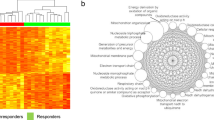
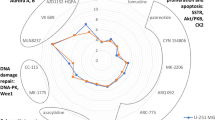
Abstract
Malignancies are reliant on glutamine as an energy source and a facilitator of aberrant DNA methylation. We demonstrate preclinical synergy of telaglenastat (CB-839), a selective glutaminase inhibitor, combined with azacytidine (AZA), followed by a single-arm, open-label, phase 1b/2 study in persons with advanced myelodysplastic syndrome (MDS). The dual primary endpoints evaluated clinical activity, safety and tolerability; secondary endpoints evaluated pharmacokinetics, pharmacodynamics, overall survival, event-free survival and duration of response. The dose-escalation study included six participants and the dose-expansion study included 24 participants. Therapy was well tolerated and led to an objective response rate of 70% with (marrow) complete remission in 53% of participants and a median overall survival of 11.6 months, with evidence of myeloid differentiation in responders determined by single-cell RNA sequencing. Glutamine transporter solute carrier family 38 member 1 in MDS stem cells was associated with clinical responses and predictive of worse prognosis in a large MDS cohort. These data demonstrate the safety and efficacy of CB-839 and AZA as a combined metabolic and epigenetic approach in MDS. ClinicalTrials.gov identifier: NCT03047993.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The scRNAseq, scBCRseq and scTCRseq data that support the findings of this study were deposited to the GEO under accession code GSE225352. MS data were deposited to Zenodo (https://doi.org/10.5281/zenodo.7641622)44. The full study protocol is publicly available and provided in the Supplementary Information. All other requests regarding data supporting the findings of this study or related to this paper should be addressed to [email protected]. Source data are provided with this paper.
Code availability
No custom algorithms were used in this study. Open-source software was used to analyze the data. Details of software versions are specified in Methods.
References
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 619–634 (2016).
Cantor, J. R. & Sabatini, D. M. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2, 881–898 (2012).
Schulze, A. & Harris, A. L. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature 491, 364–373 (2012).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Gross, M. I. et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol. Cancer Ther. 13, 890–901 (2014).
Goto, M. et al. Importance of glutamine metabolism in leukemia cells by energy production through TCA cycle and by redox homeostasis. Cancer Invest. 32, 241–247 (2014).
Gregory, M. A. et al. Targeting glutamine metabolism and redox state for leukemia therapy. Clin. Cancer Res. 25, 4079–4090 (2019).
Dranoff, G., Elion, G. B., Friedman, H. S., Campbell, G. L. & Bigner, D. D. Influence of glutamine on the growth of human glioma and medulloblastoma in culture. Cancer Res. 45, 4077–4081 (1985).
Jacque, N. et al. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 126, 1346–1356 (2015).
Matre, P. et al. Inhibiting glutaminase in acute myeloid leukemia: metabolic dependency of selected AML subtypes. Oncotarget 7, 79722–79735 (2016).
Thompson, R. M. et al. Glutaminase inhibitor CB-839 synergizes with carfilzomib in resistant multiple myeloma cells. Oncotarget 8, 35863–35876 (2017).
Zacharias, N. M. et al. Assessing metabolic intervention with a glutaminase inhibitor in real-time by hyperpolarized magnetic resonance in acute myeloid leukemia. Mol. Cancer Ther. 18, 1937–1946 (2019).
Baran, N. et al. Inhibition of mitochondrial complex I reverses NOTCH1-driven metabolic reprogramming in T-cell acute lymphoblastic leukemia. Nat. Commun. 13, 2801 (2022).
Harding, J. J. et al. A phase I dose-escalation and expansion study of telaglenastat in patients with advanced or metastatic solid tumors. Clin. Cancer Res. 27, 4994–5003 (2021).
Albitar, M. et al. Myelodysplastic syndrome is not merely ‘preleukemia’. Blood 100, 791–798 (2002).
Ma, X., Does, M., Raza, A. & Mayne, S. T. Myelodysplastic syndromes: incidence and survival in the United States. Cancer 109, 1536–1542 (2007).
Fenaux, P. et al. Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol. 10, 223–232 (2009).
Kantarjian, H. et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood 109, 52–57 (2007).
Prebet, T. et al. Outcome of high-risk myelodysplastic syndrome after azacitidine treatment failure. J. Clin. Oncol. 29, 3322–3327 (2011).
Jabbour, E. et al. Outcome of patients with myelodysplastic syndrome after failure of decitabine therapy. Cancer 116, 3830–3834 (2010).
Gerstung, M. et al. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat. Commun. 6, 5901 (2015).
Nguyen, T. T., Ramachandran, S., Hill, M. J. & Cerione, R. A. High-resolution structures of mitochondrial glutaminase C tetramers indicate conformational changes upon phosphate binding. J. Biol. Chem. 298, 101564 (2022).
Benito, J. et al. Hypoxia-activated prodrug TH-302 targets hypoxic bone marrow niches in preclinical leukemia models. Clin. Cancer Res. 22, 1687–1698 (2016).
Daemen, A. et al. Pan-cancer metabolic signature predicts co-dependency on glutaminase and de novo glutathione synthesis linked to a high-mesenchymal cell state. Cell Metab. 28, 383–399 (2018).
Wise, D. R. et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of α-ketoglutarate to citrate to support cell growth and viability. Proc. Natl Acad. Sci. USA 108, 19611–19616 (2011).
Figueroa, M. E. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
De Los Santos-Jimenez, J. et al. Metabolic adjustments following glutaminase inhibition by CB-839 in glioblastoma cell lines. Cancers (Basel) 15, 531 (2023).
Shastri, A. et al. Antisense STAT3 inhibitor decreases viability of myelodysplastic and leukemic stem cells. J. Clin. Invest. 128, 5479–5488 (2018).
Chen, J. et al. Myelodysplastic syndrome progression to acute myeloid leukemia at the stem cell level. Nat. Med. 25, 103–110 (2019).
Yoo, H. C. et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 31, 267–283 (2020).
Li, Y., Shao, H., Da, Z., Pan, J. & Fu, B. High expression of SLC38A1 predicts poor prognosis in patients with de novo acute myeloid leukemia. J. Cell. Physiol. 234, 20322–20328 (2019).
Rais, R. et al. Discovery of DRP-104, a tumor-targeted metabolic inhibitor prodrug. Sci. Adv. 8, eabq5925 (2022).
Yokoyama, Y., Estok, T. M. & Wild, R. Sirpiglenastat (DRP-104) induces antitumor efficacy through direct, broad antagonism of glutamine metabolism and stimulation of the innate and adaptive immune systems. Mol. Cancer Ther. 21, 1561–1572 (2022).
Gao, R. D. et al. Model studies towards prodrugs of the glutamine antagonist 6-diazo-5-oxo-l-norleucine (DON) containing a diazo precursor. Bioorg. Med. Chem. Lett. 50, 128321 (2021).
Nedelcovych, M. T. et al. N-(pivaloyloxy)alkoxy-carbonyl prodrugs of the glutamine antagonist 6-diazo-5-oxo-l-norleucine (DON) as a potential treatment for HIV associated neurocognitive disorders. J. Med. Chem. 60, 7186–7198 (2017).
Chou, T. C. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res. 70, 440–446 (2010).
Panina, S. B. et al. Novel mitochondria-targeting compounds selectively kill human leukemia cells. Leukemia 36, 2009–2021 (2022).
Bejar, R. et al. Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J. Clin. Oncol. 30, 3376–3382 (2012).
Greenberg, P. et al. International scoring system for evaluating prognosis in myelodysplastic syndromes. Blood 89, 2079–2088 (1997).
Cheson, B. D. et al. Clinical application and proposal for modification of the International Working Group (IWG) response criteria in myelodysplasia. Blood 108, 419–425 (2006).
Lu, X. et al. Metabolomics-based phenotypic screens for evaluation of drug synergy via direct-infusion mass spectrometry. iScience 25, 104221 (2022).
Stanford, S. M. et al. The low molecular weight protein tyrosine phosphatase promotes adipogenesis and subcutaneous adipocyte hypertrophy. J. Cell. Physiol. 236, 6630–6642 (2021).
Wishart, D. S. et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 50, D622–D631 (2022).
Tiziani, S. et al. Metabolomics data: glutaminase inhibition in combination with azacytidine in myelodysplastic syndromes. Zenodo https://doi.org/10.5281/zenodo.7641622 (2023).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 37, 38–44 (2019).
Buttner, M., Ostner, J., Muller, C. L., Theis, F. J. & Schubert, B. scCODA is a Bayesian model for compositional single-cell data analysis. Nat. Commun. 12, 6876 (2021).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Agarwal, B., Das, P., Naresh, K. N. & Borges, A. M. Angiogenic ability of metastatic squamous carcinoma in the cervical lymph nodes from unknown primary tumours. J. Clin. Pathol. 64, 765–770 (2011).
Acknowledgements
We thank the participants in this trial and their families, the study coordinators and the support staff at the clinical sites and the Calithera team members. We also thank Calithera Biosciences for providing CB-839 for both the preclinical studies and the clinical trial. This work was supported by the National Institutes of Health (NIH) National Cancer Institute (NCI) (R01 CA206210 to M.K., A.V. and S.T.), an NIH/NCI Cancer Center Support Grant (P30 CA016672 to M.K., N.P. and C.D.D.), the MD Anderson Cancer Center Leukemia SPORE (DRP 5 P50 CA100632-12) and Calithera Biosciences. M.R.G. is supported by a Leukemia and Lymphoma Society Scholar award. C.D.D. is supported by the Leukemia and Lymphoma Society Scholar in Clinical Research Award. D.V. is supported by a K12CA279871 NCI Paul Calabresi Award.
Author information
Authors and Affiliations
Contributions
Conceptualization and design, A.V., S.T., M.K., D.V. and C.D.D. Development of methodology, C.D.D., A.V., S.T., M.K., S.D. and D.V. Preclinical experiments and studies, T.C., V.M.K., D.V., A.V., A.S., G.P. and S.C. Acquisition of data (provided animals, acquired and managed participants, provided facilities, etc.), C.D.D., T.D.B., N.B., A.L., K.S., T.C., A.S., D.V., V.A.G., G.P., V.M.K., S. Konoplev, S.G.-M., K.P., S.A., G.L.H., S.C., M.C., S.R.S., J. Busquets, A.S.R., Q.D., M.R.G., S.G., B.A., G.S.C., S.S., M.S., V.T., B.W., U.S., G.D.T., J. Burger, G.B., E.J., N.P., T.K., S. Kornblau, N.G.D., K.N., N.J.S. and G.G.-M. Analysis and interpretation of data (for example, statistical analysis, biostatistics and computational analysis), C.D.D., T.D.B., N.B., A.L., K.S., T.C., X.S., A.S., D.V., S. Konoplev, S.G.-M., K.P., S.A., G.L.H., M.C., S.R.S., J. Busquets, A.S.R., Q.D., M.R.G. and B.A. Writing, review and/or revision of the paper, C.D.D., N.B., A.L., K.S., T.C., X.S., S. Konoplev, D.V., A.S., S.G.-M., K.P., S.A., M.C., S.R.S., J. Busquets, A.S.R., M.G., Q.D. and S.D. Administrative, technical or material support (that is, reporting or organizing data and constructing databases), T.D.B., N.B., A.L., K.S., T.C., X.S., A.S., D.V., V.A.G., V.M.K., K.P., S.A. and S.D. Study supervision, A.V., S.T. and M.K.
Corresponding authors
Ethics declarations
Competing interests
A.V. has received research funding from Prelude, Ryvu, BMS, GSK, Incyte, Medpacto, Curis and Eli Lilly, is a scientific advisor for Stelexis, Aurigene, Acceleron and Celgene, receives honoraria from Stelexis, Aurigene and Janssen and holds equity in Stelexis and Clinstreet. K.S. is a current employee of Gilead Sciences, Inc. All work contributing to the paper was performed while at the University of Texas MD Anderson Cancer Center. N.J.S. has received consulting fees from Pfizer Inc., GSK, NKARTA, Autolus, Adaptive Biotechnologies and Sanofi and research funding from Takeda Oncology, Astellas Pharma Inc., Xencor, GSK, NextCure, Ascentage and Novartis. N.J.S. receives honoraria from Adaptive Biotechnologies, Amgen, Takeda, Pfizer Inc., Astellas Pharma Inc. and Sanofi. C.D.D. is a consultant and/or on the advisory board of Abbvie, AstraZeneca, Astellas, BMS, Genentech, GenMab, GSK, Notable Labs, Rigel, Ryvu, Schrodinger and Servier. S. Konoplev is consultant for Cairo Diagnostics Laboratory. G.S.C. has stock options in privately held company Curis. M.R.G. reports research funding from Sanofi, Kite/Gilead, Abbvie and Allogene; consulting for Abbvie and Allogene; advisory board for Bristol Myers Squibb, Arvinas and Johnson & Johnson; honoraria from BMS, Daiichi Sankyo and DAVA Oncology; and stock ownership of KDAc Therapeutics. S.D. is employed at Calithera. M.K. received research support from Calithera. U.S. has received research funding from GlaxoSmithKline, Bayer Healthcare, Aileron Therapeutics and Novartis; has received compensation for consultancy services and for serving on scientific advisory boards from GlaxoSmithKline, Bayer Healthcare, Celgene, Aileron Therapeutics, Stelexis Therapeutics and Pieris Pharmaceuticals; and has equity ownership in and has served on the board of directors of Stelexis Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Cancer thanks James DeGregori, Alan Hutson and Jerome Tamburini for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 In vitro evaluation of GLS KO on cellular respiration.
(a) Validation of knockout efficiency in stable transfected OCI-AML3 cells, generated using shRNA control, puro-GAC plasmid, puro-KGA plasmid, puro-D3 (both isoforms) plasmid with puromycin incubation (3 days, 1 µg/ml). The expression of GAC and KGA in OCI-AML3 cells was determined by immunoblotting. Data were quantified in reference to loading control (n = 2 independent experiments, in 3 cell lines tested with similar level of knockdown). (b) Representative Seahorse Mito Stress Test assay graphs obtained from OCI-AML3 cells subjected to inducible knockout of KAG, GAC or GAC/KGA; Results were normalized to viable cell count. (n = 3-4 technical replicates, mean±SD, n = 3 independent experiments). (c) Basal, maximal oxygen consumption rate, OCR-linked ATP production, extracellular acidification rate (ECAR) and global metabolic phenotype (OCR/ECAR) were measured and calculated for OCI-AML3 cells subjected to doxycycline inducible knockout of KGA, GAC or GAC/KAG (mean ± SD, n = 3-4 technical replicates, n = 3 independent experiments, ordinary one-way ANOVA); (d) Representative Seahorse Mito Stress Test assay graphs obtained from four AML cell lines: MV4-11, MOLM13, OCI-AML3, and U937 treated with DMSO or telaglenastat (CB-839) at 1µM, for 12 hours; Results were normalized to viable cell count (n = 3–6 technical replicates, mean±SD, n = 3 independent experiments). (e) Basal, maximal oxygen consumption rate, OCR-linked ATP production, extracellular acidification rate (ECAR) and global metabolic phenotype (OCR/ECAR) were measured and calculated for AML cells as in (D) (n = 3–6 technical replicates, mean ± SD, n = 3 independent experiments, ordinary one-way ANOVA). (f) Seahorse Mito Stress Test assay graphs in OCI-AML3 cells treated with DMSO or BTES at 20µM. Results were normalized to viable cell count (n = 4 technical replicates, mean±SD, n = 3 independent experiments). (g) Mass spectrometry measurement of UMP and AMP in OCI-AML3 cells treated with DMSO or CB-839 1µM for 24 hr (n = 1 biological experiment with n = 4 technical replicates, mean±SD, unpaired t-test).
Extended Data Fig. 2 Evaluation of CB-839/AZA drug synergy in AML cell lines in vitro.
Representative graphs demonstrating synergistic effect of CB-839 and azacytidine (AZA) combination in AML cells. Cells were exposed to increasing doses of CB-839 and AZA alone or at constant drugs’ ratios for 5 days. Cell viability was determined by CellTiter-Glo luminescence assay. Combination index (CI) is the average ±SD of CIs at effective doses EC50, EC75, and EC90 calculated using CalcuSyn software. The CI < 1 = synergism; CI > 1 = antagonism (n = 4 technical replicates, mean ± SD).
Extended Data Fig. 3 Patient outcome analysis and biomarker studies.
(a) Overall survival of patients with complex cytogenetics. (b) Overall survival of patients with TP53 mutation. (c) Overall survival of patients harboring mutations in receptor tyrosine kinase gene (RTK). (d) Level of telaglenastat measured in plasma by LC-MS at indicated timepoints as compared between responders: CR and mCR, n = 7, mean ± SD, and patients with HI and NR n = 9, mean ± SD, ns-no significance. (e) Dynamic of changes in telaglenastat level measured in responders and non-responders at day15 of cycle1, day 1 of cycle 2 and at cycle 4 or later, (CR and mCR, n = 7, HI and NR n = 9, mean ± SD, ns-no significance).
Extended Data Fig. 4 scRNA UMAP presentation of selected biomarkers.
scRNAseq was conducted on bone marrow cells from CB839 treated patients. 5 clinical responders and 3 non responders were analyzed, and 11 distinct cell populations identified based on gene expression patterns. Representative UMAP figures show enrichment of CXCL8 (A), S100A9 (B) and S100A8 (C) expression shown between responders and non-responders.
Extended Data Fig. 5 Results for secondary transplantation of MDS PDX models.
(a-b) Percentage of mCD45−hCD45+ (a) and percentage of hCD45 + hCD33+ myeloid (b) cells in the bone marrow aspirate of NSG mice 5 weeks after the transplantation of human AML cells. Mice were randomly and blindly assigned to the vehicle and CB-839 treatment groups (mean ± SD, n = 5, unpaired t-test, ns-no significant). (c) Representative Flow cytometer on serial bone marrows from patients in complete remission (CR) before and after the combined treatment with AZA/Telaglenastat, showing a reduction in leukemic stem cells (LSCs) (CD34+/CD38-/Lin−ve with IL1RAP+).
Extended Data Fig. 6 Validation of SLC38A1 KO in vitro.
(a) Evidence of successful knockdown of SLC38A1 with siRNAs measured by RT-PCR in HEL cells. (mean ± SD, n = 3, unpaired t-test). (b) The intracellular levels of glutamine, glutamate and aspartate decrease in leukemic HEL cells following knockdown of the glutamate transporter SLC38A1 with siRNA. Knockdown of SLC38A1 led to reduced glutamine transport in leukemic HEL cells as measured by LCMS (mean ± SD, n = 4 replicates per condition, unpaired t-test). (c) Evidence of successful knockdown of SLC38A1 with lentiviral shRNA measured by RT-PCR in MOLM13 and THP1 cells (n = 3, mean ± SD, unpaired t-test). SLC38A1 KO GFP+ cells were FACS-sorted and used for growth and viability monitoring (D). (d) Cell growth and viability of MOLM13 and THP1 cells transfected with scrambled or SLC38A1 shRNA lentiviral vectors (n = 3, mean ± SD, unpaired t-test).
Supplementary information
Supplementary Information
Study protocol.
Supplementary Tables
Supplementary Tables 1 and 2.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Files
Source Data Figs. 1 and 7 and Extended Data Fig. 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
DiNardo, C.D., Verma, D., Baran, N. et al. Glutaminase inhibition in combination with azacytidine in myelodysplastic syndromes: a phase 1b/2 clinical trial and correlative analyses. Nat Cancer 5, 1515–1533 (2024). https://doi.org/10.1038/s43018-024-00811-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s43018-024-00811-3
This article is cited by
-
Targeting glutamine metabolism as a potential target for cancer treatment
Journal of Experimental & Clinical Cancer Research (2025)
-
Mitochondrial-cytochrome c oxidase II promotes glutaminolysis to sustain tumor cell survival upon glucose deprivation
Nature Communications (2025)
-
Taurine from tumour niche drives glycolysis to promote leukaemogenesis
Nature (2025)