °C for 3 to 4 days until viral plaques formed.
Following incubation, 10% formalin was added to each well for 2 h to fix the cells and inactivate the virus. The wells were then stained with 0.1% crystal violet dissolved in distilled water to visualize the plaques. Control wells were included, where the untreated virus was incubated with Vero E6 cells. The number of plaques was counted, and the percentage reduction in plaque formation was calculated in comparison to the control wells using the following equation, as previously described by Tobita (1975)25:
To ensure complete removal of unabsorbed viral particles, the cell monolayers were washed multiple times with supplement-free medium following the adsorption period. This step is essential to eliminate any non-adherent virus particles, ensuring that only the adsorbed virus remains on the cell surface. Additionally, a control group, in which no washing was performed, can be included to validate the effectiveness of the washing process.
Mode of action for virus inhibition
The potential mode of action of the selected potent aqueous extract against viral inhibition was examined at three distinct stages of the virus propagation cycle, based on the following mechanisms: (i) inhibition of viral budding and replication. (ii) Blocking viral entry (viral adsorption) by preventing the attachment of the virus to host cell membranes and inhibiting membrane fusion. (iii) direct virucidal activity, where the extract inactivates viral viability. These mechanisms may contribute to the observed antiviral activity either independently or in synergistic combinations.
Viral adsorption assay
The viral adsorption assay was performed according to Zhang et al.. (2020)27. A six-well plate containing various concentrations of the plant extract (100 µL per well) was prepared, and the cell monolayers were incubated at 37 °C in a 5% CO2 atmosphere for 2 h. After incubation, the remaining unabsorbed extract was removed, and the cells were washed twice with 1 mL of supplement-free medium. Following this, 100 µL of virus suspension at a 10−4 plaque-forming unit (PFU) per well was added to the pretreated cells and incubated for 1 h to allow viral adsorption. After this period, 3 mL of DMEM supplemented with 2× infection medium and 2% agarose was added to each well, then the plates were left undisturbed to allow solidification. The plates were then incubated at 37 °C for up to 3 days until viral plaques formed. The cells were fixed using 10% formalin and stained with 0.1% crystal violet to visualize the plaques. The viral inhibition rate (IR, %) was calculated using the following formula:
Viral replication assay
The viral replication assay was conducted following the method described by Kuo et al.28. A six-well plate containing confluent cells was inoculated with 10⁻4 PFU/well of the virus and incubated at 37 °C in a 5% CO2 atmosphere for 1 h to allow for viral adsorption. After incubation, the unabsorbed virus was removed, and the cell monolayers were washed twice with 1 mL of supplement-free medium to eliminate residual viral particles. Following the washing steps, 100 µL of different concentrations of the aqueous plant extract was added to each well, and the plate was incubated for 1 h. Subsequently, 3 mL of DMEM supplemented with 2% agarose was added to each well and left to solidify. The plates were incubated at 37 °C for 2 days, after which the viral plaques were visualized as previously described.
Virucidal assay
The virucidal assay was performed according to the method described by Schuhmacher et al.29. , to evaluate the direct effect of the plant extract on viral infectivity. In this assay, 100 µL of the virus suspension at a 104 PFU/well dilution was mixed with 100 µL of different concentrations of the plant extract in a 96-well plate and incubated at 37 °C for 1 h. Meanwhile, a six-well plate containing confluent cells was prepared two days prior. The growth medium was removed, and 200 µL of each virus-extract mixture was added to the cell monolayers. Following the incubation period, 3 mL of overlay medium (comprising 50% 2× DMEM and 50% 2% agarose) was added to each well and left to solidify. The plates were then incubated at 37 °C until viral plaques formed. The plaques were fixed for 2 h using 10% formalin solution and then stained with 0.1% crystal violet for visualization. To ensure accurate assessment of the antiviral effects, the experiments included untreated cell controls, virus-only controls, and solvent controls. The data were normalized against the control values, and statistical significance was determined using one-way ANOVA. To differentiate viral plaques from other cell changes that may occur during the two-day incubation period, a control group of uninfected cells was included in each experiment. This control group allowed for direct comparison, ensuring that the observed plaques were specifically due to viral infection rather than spontaneous cell death, environmental factors, or morphological changes unrelated to viral replication.
Docking studies
Preparation of protein
The atomic coordinates of the MERS-CoV receptor-binding ___domain (RBD) (PDB ID: 4L72) and the SARS-CoV-2 spike RBD (PDB ID: 6LZG) were retrieved from the RCSB Protein Data Bank (PDB)30,31,32. Protein preparation was carried out by removing co-crystallized ligands, water molecules, and unnecessary atomic coordinates from the complex structure using Discovery Studio Visualizer, as illustrated in (Fig. 1)33. Subsequently, polar hydrogen atoms and Kollman charges were added to each protein, and the structures were saved in PDBQT format for docking analysis.
Ligand preparation
Phytochemical components from M. officinalis were identified through GC-MS analysis, and more than 200 compounds previously reported in the literature were retrieved from the PubChem database in SDF format34. The ligand SDF files were then converted to PDB format for molecular docking studies. AutoDock 4.2 (MGL Tools 1.5.6) was used to prepare the ligands, identifying aromatic carbon rings to form a torsion tree35. For further screening, the prepared ligand structures were saved in PDBQT format, as shown in (Fig. 2).
Active site analysis and grid parameter setup
To identify the active region of the protein where a substrate binds and undergoes a chemical reaction, molecular docking analysis was conducted. These predictions help in understanding molecular mechanisms and enable researchers to focus on specific active sites for therapeutic interventions. For SARS-CoV-2 RBD, the active site was determined in the RBD, specifically the region interacting with the ACE2 receptor, comprising the following key residues: Tyr505, Asn501, Thr500, Gly496, Tyr489, Phe486, Phe456, Tyr453, Tyr449, Gln498, Arg403, and Leu455. For MERS-CoV RBD, the active site was identified in the region interacting with the DPP4 receptor, including the residues: Tyr499, Asn501, Lys502, Leu506, Asp510, Arg511, Glu513, Asp537, Gly538, Asp539, Tyr540, Arg542, Trp553, and Val555. A grid box configuration file was created for molecular docking using AutoDock 4.2, with the following center coordinates and dimensions: MERS-CoV RBD: Center: X = -26.791, Y = -21.541, Z = -21.050; Dimensions: X = 80 Å, Y = 52 Å, Z = 48 Å SARS-CoV-2 Spike RBD: Center: X = -40.148, Y = 30.211, Z = 8.451; Dimensions: X = 40 Å, Y = 90 Å, Z = 50 Å.
Molecular docking
AutoDock Vina, a molecular docking software, was used to determine the optimal binding configurations between the selected proteins and chemical compounds36. Docking simulations were performed for 277 compounds against the RBD of MERS-CoV and SARS-CoV-2. The best compounds were selected based on their binding energy scores, and up to 10 clusters were analyzed for each ligand to determine the optimal binding interactions. PyMOL and LigPlot + were used for visualization and interaction analysis of RBD residues and ligand binding sites37,38.
Drug-likeness analysis
Lipinski’s Rule of Five states that an orally active drug should not violate more than two of the following criteria: molecular weight ≤ 500 Da, hydrogen bond acceptors (HBA) ≤ 10, hydrogen bond donors (HBD) ≤ 5, and lipophilicity (logP) ≤ 5. If more than two of these criteria are violated, bioavailability concerns may arise39. The SwissADME web server was used to evaluate the physicochemical properties of the selected compounds and to assess their drug-likeness based on these parameters40.
Toxicity risk
The ProTox web server was used to predict median oral lethal doses (LD50 values) and toxicity classes in rodents. Toxicity prediction is a crucial step in the drug and chemical development process, in order to prioritize compounds with the best potential for safe and effective use in humans while simultaneously lowering the chance of expensive late-stage failure. It is based on a two-dimensional (2D) similarity analysis prediction method for vehicles with an LD50 value. Canonical smileys of the M. officinals compounds were uploaded from the Pubchem server, which returned results indicating the LD50.
Statistical analysis
GraphPad Software (USA), Prism version 10, was utilized in this study for data analysis. All experiments were performed in triplicate. Data were expressed as mean ± standard deviation (SD). Statistical significance was analyzed using one-way ANOVA with post hoc Tukey’s test, and p-values < 0.05 were considered significant.
Results and discussion
Cytotoxicity and antiviral activity assay
A cytotoxicity assay was performed to determine the cytotoxic concentration (CC50) of the extracts, while an antiviral assay was conducted to establish the inhibitory concentration (IC50). The antiviral potential of the water extract of M. officinalis was evaluated in vitro, demonstrating significant inhibitory effects against the tested viruses. IC50 and CC50 values were utilized to calculate the selectivity index (SI), which serves as a crucial parameter for assessing the therapeutic potential of the extracts. The results, illustrated in (Fig. 3), indicate that the water extract exhibited the highest antiviral potency against the H1N1 virus, with an SI of 53.6, compared to MERS-CoV and SARS-CoV-2, which had SI values of 19 and 10, respectively. To further explore the antiviral efficacy of M. officinalis, additional extracts—including acetone, alkaloid, non-alkaloid, ethanol, and methanol—were assessed for their activity against SARS-CoV-2 and MERS-CoV (Fig. 4) and H1N1 (Fig. 5). Among all extracts, methanol extract demonstrated the most potent antiviral activity across all tested viruses. Against H1N1, the methanol extract exhibited a CC50 of 588.1 µg/mL and an IC50 of 2.549 µg/mL, resulting in an SI of 230 in MDCK cells. Furthermore, the methanol extract displayed strong antiviral efficacy against SARS-CoV-2 and MERS-CoV, with IC50 values of 10.83 µg/mL and 9.82 µg/mL, respectively, and a CC50 of 587 µg/mL in Vero E6 cells (Table 1). The calculated SI values for SARS-CoV-2 and MERS-CoV were 54.2 and 59.77, respectively, suggesting that the methanol extract of M. officinalis exhibits promising antiviral activity and potential for further pharmacological development. These results highlight the significant antiviral potential of M. officinalis, particularly its methanol extract, against a broad spectrum of respiratory viruses. In contrast, ethanol and alkaloid extracts showed moderate activity, while non-alkaloid and acetone extracts had relatively weaker effects.
The antiviral potential of M. officinalis against influenza (H1N1), SARS-CoV-2, and MERS-CoV observed in our study aligns with previous research findings on the antiviral effects of M. officinalis extracts, particularly its methanol and aqueous extracts. When comparing our results with studies on other viruses such as Enterovirus 71 (EV71)41 and herpes simplex virus (HSV-1)42, a common trend emerges: the strong antiviral activity of M. officinalis is primarily associated with phenolic compounds such as rosmarinic acid, caffeic acid, and p-coumaric acid. The study by Chen et al. (2017)43 demonstrated that a methanol extract of M. officinalis had potent antiviral effects against EV71, a major cause of hand-foot-and-mouth disease. The methanol extract inhibited plaque formation, cytopathic effects, and viral protein synthesis in EV71-infected cells. The identified active compound, rosmarinic acid (RA), significantly reduced viral attachment and entry while interfering with reactive oxygen species (ROS)-mediated viral replication. Our study showed that the methanol extract was the most potent against H1N1 (IC50 = 2.549 µg/mL, SI = 230) and had significant activity against SARS-CoV-2 (IC50 = 10.83 µg/mL, SI = 54.2) and MERS-CoV (IC50 = 9.82 µg/mL, SI = 59.77). This suggests that rosmarinic acid and other phenolics may be responsible for the observed antiviral effects in our study as well. Furthermore, our results corroborate the mechanism of action proposed by Chen et al.., suggesting that M. officinalis may exert its antiviral effects by disrupting viral attachment and replication, potentially through oxidative stress modulation.
Astani et al.42 investigated the antiviral activity of M. officinalis against acyclovir-resistant herpes simplex virus type 1 (HSV-1). The study found that the extract exhibited strong inhibition of HSV-1 attachment and penetration, with IC50 values as low as 0.13 µg/mL and selectivity indices (SI) reaching 2692. This is significantly higher than the SI values reported in our study for coronaviruses and influenza, suggesting that HSV-1 may be more susceptible to M. officinalis extracts than respiratory viruses. The authors attributed the antiviral effect primarily to rosmarinic acid, which was found to disrupt viral entry and penetration. This aligns with our study, where the methanol extract had the highest SI values, supporting the hypothesis that the antiviral efficacy is linked to phenolic constituents. Vilhelmova-Ilieva et al.43 evaluated Rosa damascena and Rosa alba essential oils for antiviral activity against HSV-1. The study found that while these oils did not directly affect viral replication, they inhibited viral adsorption and extracellular virions, similar to the proposed mechanism for M. officinalis. Moreover, the synergistic effects of plant-derived compounds with conventional antivirals were highlighted, suggesting that combinations of M. officinalis extracts with existing antiviral drugs may enhance efficacy. Our results also indicate that M. officinalis extracts, especially methanol extract, could be potent candidates for combinational antiviral therapy.
Inhibitory potential of M. officinalis extract against to MERS-CoV and SARS-CoV-2 viruses
The plaque infectivity titer of MERS-CoV-NRCE-HKU270/2013 and hCoV-19/Egypt/NRC-03/2020 SARS-CoV-2 was initially determined to be 20 × 102 PFU/mL. The water extract of M. officinalis was assessed for its in vitro antiviral activity against MERS-CoV and SARS-CoV-2 infections in Vero E6 cells using the plaque reduction assay at four different concentrations (500, 1000, 2000, and 4000 µg/mL). The results (Fig. 6a) demonstrated a dose-dependent antiviral effect, with inhibition of MERS-CoV not exceeding 70%, while SARS-CoV-2 inhibition exceeded 90% at the highest concentration. This suggests that SARS-CoV-2 is more susceptible to M. officinalis extract than MERS-CoV, potentially due to differences in viral entry mechanisms or variations in envelope protein interactions with host receptors. Additionally, the water extract exhibited antiviral activity against H1N1 in MDCK cells at concentrations of 375, 750, 1500, and 3000 µg/mL (Fig. 6b). The highest concentration (3000 µg/mL) achieved a maximum inhibition of 80%, suggesting moderate efficacy against influenza virus. These results align with previous studies highlighting the antiviral effects of M. officinalis against various RNA viruses, including influenza44, herpes simplex virus45, and coronaviruses46. He stronger inhibition observed against SARS-CoV-2 compared to MERS-CoV and H1N1 suggests that the bioactive compounds in M. officinalis, particularly rosmarinic acid, may interfere with viral entry and replication processes.
Mechanism of action
The antiviral activity of M. officinalis extract against MERS-CoV, SARS-CoV-2, and H1N1 was evaluated based on its effects on virucidal activity, replication inhibition, and adsorption inhibition, as illustrated in (Fig. 7a–c). The results suggest that the primary mode of action varies among the tested viruses, indicating potential differences in the susceptibility of viral structures and entry mechanisms to M. officinalis bioactive compounds. For MERS-CoV, M. officinalis demonstrated its antiviral effect primarily through a virucidal mechanism, achieving an inhibition of up to 97%by targeting the spike protein, leading to direct viral inactivation (Fig. 7a). Additionally, the extract showed moderate inhibition of viral replication, suggesting interference with viral genome replication. However, adsorption inhibition was minimal (~ 10–15%), indicating that M. officinalis was ineffective at preventing MERS-CoV from attaching to host cells. This results contrasts with previous studies showing that polyphenolic compounds in M. officinalis can block viral entry. Astani et al. (2012)45 and Mazzanti et al.. (2008)46 reported that M. officinalis interferes with viral adsorption by disrupting envelope glycoproteins, particularly in HSV-1 and other enveloped viruses. The lower adsorption inhibition observed in MERS-CoV may be due to differences in receptor usage, as MERS-CoV utilizes DPP4 (CD26) receptors, unlike SARS-CoV-2, which binds to ACE2.
For SARS-CoV-2, M. officinalis exerted a strong virucidal effect (90% inhibition), confirming its ability to inactivate viral particles before entry (Fig. 7b). Additionally, moderate inhibition was observed in both replication (60%) and adsorption (30–40%), suggesting that M. officinalis may partially interfere with spike protein binding to ACE2 receptors. These results align with results from Astani et al.. (2012)45, which demonstrated that M. officinalis inhibits HSV-1 attachment and entry by interacting with viral envelope proteins. The moderate adsorption inhibition against SARS-CoV-2 further supports the idea that M. officinalis polyphenols, such as rosmarinic acid, could bind to viral spike proteins and alter their interaction with host receptors. This mechanism is consistent with previous studies on plant-derived polyphenols, which have been shown to block ACE2-spike interactions in SARS-CoV-2. For H1N1, M. officinalis primarily exhibited a virucidal effect (85% inhibition), with lower inhibition observed in replication (40–60%) and adsorption (35%) (Fig. 7c). The observed effect on adsorption inhibition can be attributed to the ability of M. officinalis to interfere with hemagglutinin (HA) receptors, thereby preventing viral binding to host cells. This result is strongly supported by Mehrbod et al.. (2021)44, who reported that plant-derived polyphenols inhibit hemagglutinin-mediated viral attachment in influenza A virus (IAV). Moreover, the moderate inhibition of H1N1 replication aligns with previous findings indicating that M. officinalis suppresses viral genome replication by modulating host oxidative stress pathways.
GC-MS determination of alcoholic plant extract
The GC-MS analysis of M. officinalis extracts revealed a variety of bioactive compounds with potential antiviral, anti-inflammatory, and antioxidant properties (Table 2). Notably, the methanol extract contained the highest number of compounds (25), followed by ethanol (20) and acetone (9). Key identified compounds include docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA), both known for their anti-inflammatory and immunomodulatory effects, which may contribute to viral inhibition. Paromomycin, an aminoglycoside antibiotic detected in both methanol and ethanol extracts, suggests possible antimicrobial synergy. Additionally, phenolic acids such as 3,4-dihydroxycinnamic acid (Caffeic Acid) and (R)-2-(3,4-dihydroxytranscinnamoyloxy)-3-(3,4-dihydroxyphenyl) propionic acid (Trans-rosmarinic acid) align with the common trend that associates the strong antiviral activity of M. officinalis with phenolic compounds. These results reinforce M. officinalis as a rich source of bioactive compounds with potential therapeutic applications against viral infections.
Docking studies
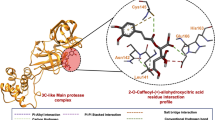
Molecular docking analysis was performed on over 270 phytoconstituents from M. officinalis to identify potential compounds capable of interacting with the receptor-binding domains (RBDs) of MERS-CoV and SARS-CoV-2 spike proteins (PDB ID: 4L72 and 6LZG). These compounds were evaluated for their binding interactions and docking scores, with those exhibiting binding energy values of −7.2 kcal/mol or lower considered promising candidates for antiviral activity (Table 3). Five key compounds demonstrated strong binding affinities with low binding energy values, indicating potential stability and effective interactions with the MERS-CoV RBD. Among them, 5-Methyl-5 H-naphtho[2,3-c]carbazole exhibited the lowest binding energy (−7.9 kcal/mol) and formed hydrophobic interactions with bond lengths < 5 Å, suggesting a strong and stable complex formation. Additionally, 7 H-Dibenzo[b, g]carbazole, 7-methyl also had a binding energy of −7.9 kcal/mol, forming one hydrogen bond with Ser557, reinforcing its potential antiviral activity. Furthermore, hesperidin displayed a strong affinity by establishing nine hydrogen bonds with key residues including Ser532, Thr392, Tyr541, Asp539, and Pro394 (Table 4), enhancing its binding stability. For SARS-CoV-2 RBD, Luteolin-7-glucoside-3′-glucuronide demonstrated the strongest binding, with a minimum binding energy of −9.2 kcal/mol. This compound formed nine hydrogen bonds with Arg403, Gln498, Asn501, Gly496, Gln409, Lys417, and Gly416, indicating a highly stable interaction. Additionally, hesperidin showed a binding energy of −8.6 kcal/mol, forming seven hydrogen bonds with Tyr449, Gln498, Gly496, Asn501, Gln409, and Arg403, reinforcing its potential inhibitory activity against SARS-CoV-2 RBD. Moreover, Melitric acid A exhibited a binding energy of −8.4 kcal/mol and formed six hydrogen bonds with Arg403, Asn501, Gly496, Tyr453, and Gln409 (Table 5), further supporting its therapeutic relevance. Beyond hydrogen bonding, these compounds also established electrostatic interactions, hydrophobic interactions, and van der Waals forces, which contribute significantly to binding stability and inhibition potential (Figs. 8 and 9). These interactions enhance molecular docking affinities and suggest that phytoconstituents from M. officinalis could effectively target MERS-CoV and SARS-CoV-2 RBDs, potentially interfering with viral entry and host receptor interactions. These docking results align with previous studies demonstrating that phenolic compounds and flavonoids can interfere with viral entry, fusion, and replication processes44,46.
Molecular docking studies of top four components with MERS-CoV RBD. (A) On the left is a 3d structure of the receptor ligands complex receptor binding ___domain protein (light teal), 5-Methyl-5 H-naphtho[2,3-c] carbazole (yellow), 7 H-Dibenzo[b, g]carbazole, 7-methyl(pink), Hesperidin(blue), and Luteolin-7-glucoside-3′-glucuronide(green) generated using PyMOL software (version 2.5.2). At the right, there is 2D Interaction of ligands with MERS-CoV RBD, (B) 5-Methyl-5 H-naphtho[2,3-c] carbazole, (C) 7 H-Dibenzo[b, g]carbazole, 7-methyl, (D) Hesperidin, (E) Luteolin-7-glucoside-3′-glucuronide using discovery studio software (v21.1.0.20298).
Molecular docking studies of top four components with SARS-CoV-2 spike RBD. On the left is a (A) 3d structure of the receptor ligands complex receptor binding ___domain protein (light orang), Luteolin-7-glucoside-3′-glucuronide (pink), Hesperidin (blue), Melitric acid A (yellow), and 7 H-Dibenzo[b, g]carbazole, 7-methyl (green) using PyMOL software (version 2.5.2). At the right, there is 2D Interaction of ligands with SARS-CoV-2 spik RBD, the black ball indicates the carbon atom, the red ball indicates the oxygen atom, the blue ball indicates the nitrogen atom, the red eyelash lines indicate the hydrophobic interaction between ligands and, and the green dashed string indicates the hydrogen bond formed between ligands and the amino acid residues of RBD. (B) Luteolin-7-glucoside-3′-glucuronide, (C) Hesperidin, (D) Melitric acid A, (E) 7 H- Dibenzo[b, g]carbazole, 7-methyl, were generated using LigPlot + software (v.2.2), developed by the European Bioinformatics Institute.
Drug-likeness analysis
The SwissADME web server was used to evaluate the physicochemical properties of the bioactive compounds identified in M. officinalis extracts, focusing on their drug-likeness and oral bioavailability. The analysis was based on Lipinski’s Rule of Five, a widely accepted criterion for assessing oral drug candidates.
Among the analyzed compounds, Nepetoidin A, Rosmarinic acid, Pregan-20-one, 2-hydroxy-5,6-epoxy-15-methyl, N,N’-Bis(Carbobenzyloxy)-lysine methyl(ester), and1,3,5Triazino[1,2-a]1,3benzimidazol-2-amine, 1,4-dihydro-4-(2-methylphenyl) conformed to Lipinski’s Rule of Five, indicating that these compounds are likely to be orally bioavailable (Table 6). However, some compounds violated one or more of these rules, which may impact their oral bioavailability. Notably, high molecular weight, excessive hydrogen bond donors/acceptors, or extreme lipophilicity could limit absorption and permeability. Nevertheless, previous studies suggest that Lipinski’s rule is not an absolute determinant of drug efficacy, as several clinically used drugs violate these rules but are still effective via alternative administration routes47. For compounds that failed Lipinski’s criteria, structural modifications or alternative formulations, such as prodrugs, nanoencapsulation, or lipid-based drug delivery systems, could enhance their bioavailability and therapeutic potential48,49.
Toxicity prediction
The LD50 value is an important step in the drug molecule screening process since it indicates the toxicity level of the compounds; the higher LD50 value, the safer drug compound. To calculate the LD50 dose, the toxicity of each chemical component was examined; all of the compounds fell into non-toxic classes. Toxicity predicted by PROTOXII is summarized in (Table 7). 5-Methyl-5 H-naphtho[2,3-c] carbazole, 7 H-Dibenzo[b, g]carbazole, 7-methyl, Sagerinic acid, Ursolic acid, Oleanolic acid, Melitric acid A, [5,9-Dimethyl-1-(3-phenyl-oxiran-2-yl)-deca-4,8-dienylidene]-(2-phenyl-aziridin-1-yl)-amine, Z,Z, Z-1,4,6,9-Nonadecatetraene, N,N’-Bis(Carbobenzyloxy)-lysine methyl(ester), and1,3,5Triazino[1,2-a]1,3benzimidazol-2-amine, 1,4-dihydro-4-(2-methylphenyl)- are classified in class 4 (300 mg/kg < LD50 ≤ 2000 mg/kg) which indicates that these compounds have acceptable toxicity. Luteolin-7-glucoside-3′-glucuronide, Betulinic acid, Nepetoidin A, Rosmarinic acid, and Pregan-20-one, 2-hydroxy-5,6-epoxy-15-methyl are predicted to be a low acute toxicity compound since it is classified in class 5 (2000 mg/kg < LD50 ≤ 5000 mg/kg). Hesperidin, 2-[4-methyl-6-(2,6,6-trimethylcyclohex-1-enyl)hexa-1,3,5-trienyl]cyclohex-1-en-1-carboxaldehyde, and Methyl arachidonate are classified in class 6 (LD50 > 5000 mg/kg) this result indicates that this compound is considered a nontoxic compound.
Limitations
This study utilized a single batch of extracts, which limits the reproducibility of findings. Additionally, while in silico docking provides theoretical insights, experimental validation is essential to confirm the proposed mechanisms. Future research should focus on isolating active compounds and evaluating their efficacy in vivo.
Future perspectives
Future research should focus on isolating active compounds and evaluating their efficacy in vivo. Additionally, optimizing extraction methods and exploring synergistic effects with existing antiviral treatments could enhance the therapeutic potential of M. officinalis extracts.
Conclusions
This study demonstrates that M. officinalis extracts exhibit significant antiviral activity against influenza A (H1N1), SARS-CoV-2, and MERS-CoV, highlighting their potential as natural antiviral agents. The bioinformatics-driven molecular docking analysis identified key phytoconstituents with strong binding affinities to the spike proteins of SARS-CoV-2 and MERS-CoV, suggesting their potential to interfere with viral entry and replication mechanisms. Our results indicate that these bioactive compounds, particularly phenolic acids, flavonoids, and polyunsaturated fatty acids, could serve as promising candidates for antiviral drug development. However, further validation is required through in vitro antiviral assays, mechanistic studies, and in vivo evaluations to determine their therapeutic efficacy, pharmacokinetics, and safety profiles. This research supports the continued exploration of plant-derived antiviral agents and underscores the need for clinical translation of M. officinalis-derived compounds as potential therapeutic interventions against emerging respiratory viruses. Future studies should focus on isolating and characterizing active compounds, optimizing their formulations, and evaluating their synergistic effects with existing antiviral treatments.
Data availability
Data generated and utilized for analyses of results presented in this manuscript are available from the corresponding author on reasonable requests.
Change history
16 June 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-06237-w
References
Terrier, O. et al. Influenza viruses and coronaviruses: knowns, unknowns, and common research challenges. PLoS Pathog. 17 (12), e1010106. (2021).
Xu, R. H. et al. Epidemiologic clues to SARS origin in China. Emerg. Infect. Dis. 10 (6), 1030–1037. https://doi.org/10.3201/eid1006.030852 (2004).
Wilder-Smith, A. The severe acute respiratory syndrome: impact on travel and tourism. Travel Med. Infect. Dis. 4 (2), 53–60 (2006).
WHO. Middle East respiratory syndrome. World Health Organization. https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html (2024).
Banik, G. R., Khandaker, G. & Rashid, H. Middle East respiratory syndrome coronavirus MERS-CoV: current knowledge gaps. Paediatr. Respir. Rev. 16 (3), 197–202 (2015).
Almuhrzi, H., Alriyami, H. & Scott, N. Tourism in the Arab World: an Industry Perspective 78 (Channel View, 2017).
Zhu, N. et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 382 (8), 727–733 (2020).
Gregorczyk, M. & Roskal-Wałek, J. Ocular symptoms in SARS-CoV-2 infection. Polski Merkuriusz Lekarski: Organ. Polskiego Towarzystwa Lekarskiego 50 (296), 86–93 (2022).
Struyf, T. et al. Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19. Cochrane Database Syst. Rev. (5). (2022).
Ryu, S. & Cowling, B. J. Human influenza epidemiology. Cold Spring Harbor Perspect. Med. 11 (12), a038356 (2021).
Timothy, M. et al. Influenza. Lancet 400 (10353), 693–706. https://doi.org/10.1016/S0140-6736(22)00982-5 (2022).
Del Prete, A. et al. Herbal products: benefits, limits, and applications in chronic liver disease. Evid. Based Complement. Altern. Med. 2012 (1), 837939 (2012).
Boneza, M. M. & Niemeyer, E. D. Cultivar affects the phenolic composition and antioxidant properties of commercially available lemon balm (Melissa officinalis L.) varieties. Ind. Crops Prod. 112, 783–789 (2018).
Bogdanovic, A. et al. Supercritical and high pressure subcritical fluid extraction from lemon balm (Melissa officinalis L., Lamiaceae). J. Supercrit. Fluids 107, 234–242 (2016).
Shakeri, A., Sahebkar, A. & Javadi, B. Melissa officinalis L. A review of its traditional uses, phytochemistry and pharmacology. J. Ethnopharmacol. 188, 204–228 (2016).
Said-Al Ahl, H. A., Sabra, A. S., Gendy, A. S. & Astatkie, T. Essential oil content and concentration of constituents of lemon balm (Melissa officinalis L.) at different harvest dates. J. Essent. Oil Bearing Plants 21 (5), 1410–1417 (2018).
Elkady, A. I. Crude alkaloid extract of rhazya stricta inhibits cell growth and sensitizes human lung cancer cells to cisplatin through induction of apoptosis. Genet. Mol. Biol. 36, 12–21 (2013).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65 (1–2), 55–63 (1983).
Wongchitrat, P. et al. Melatonin inhibits Zika virus replication in Vero epithelial cells and SK-N-SH human neuroblastoma cells. Melatonin Res. 5 (2), 171–185 (2022).
Chiamenti, L. et al. Cytotoxicity and antiviral activity evaluation of cymbopogon spp hydroethanolic extracts. Brazilian J. Pharm. Sci. 55, e18063. (2019).
Finkel, Y. et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 594 (7862), 240–245 (2021).
Li, Y. et al. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 Spike. Iscience 23 (6). (2020).
Guo, H. et al. Kinetic analysis of the influenza A virus HA/NA balance reveals contribution of NA to virus-receptor binding and NA-dependent rolling on receptor-containing surfaces. PLoS Pathog. 14 (8), e1007233. (2018).
Feoktistova, M., Geserick, P. & Leverkus, M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harb. Protoc. 2016 (4), pdb-prot087379. (2016).
Tobita, K. Permanent canine kidney (MDCK) cells for isolation and plaque assay of influenza B viruses. Med. Microbiol. Immunol. 162 (1), 23–27 (1975).
Hayden, D. What would a non-sexist city be like? Speculations on housing, urban design, and human work. Signs J. Women Cult. Soc. 5 (S3), S170–S187 (1980).
Zhang, L. et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science 368 (6489), 409–412 (2020).
Kuo, Y. C. et al. Samarangenin B from limonium sinense suppresses herpes simplex virus type 1 replication in Vero cells by regulation of viral macromolecular synthesis. Antimicrob. Agents Chemother. 46 (9), 2854–2864 (2002).
Schuhmacher, A., Reichling, J. & Schnitzler, P. J. P. V. Virucidal effect of peppermint oil on the enveloped viruses herpes simplex virus type 1 and type 2 in vitro. Phytomedicine 10 (6–7), 504–510 (2003).
Bittrich, S. et al. RCSB protein data bank: efficient searching and simultaneous access to one million computed structure models alongside the PDB structures enabled by architectural advances. J. Mol. Biol. 435 (14), 167994 (2023).
Wang, N. et al. Structure of MERS-CoV Spike receptor-binding ___domain complexed with human receptor DPP4. Cell Res. 23 (8), 986–993 (2013).
Wang, Q. et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181 (4), 894–904 (2020).
Biovia, D. S. & Dsme, R. San Diego: Dassault systèmes. Discovery Studio Visualizer. (Release 2021) (2015).
Kim, S. et al. PubChem substance and compound databases. Nucleic Acids Res. 44 (D1), D1202–D1213 (2016).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30 (16), 2785–2791 (2009).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31 (2), 455–461 (2010).
Janson, G., Zhang, C., Prado, M. G. & Paiardini, A. PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 33 (3), 444–446 (2017).
Laskowski, R. A. & Swindells, M. B. LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. (2011).
Lipinski, C. A., Lombardo, F., Dominy, B. W. & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 64, 4–17 (2012).
Daina, A., Michielin, O. & Zoete, V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7 (1), 42717 (2017).
Chen, S. G. et al. Anti-enterovirus 71 activities of Melissa officinalis extract and its biologically active constituent rosmarinic acid. Sci. Rep. 7 (1), 12264 (2017).
Astani, A., Heidary Navid, M. & Schnitzler, P. Attachment and penetration of acyclovir-resistant herpes simplex virus are inhibited by Melissa officinalis extract. Phytother. Res. 28 (10), 1547–1552 (2014).
Vilhelmova-Ilieva, N., Dobreva, A., Doynovska, R., Krastev, D. & Mileva, M. Antiviral activity of Rosa Damascena mill. And Rosa Alba L. Essential oils against the multiplication of herpes simplex virus type 1 strains sensitive and resistant to acyclovir. Biology 10 (8), 746 (2021).
Mehrbod, P. et al. Potential antiviral effects of some native Iranian medicinal plants extracts and fractions against influenza A virus. BMC Complement. Med. Ther. 21, 1–12 (2021).
Astani, A., Reichling, J. & Schnitzler, P. Melissa officinalis extract inhibits attachment of herpes simplex virus in vitro. Chemotherapy 58 (1), 70–77 (2012).
Mazzanti, G. et al. Inhibitory activity of Melissa officinalis L. extract on herpes simplex virus type 2 replication. Nat. Prod. Res. 22 (16), 1433–1440 (2008).
Benet, L. Z., Hosey, C. M., Ursu, O. & Oprea, T. I. BDDCS, the rule of 5 and drugability. Adv. Drug Deliv. Rev. 101, 89–98 (2016).
Aungst, B. J. Optimizing oral bioavailability in drug discovery: an overview of design and testing strategies and formulation options. J. Pharm. Sci. 106 (4), 921–929 (2017).
Denaro, M. et al. Antiviral activity of plants and their isolated bioactive compounds: an update. Phytother. Res. 34 (4), 742–768 (2020).
Acknowledgements
The author extends his gratitude towards the Deanship of Scientific Research (DSR) at King Abdul-Aziz University (KAU), Jeddah, Saudi Arabia has funded this project, under grant no. (RG-21-130-43). I would also like to thank Saleh M. Al-Maaqar and Eman Taibah for their invaluable assistance in the lab.
Funding
The author extends his gratitude towards the Deanship of Scientific Research (DSR) at King Abdul-Aziz University (KAU), Jeddah, Saudi Arabia has funded this project, under grant no. (RG-21-130-43).
Author information
Authors and Affiliations
Contributions
Conceptualization, T.A. and T.B. methodology, T.A. and T.B.software, A.A. and S.A.validation, A.A., Y.A. formal analysis, M.A., Y.M.investigation, T.A., T.B., O.K.,and Y.A.data curation, M.G., F.A.,and I.Avisualization, T.A. and T.B. writing—original draft preparation, E.M., F.S., R.B., and R.B.writing—review and editing, T.A. and T.B. and Y.Asupervision, T.A. and T.B. project administration, T.A.All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in Affiliation 4. Full information regarding the correction made can be found in the correction for this Article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alsahafi, T., Bouback, T., Albeshri, A. et al. Antiviral potential of Melissa officinalis extracts against influenza and emerging coronaviruses. Sci Rep 15, 12118 (2025). https://doi.org/10.1038/s41598-025-96417-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-96417-5