Abstract
Understanding biological activities in cells or deep tissues requires high-speed three-dimensional (3D) imaging. Substantial progress has been made with the emergence of 3D random-access microscopy. However, current solutions for fast 3D random-access imaging remain complex and costly. Herein we propose a simple, cost-effective, and fast 3D random-access confocal microscopy with remote focusing system. Our system shows isotropic response times across the x, y, and z axes, with a 34-fold improvement in axial response time over traditional piezo stages. We demonstrate its volumetric imaging performance with fluorescent particles and live cells. Furthermore, we validate the 3D random-access imaging capability of this system by continuously monitoring the signals in three different planes, showing a refresh rate of 500 Hz on two different positions in 3D. The simplicity, versatility, and affordability of our system promise widespread applications in research and industry.
Similar content being viewed by others
Introduction
Optical microscopy has become an indispensable tool in biological research by providing essential insights into the structures and functions of biological systems at the cellular and molecular levels1,2,3,4,5,6,7,8,9,10,11. Among various optical microscopies, confocal laser scanning microscopy (CLSM) is one of the most popular imaging techniques in biological research due to its high spatial resolution and versatility in examining various types of biological samples12,13,14,15. The optical sectioning capability of CLSM enables it to study biological events in different depths in thick samples. However, CLSM builds images by scanning one point at a time through a focused laser beam, which must be precisely moved across the sample in a raster pattern. This point-by-point scanning process is inherently time-consuming, especially when high resolution is required. Moreover, to acquire a 3D image of the sample, multiple optical sections at different depths of the sample are needed, which further increases the image acquisition time. The low temporal resolution of CLSM in 3D imaging makes it challenging to study dynamical biological events in 3D.
In recent decades, random-access microscopy has been developed to monitor neuronal activities at multiple locations16,17,18,19,20,21,22,23,24,25,26,27. Random-access microscopy typically employs two-dimensional acousto-optic deflection (AOD) components to drive the laser focus in x and y directions16,17,22,23. For z direction, another pair of AODs, which form two orthogonal electric cylinder lenses filled with chirped acoustic waves, can be used to shift the axial laser focus positions23. While these AOD-based random-access microscopies possess high spatiotemporal resolution and large scan range, there are still limitations that hinder their widespread applications. For instance, the AOD unit is sensitive to wavelength variations, which restricts its multicolor imaging capabilities28. Additionally, the AOD induced optical aberrations could degrade the resolution and overall image quality. Furthermore, AOD-based random-access imaging systems are sophisticated and costly. The requirement for specialized optics expertize and a substantial budget to build such systems could restrict their use in smaller laboratories.
Remote focusing imaging, developed over a decade ago, is a technique that adjusts the focal plane of an optical system without requiring the mechanical movement of the sample29,30,31,32. This method employs auxiliary optical elements, such as remote objective with translational mirror, to dynamically change the imaging focal plane with minimal aberration29,30,31,33. Here we propose a simple, cost-effective, and fast 3D random-access confocal microscopy, termed 3D Dynamical Focusing Imaging (3D-DyFI). 3D-DyFI utilizes a dynamical focusing system to perform 3D remote focusing imaging. The 3D-DyFI eliminates the requirement of physically moving the sample or the objective lens, facilitating fast 3D imaging over various depths without the drawbacks associated with mechanical movement. Additionally, the de-scan configuration of 3D-DyFI allows the use of more economic highly sensitive detectors with small active detection areas, such as avalanche photodiode (APD). We validate the performance of 3D-DyFI with 3D imaging of fluorescent beads and live cells. The spatial resolution and response times were characterized, which shows isotropic scanning speeds across all three dimensions. We also scrutinized the random-access imaging capability of 3D-DyFI by monitoring the fluorescence intensity of beads in different axial planes. The refreshing rate of 3D-DyFI on two different detection positions in 3D can reach 500 Hz, about 25 times faster than the piezoelectric stage-based random-access imaging. Although in this work 3D-DyFI is configured with single-photon excitation, it is also compatible with multiphoton microscopy. Additionally, 3D-DyFI can perform multicolor imaging, offering an advantage over AOD-based random-access microscopy systems. Moreover, the affordability and simplicity of 3D-DyFI allow for its straightforward implementation in most laboratories.
Results
Configuration and 3D resolution characterization of 3D-DyFI
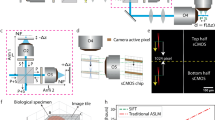
The schematic design of 3D-DyFI is shown in Fig. 1a. An economically efficient 635 nm laser (LWRL635, 100 mW, Laserwave), a 561 nm laser (LCX-561L-50-CSB-PPA, 50 mW, OXXIUS) and a 488 nm laser (LBX-488-60-CSB-PPA, 60 mW, OXXIUS) were used for excitation. Beam collimation and expansion were realized with a pair of achromatic doublet lenses (f = 50 mm, AC254-050-A-ML & f = 200 mm, AC254-200-AB-ML, Thorlabs). This beam expansion, quadrupling the spot size, optimally fills the back aperture of the objective lens (100x, 1.35NA, Nikon). A pinhole (Φ = 30 µm, P30K, Thorlabs) was placed in the focus of the first achromatic doublet lens. After being reflected by a dichroic mirror (for 635 nm excitation: ZT635dcrb; for 561 nm excitation: ZT561rdc; for 488 nm excitation: ZT488rdc, Chroma), the beam was then directed into the 3D dynamic focusing system (FL8220-3D-500, Sino-Galvo), which was used to manipulate the laser focus in 3D and de-scan fluorescence from the sample. The detailed information about remote focusing system can be found in Supplementary Fig. S1-S3 and Supplementary Note 1. The 3D dynamic focusing system incorporates two orthogonal one-dimensional galvanometers, two opposing plano-convex lenses, and a plano-concave lens mounted on a linear actuator, with its concave side facing the incident light. Changing the position of the plano-concave lens enables the movement of focused spot along the z-axis after objective lens. Lateral plane scanning was achieved through changing the angles of two one-dimensional galvanometers. A laser scanning tube lens (f = 200 mm, TTL200MP, Thorlabs) was precisely positioned in front of the objective lens. The emitted fluorescence from the sample was de-scanned by the 3D dynamic focusing system, passed through the dichroic mirror, and subsequently relayed and spatially filtered by a pair of dual achromatic doublet lenses (f = 50 mm, AC254-50-AB-ML, Thorlabs) and a pinhole (Φ = 25 µm, P25K, Thorlabs). A band pass filter (for 635 nm excitation: ET706/95 m, 25 mm diameter, Chroma; for 561 nm excitation: ET610/75 m, 25 mm diameter, Chroma; for 488 nm excitation: FF01-535/50-25, 25 mm diameter, Semrock) was used to reject the laser and a highly sensitive avalanche photodiode (APD, SPCM-AQRH-15, 65% quantum yield in 650 nm, Excelitas Technologies) was used for collecting the fluorescence signal. With the use of laser scanning tube lens, the field distortion and aberration can be reduced (Supplementary Note 2). To characterize the spatial resolution of 3D-DyFI, 50 nm green fluorescent microspheres were scanned in 3D with both piezoelectric stage and 3D-DyFI (Fig. 1b, c). The full widths at half maximum (FWHM) of intensity profiles of the fluorescent microspheres were subsequently measured (Supplementary Fig. S4), revealing resolutions of 301 nm × 330 nm × 0.96 µm for 3D-DyFI and 311 nm × 322 nm × 0.93 µm for piezoelectric stage scanning in x, y and z, respectively. The resolution of 3D-DyFI shows slightly worse than piezoelectric stage scanning result, which could be attributed to the spherical aberrations from one plano-concave lens and two convex lenses in dynamic focusing system. This can be improved by replacing these lenses with achromatic doublets lenses in the future. The resolution discrepancy in x and y could come from the alignment and the poor quality of the laser source34. These results indicate that under similar measurement conditions, the resolution of 3D-DyFI is comparable to that of piezoelectric stage based confocal microscopy (Supplementary Figs. S5–S6).
a Schematic of 3D-DyFI. OL: Objective Lens, M: Mirror, F: fluorescence filter, DM: Dichroic Mirror, PH: Pinhole, PCL: Plano-Concave Lens, APD: Avalanche Photodiode. The x-y scanning images (up panel) and x-z scanning images (low panel) of 50 nm green fluorescent microsphere with (b) piezoelectric stage scanning and (c) 3D-DyFI. The color indicates fluorescence intensity. Scale bar: 500 nm. The laser focus positions as a function of applied voltage in x (d), y (e), and z (f) axes. Measured data are represented by blue circles, while the fitting results are shown with green lines. n = 3.
Linearly focus position changing with the applied voltage on 3D dynamic focusing system
Next, we validated that the focus position is linearly changed with the voltage applied on 3D dynamic focusing system (Fig. 1d–f). The positions of the piezoelectric stage were used as references. Initially, the laser was focused on a 200 nm fluorescent bead. Subsequently, the sample was incrementally moved at predefined distances along the x, y, and z axes, displacing the bead from the laser’s focus. To recenter the laser on the bead, voltage adjustments were made to the 3D dynamic focusing system along the respective axes. By correlating the applied voltage with the displacement distances of the piezoelectric stage, we established a linear relationship between the change in laser focus distance and the voltage applied in the x, y, and z directions (Fig. 1d–f). The linear response of the 3D dynamic focusing system ensured the imaging accuracy of the 3D-DyFI and avoided further data processing.
Spatial resolution distribution in 3D range
The uniform spatial resolution over a large 3D range is important for 3D imaging microscopy13. Here we examined the spatial resolution of 3D-DyFI in various lateral and axial positions to provide a comprehensive assessment of its imaging performance. To investigate resolution consistency across an extensive scanning range (40 µm × 40 µm × 10 µm in x, y, and z), four locations (18, 0), (−18, 0), (0, 18), and (0, −18) were chosen for lateral resolution analysis. Although the dynamic range for the x and y axes can extend up to (−50 µm, 50 µm), as shown in Fig. 1e, f, aberrations were observed when the positions deviated substantially from the center. Consequently, we limited the operational range of x and y to (-20 µm, 20 µm) during imaging. Microspheres were initially positioned using the piezoelectric stage, followed by focused scanning at these locations with 3D-DyFI, as shown in Fig. 2a. The results indicate that even at the lateral boundary positions, the spatial resolution of 3D-DyFI remains nearly consistent with that at the center position of the dynamical focusing system. The relative uniform spatial resolution of 3D-DyFI across the entire x-y plane should be attributed to the distortion-correcting capability of the laser scanning tube lens (see Supplementary Note 2 for detailed information). A statistically spatial resolution evaluation was conducted with 39 measurements in 3D ( ± 18 µm, ±18 µm, ±4 µm), resulting average resolution of 319 ± 55 nm, 405 ± 56 nm, and 1.10 ± 0.11 µm in x, y and z respectively (Fig. 2b). Additionally, axial resolution consistency along the z-axis was further examined by measuring a microsphere at various depths (−4 µm, −2 µm, 0, 2 µm, and 4 µm), as shown in Fig. 2c, d. Notably, the best axial resolution was obtained at the z = 0 position, with a slight resolution decrease as the axial position deviated from this central position, which can be improved by using long working distance objective lenses35. These results demonstrate that 3D-DyFI maintains relatively uniform resolution across a decent 3D scanning volume (40 µm × 40 µm × 10 µm). The spatial resolution distribution in 3D range has also been characterized by red fluorescent microspheres with 635 nm excitation, showing similar result with slightly lower resolution due to the larger wavelength (Supplementary Figs. S6–S7). It is important to note that optimal resolution is achieved at the center of the scanning range of the 3D dynamic focusing system. Deviations from the center lead to resolution degradation, an effect that is more pronounced in the axial direction. Consequently, for optimal 3D imaging using 3D-DyFI, it is advisable to position the imaging target near the center of the scanning range of the 3D dynamic focusing system.
a Images of 50 nm fluorescent green microspheres located at the positions of (18, 0), (−18, 0), (0, 18), and (0, −18) in the x-y plane. The upper panel shows the PSF in x-y plane, while the lower panel shows the PSF in x-z plane. Scale bar = 500 nm. b The statistical histogram of spatial resolution distribution in x, y, and z directions, which shows mean values of 319 ± 55 nm, 405 ± 56 nm, and 1.10 ± 0.11 µm in x, y, and z. n = 39. c Fluorescent microsphere images at different z-depths (−4 μm, −2 μm, 0 μm, 2 μm, and 4 μm). Scale bar = 500 nm. d Plots of the intensity distribution of the PSFs in different axial positions in (c). The axial resolutions were determined by fitted FWHM. The color indicates fluorescence intensity in (a) and (c).
Temporal resolution assessment
The response time of the scanning unit is crucial for achieving high temporal resolution in microscopy, especially in 3D random-access imaging microscopy. To assess the temporal resolution of 3D-DyFI, we examined the response times of 3D dynamic focusing system and 3D piezo stage across the x, y, and z axes, as shown in Fig. 3. It is worthy to note that both the 3D dynamic focusing system and 3D piezo stage can perform addressable laser focus positioning and thereby realize 3D random-access imaging. Therefore, in this work we use the piezo stage-based confocal microscopy as the benchmark reference.
The response times of the piezo stage were determined by the position feedback signals from the controller. Specifically, a displacement of 1 µm was induced to the piezo stage with simultaneously recording the feedback signals from the piezo stage controller. The obtained response times of x, y, and z are 13.6 ms, 9.3 ms, and 22.3 ms, respectively (Fig. 3a). In contrast, the 3D dynamic focusing system lacks direct feedback signal output. To determine the response time, a fluorescent microsphere was positioned at the focal point, and the laser focus was then displaced by 1 µm using the 3D dynamic focusing system. The response time was quantified by analyzing the fluorescence intensity changing time. Here, the measured response times of 3D dynamic focusing system on x, y, and z are 0.74 ms, 0.82 ms, and 0.65 ms, respectively (Fig. 3b). These results show that the 3D dynamic focusing system has a substantial improvement in response speed, with approximately 34 times faster on the z-axis, 18 times faster on the X-axis, and 11 times faster on the y-axis compared to the piezoelectric stage. Moreover, the galvo system demonstrated near isotropic response times across all three dimensions, which is advantageous for random-access imaging. Additionally, the use of 3D dynamic focusing system not only facilitates faster imaging but also avoids potential disturbances caused by moving samples with piezo stage.
Live cell 3D imaging with 3D-DyFI
To demonstrate the 3D imaging performance of 3D-DyFI with biological samples, we performed volumetric cellular imaging in live cells. The cell membrane was stained with DiD (C1995S, Beyotime), a red membrane dye. The cells were first imaged with piezo stage-based confocal microscopy (Fig. 4a), followed by 3D imaging with 3D-DyFI in the same area (Fig. 4b). By examining the three-dimensional volume images and the section images in x-y, x-z, and y-z planes of the same cell, we found that the two systems have consistent 3D imaging performance (Fig. 4a, b).
a 3D live cell imaging with piezo stage-based confocal microscopy. b 3D live cell imaging with 3D-DyFI. The corresponding section images in the x-y, x-z, and y-z planes are shown in the right panel. The x-y plane section images were taken at the axial position of z = 0.4 μm; the x-z plane section images were taken at the position of y = 15 μm; and the y-z plane section images were taken at the position of x = 24 μm. Dashed lines at x = 24 μm (cyan) and y = 15 μm (yellow) are marked on the x-y plane image. The color indicates fluorescence intensity in (a) and (b). The intensity profiles along the y = 15 μm (c) and x = 24 μm lines (d), respectively. The relative root means squared errors (RRMSE) between the two intensity profiles were calculated, resulting in 0.031 and 0.082, respectively. Pixel size in x and y: 0.2 μm; Pixel size in z: 0.4 μm.
To quantitatively evaluate the 3D imaging performance of 3D-DyFI, we compared two intensity profiles on x and y axes (Fig. 4c, d). The intensity profiles show a high degree of consistency in the two systems. The relative root mean squared errors (RRMSE) is a commonly used method to quantify the similarity of two sets of data36,37. Here we measured the RRMSE of intensity profiles in Fig. 4c and Fig. 4d, resulting values of 0.031 and 0.082, which indicate minimal differences between the image data collected by the two systems. These results show that the spatial resolution and accuracy of 3D-DyFI are comparable with the piezo stage-based confocal microscopy.
To further demonstrate the 3D cellular imaging capability of 3D-DyFI. We imaged mitochondria (Fig. 5), cell microtubule (Supplementary Fig. S8), cell membrane (Supplementary Fig. S9), and lipid droplets in live cell (Supplementary Fig. S10). Additionally, the 3D-DyFI can also be used for 3D imaging in thick cells (Supplementary Fig. S11). These results demonstrate that the 3D-DyFI can be employed for 3D volume cellular imaging.
The mitochondria were labeled with mGold fluorescent protein. The left panel shows 3D live cell imaging. The right panel shows the section image in the x-y, x-z, and y-z planes. The sectioning positions are labeled in the upper right corner of the images. a, b are obtained from two different cells. The color indicates fluorescence intensity.
3D Random-access imaging
Random-access imaging allows for rapid and targeted imaging of multiple regions of interest, bypassing the need for time-consuming raster scanning16,17,18,19,20,21,22,23,24,25,26,27. This unique feature is particularly advantageous for dynamic studies where high temporal resolution is required. To demonstrate the 3D random-access imaging capability of the 3D-DyFI, we employed 3D-DyFI to monitor the fluorescence signals of three fluorescent beads in different locations with fast-switching rate (Fig. 6a–d).
a Three-dimensional image of three fluorescent beads embedded in agarose obtained with 3D-DyFI. The particles are labeled with A, B, and C, respectively. b The segmentary intensity trace of the three fluorescent beads in (a). The illumination time in each period is 2 ms. The lines in different colors indicate different particles as labeled in (a). The continuous intensity trace from three fluorescent beads with 2 ms illumination time (c) and 5 ms illumination (d) in each imaging period. The magenta lines show the z position of laser focus, obtained from the input of 3D dynamics focusing system. The fluorescence intensity is bin in 400 μs, considered the about 800 μs response time of 3D dynamic focusing system. e Three-dimensional image of three fluorescent beads embedded in agarose obtained with piezo stage-based confocal system. The particles are labeled with A, B, and C, respectively. f The segmentary intensity trace of the three fluorescent beads in (e). The illumination time in each period is 50 ms. The lines in different colors indicate different particles as labeled in (e). The continuous intensity trace from three fluorescent beads with 10 ms illumination time (g) and 50 ms illumination (h) in each imaging period. The magenta lines show the readout position from the piezoelectric stage controller. The yellow line indicates the input position of the piezoelectric stage controller. The color indicates fluorescence intensity in (a) and (e).
Initially, fluorescent beads were embedded in agarose and imaged in 3D (Fig. 6a, with beads labeled A, B, and C). Subsequently, the fluorescence intensities of these three beads were recorded by sequentially focusing laser on each bead at a fast-switching rate (Fig. 6b–d). An integration time of 400 μs was employed for each data point to average out the fluorescence noise, considering the ~800 μs response time of the 3D dynamic focusing system. Different illumination times were applied to each particle in successive imaging period to assess the random-access imaging performance of 3D-DyFI (Fig. 6b–d). With 2 ms illumination time, the intensity difference among the three particles can be resolved, although only a few data points were presented in each stage (Fig. 6b, c). Figure 6b shows the segmentary intensity trace of the three fluorescent beads. The lines with different colors indicate different particles, which also show different fluorescent intensities. Figure 6c, d shows the continuous intensity trace from three fluorescent beads, which reveals the intensity changes as the laser switches between particles. When the illumination time is increased to 5 ms, a more pronounced stair-stepping intensity profile is observed (Fig. 6d). Additionally, the 3D imaging of fluorescent microspheres were conducted with the pixel illumination time of 2 ms, 5 ms, and 10 ms, respectively (Supplementary Fig. S12). These results demonstrate that the refreshing rate of 3D-DyFI on two different detection positions in 3D can reach up to 500 Hz.
For comparison, we also performed 3D random-access imaging using piezo stage-based imaging method (Fig. 6e–h). Similar with 3D random-access imaging by 3D-DyFI, the 3D image of three fluorescent beads was presented, and their intensity traces were recorded with 10 ms and 50 ms illumination time. The readout position and the input position of the piezo stage can be obtained from the controller of piezo stage (magenta line and the yellow line in Fig. 6g, h). It is evident that the piezo stage’s position lags behind the input signal due to its slower response speed. Moreover, due to the ~20 ms response time of piezoelectric stage, the fluorescence signals of the three particles cannot be resolved with 10 ms illumination time (Fig. 6g). When using 50 ms illumination time, the three particles can be identified, showing a refreshing rate of 20 Hz (Fig. 6f, h). However, there are signal drops caused by the long transition time between particles (Fig. 6h). Compared with the piezo-stage based random-access imaging method, 3D-DyFI shows 25 times improvement on refreshing rate, enabling milliseconds dynamics observation over 3D.
Conclusion and discussion
In this study, we developed a 3D random-access confocal microscopy with remote-focusing system, termed 3D-DyFI. This system not only can perform fast 3D confocal raster scanning imaging, but also able to realize 3D random-access imaging. The 3D spatial resolution, imaging field range, and the response time of 3D-DyFI were fully characterized. 3D-DyFI was employed to capture the 3D images of live HeLa cells, demonstrating its compatibility with biological sample imaging. Additionally, the random-access imaging capability of 3D-DyFI was validated by monitoring the fluorescence intensity of nanoparticles in different locations in 3D, showing a refreshing rate up to 500 Hz.
Compared with 3D piezo stage-based confocal microscopy, 3D-DyFI utilizes remote-focusing system to perform fast and accurate laser focus manipulation in 3D. The 3D remote dynamical focusing system has several advantages over piezo stage-based method. The first one is that it avoids the physical movement of the sample or the objective lens, thereby eliminating the potential disturbances caused by mechanical movements. Secondly, while maintaining the spatial resolution comparable to piezo stage-based confocal microscopy, its 3D random-access imaging temporal resolution is improved by approximately 25-fold. Additionally, the cost of dynamical focusing system is also reduced compared with 3D piezo stage. The more affordable cost of dynamical focusing system increases the feasibility of 3D-DyFI in a broader range of research laboratories. In the remote objective based-remote focusing methods29,30,31,32,33, a translational mirror and a polarization beam splitter are utilized in the detection path. The use of polarization beam splitter results in 50 % fluorescence loss in these methods. As contrast, 3D-DyFI doesn’t suffer from this problem and has higher photon collection efficiency.
In previously reported 3D random-access microscopy, two pairs of AODs are employed to manipulate the laser focus manipulation in 3D17,23,24. While this kind of 3D random-access microscopy shows great imaging speed and large imaging field, the reliance of AOD limited its multicolor applications. In contrast, 3D-DyFI is not wavelength sensitive and is able to perform multicolor imaging. Additionally, in AOD-based 3D random-access microscopy, the fluorescence does not go through the de-scan process, which means that the system requires detectors with large detection area, such as photomultiplier tubes (PMTs). This limitation precludes the use of more economic single photon counting avalanche photodiode (APDs), of which detection area is usually less than 200 μm. Moreover, the high complexity and high cost of AOD-based 3D random-access microscopy necessitate specialized optics expertize and a substantial budget to construct it, precluding the widespread application of 3D random-access microscopy. In contrast, 3D-DyFI provides a more economic, simple, and versatile solution for 3D random-access imaging.
The spatial light modulator (SLM) and digital micromirror device (DMD) have also been used to generate 3D scanning patterns32,38,39. However, the number of degrees of freedom (DoF) in these systems inherently limits the number of accessible voxels, despite improvements with the recently reported three-dimensional multi-site random access photostimulation (3D-MAP) method38. In contrast, in 3D-DyFI, the laser focus position is determined by the input voltage to the 3D dynamic focusing system, which eliminates this limitation.
Another commonly used optical device for rapid axial scanning is the tunable acoustic gradient index of refraction (TAG) lens40,41,42,43. The TAG lens can oscillate the laser focus sinusoidally along the axial direction at frequencies reaching hundreds of kilohertz. However, the TAG lens is unable to maintain the laser focus at a specific focal plane for extended periods, and pulsed light is required for selective focal plane excitation. In 3D-DyFI, the focal position is controlled through analog voltage, allowing for more straightforward and precise manipulation of the excitation focus.
It should be noted that although in this work only single-photon 3D imaging was presented, 3D-DyFI is inherently compatible with multiphoton imaging. Moreover, by using lower magnification objective lens, the imaging field and imaging depth of 3D-DyFI can be further increased. Additionally, the remote-focusing configuration enables 3D-DyFI to perform multimode imaging, such as combined 3D fluorescence imaging with atom force microscopy, optical tweezers imaging or manipulation, fluorescence lifetime imaging, and electrophysiological measurement with patch clamp44,45,46,47,48,49,50,51. Furthermore, as the sample is stationary during imaging process as in 3D-DyFI, it has the potential to achieve isotropic resolution imaging via dual-view plane illumination method with switching illumination and detection between two perpendicular objectives52. The recently developed deep-learning methods can also be adopted to improve the axial resolution53,54,55,56. It can be expected that the simplicity, versatility, and affordability of 3D-DyFI promise its widespread applications in both the research community and industry.
Methods
Optical alignment and calibration
Prior to collimation, the position of the plano-convex lens within the galvanometer was set to z = 0. Subsequently, the laser scanning tube lens was aligned to be conjugate with the galvanometer system, ensuring that the emitted light remained parallel. The back focal plane of the objective lens was conjugated with the laser scanning tube lens. This stringent arrangement is crucial for achieving high quality imaging. The beam expansion ratio has been meticulously engineered so that the spot size before entering the objective lens fully fills the entrance pupil of the objective lens. To facilitate system evaluation and comparative performance studies, a piezoelectric stage (LFHS3, Piezoconcept) was integrated into the custom-built microscope setup, allowing for direct placement of the sample on the stage. This optical setup enables 3D confocal imaging with either a 3D dynamic focusing system or a piezoelectric stage.
The 3D dynamic focusing system need to be precisely calibrated to determine the actual laser focus position after the objective lens for the correspondent control analog voltage. Moreover, the deflection range of 3D dynamic focusing system also needs to be obtained. Fortunately, nano piezoelectric stages capable of nanometer movements provide a precise “ruler.” We utilize the piezoelectric stage as a calibration standard to align the scanning parameters of the 3D dynamic focusing system, including step length and scanning range. Detailed calibration steps include:
a) Place a 50/50 beam splitter between the 3D dynamic focusing system and the laser scanning tube lens to divert part of the fluorescence signal into the camera. The images at this ___location, yet to be de-scanned, indicate the movement of the light spot after the objective lens.
b) Capture images of fixed 200 nm red fluorescent microspheres (PSRF00200, Beijing Zhongkeleiming Technology) with camera and move the piezoelectric stage 1 µm in the x-y direction to determine pixel size of the camera produced image for subsequent displacement calculations.
c) Place a glass slide after the objective lens and record the laser reflection signal with camera. Adjust the x, y, and z axes of the 3D dynamic focusing system and measure the displacement of reflection spot on camera. Using the calibrated pixel size as a reference, the scanning step length and scanning range of the dynamic focusing system’s x-y axis can be estimated, establishing the relationship between the galvanometer’s deflection angle (or applied voltage) and the laser focus displacement after the objective lens.
d) Move the microspheres 1 µm away from the center position in x, y, and z directions with piezoelectric stage. Then use the 3D dynamic focusing galvanometer to compensate this motion to determine the calibration factors. Comparing the images scanned by the piezoelectric stage and the corrected scanning results of the 3D dynamic focusing galvanometer multiple times ensures optimal alignment across all dimensions.
Sample preparation
Fluorescent microspheres sample preparation
Red and green fluorescent microspheres with a diameter of 50 nm (FSFR001 and FSDG001, Bangs Laboratories, Inc.) and red fluorescent microspheres with a diameter of 200 nm (PSRF00200, Beijing Zhongkeleiming Technology) were used in this study. The microspheres were diluted in phosphate buffered saline (PBS, E607008, Sangon Biotech) at a ratio of 1:4000 for 200 nm microspheres and 1:12,000 for 50 nm microspheres. For resolution characterization, 2 mL of 50 nm fluorescent microsphere solution was added into a glass-bottom cell culture dish (801001, Nest). For 3D random-access imaging, 200 nm red fluorescent beads were dispersed in 0.5% agarose at a dilution ratio of 1:2000. Initially, the 0.5% agarose solution was prepared and, prior to solidification, the fluorescent beads were introduced. The mixture was then gently stirred to ensure uniform distribution and to prevent bubble formation. Afterward, this homogeneous mixture was promptly transferred to a glass-bottom cell culture dish, where it was allowed to cool and solidify, thereby embedding the fluorescent beads in a stable three-dimensional matrix.
Culture of HeLa cells
HeLa cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; PM150223-500, Pricella) supplemented with 10% fetal bovine serum (FBS; F0193, Sigma-Aldrich) and 1% penicillin-streptomycin (15140122, Gibco). The cells were maintained at 37 °C in an atmosphere containing 5% CO2 until they reached 60–80% confluency. For subsequent experiments, cells were plated on glass-bottom dishes (801001, Nest) to achieve ~70% coverage.
Label mitochondria and microtubules with fluorescent protein
Mitochondria and microtubules were labeled with fluorescent proteins using plasmid transfections performed with Lipofectamine 3000 (L3000008, Invitrogen) as per the manufacturer’s instructions. Between 14 and 18 h post-transfection, cells expressed either 2 × COX8-mGold-HaloTag or mBaojin-Tubulin, supplied by the WeKwikGene Team (0000260). The COX8 gene (synthesized from Tsingke Biotech, Beijing, China) were engineered to fuse with mGold, a yellow fluorescent protein, which was derived from pCMV-mGold-Actin-C-18 (MiaoLingBio, P50209).
Membrane staining with DiD
For live-cell membrane staining, HeLa cells were treated with DiD (C1995S, Beyotime) following the manufacturer’s guidelines. Cells were plated at a density of 1.5 × 105 cells per dish and allowed to adhere overnight. Subsequently, the culture medium was removed, and the cells were washed twice with DMEM. The staining solution was then applied, and the dishes were gently agitated to promote even dye distribution. Cells were incubated in the dark at 37 °C for 20 min. Post-incubation, the cells were washed three times with DMEM, followed by the addition of pre-warmed medium for imaging.
Stain L929 cells with Neil red
Glass-bottom culture dishes were pre-coated with fibronectin (10 μg/mL in PBS, 33016015, Gibco™) for 2 h at 37 °C. L929 cells were seeded into the coated dishes (Nest; 801001, China) at a density of 6000 cells per dish and incubated for 15 h at 37 °C in 5% CO2, allowing the cells to reach 40–50% confluence. Live L929 cells were then stained with Nile Red (1 μM) in culture medium for 2 hours in a 37 °C, 5% CO2 incubator. After incubation, the cells were washed twice with PBS and imaged under the microscope in DMEM.
To ensure cell viability, all live-cell imaging experiments were completed within 20 min, with samples being replaced as necessary if the time was exceeded.
Remote focusing system operation
The motors for driving PCL and the galvo mirrors are independently controlled via three analog voltage output channels of the data acquisition card. Custom-build software developed in LabVIEW is employed to set the analog voltages applied to the three axes of the remote focusing system. Specifically, the voltages applied to the motors for PCL and galvo mirrors determine the position of laser focus position after objective lens. In random-access imaging mode, the remote focusing system sequentially switches laser focus among several predefined positions. In confocal raster scanning imaging mode, the remote focusing system moves the laser focus in a raster scanning pattern. At the same time, the fluorescence intensity signal from avalanche photodiode (APD) detector is recorded on the digital input channel of the data acquisition card. Both the applied analog voltages to the remote focusing system and the fluorescence intensity signals are logged by the data acquisition card with precise timing tags.
Data acquisition and processing
Data acquisition and processing for 3D-DyFI were realized via custom-developed software, which integrated hardware control, data collection, and display functions. An FPGA data acquisition card (PCIe-7858, NI) was employed to precisely manipulate the laser position and record the fluorescent signal. It should be noted here that a more economic data acquisition card could also fulfill these functions. Two 20 dB attenuators (PE7000-20, Pasternack) were incorporated at the inputs of the x and y axes. Fluorescence intensity data and the corresponding 3D coordinates during imaging are recorded and stored for subsequent analysis. Imaris and ImageJ software were used for image processing.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data underlying the results presented in this paper are not publicly available at this time but may be obtained from the authors upon reasonable request.
References
Gao, Z., Li, Q., Fan, C. & Hou, S. Deciphering live-cell biomolecular dynamics with single-molecule fluorescence imaging. Sci. Bull. 69, 2095–9273 (2024).
Hou, S. G., Johnson, C. & Welsher, K. Real-time 3D single particle tracking: towards active feedback single molecule spectroscopy in live cells. Molecules 24, 2826 (2019).
Liu, Z. C. et al. Structural and functional imaging of brains. Sci. China Chem. 66, 324–366 (2023).
Zhang, X. F., Li, H. Y., Ma, Y., Zhong, D. P. & Hou, S. G. Study liquid-liquid phase separation with optical microscopy: A methodology review. Apl. Bioeng. 7, 021502 (2023).
Guo, Y. T. et al. Visualizing intracellular organelle and cytoskeletal interactions at nanoscale resolution on millisecond timescales. Cell 175, 1430–1442 (2018).
Ray, S., Widom, J. R. & Walter, N. G. Life under the microscope: single-molecule fluorescence highlights the RNA world. Chem. Rev. 118, 4120–4155 (2018).
Jing, Y. Y., Zhang, C. S., Yu, B., Lin, D. Y. & Qu, J. L. Super-resolution microscopy: shedding new light on in vivo imaging. Front. Chem. 9, 746900 (2021).
Filbrun, S. L. et al. Imaging dynamic processes in multiple dimensions and length scales. Annu. Rev. Phys. Chem. 73, 377–402 (2022).
Hou, M. D. et al. Molecular resolution mapping of erythrocyte cytoskeleton by ultrastructure expansion single-molecule localization microscopy. Small Methods 7, 2201243 (2023).
Wang, Z. J. et al. High-speed image reconstruction for optically sectioned, super-resolution structured illumination microscopy. Adv. Photonics 4, 026003 (2022).
Sha, H., Li, H. Y., Zhang, Y. B. & Hou, S. G. Deep learning-enhanced single-molecule spectrum imaging. Apl. Photonics 8, 096102 (2023).
Carlsson, K. et al. Three-dimensional microscopy using a confocal laser scanning microscope. Opt. Lett. 10, 53–55 (1985).
Jonkman, J., Brown, C. M., Wright, G. D., Anderson, K. I. & North, A. J. Tutorial: guidance for quantitative confocal microscopy. Nat. Protoc. 15, 1585–1611 (2020).
Stachelek, P., MacKenzie, L., Parker, D. & Pal, R. Circularly polarised luminescence laser scanning confocal microscopy to study live cell chiral molecular interactions. Nat. Commun. 13, 553 (2022).
Ravel, G. et al. Inferring characteristics of bacterial swimming in biofilm matrix from time-lapse confocal laser scanning microscopy. Elife 11, e76513 (2022).
Bansal, V., Patel, S. & Saggau, P. High-speed addressable confocal microscopy for functional imaging of cellular activity. J. Biomed. Opt. 11, 34003 (2006).
Duemani Reddy, G., Kelleher, K., Fink, R. & Saggau, P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat. Neurosci. 11, 713–720 (2008).
Shi, R. et al. Random-access wide-field mesoscopy for centimetre-scale imaging of biodynamics with subcellular resolution. Nat. Photonics 18, 721–730 (2024).
Salomé, R. et al. Ultrafast random-access scanning in two-photon microscopy using acousto-optic deflectors. J. Neurosci. Methods 154, 161–174 (2006).
Ashraf, M. et al. Random access parallel microscopy. Elife 10, e56426 (2021).
Iyer, V., Hoogland, T. M. & Saggau, P. Fast functional imaging of single neurons using random-access multiphoton (RAMP) microscopy. J. Neurophysiol. 95, 535–545 (2006).
Bullen, A., Patel, S. S. & Saggau, P. High-speed, random-access fluorescence microscopy .1. High-resolution optical recording with voltage-sensitive dyes and ion indicators. Biophys. J. 73, 477–491 (1997).
Katona, G. et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat. Methods 9, 201–208 (2012).
Nadella, K. M. N. S. et al. Random-access scanning microscopy for 3D imaging in awake behaving animals. Nat. Methods 13, 1001–1004 (2016).
Geng, Q., Gu, C., Cheng, J. & Chen, S.-C. Digital micromirror device-based two-photon microscopy for three-dimensional and random-access imaging. Optica 4, 674–677 (2017).
Janiak, F. K. et al. Non-telecentric two-photon microscopy for 3D random access mesoscale imaging. Nat. Commun. 13, 544 (2022).
Rózsa, B. et al. Random access three-dimensional two-photon microscopy. Appl. Opt. 46, 1860–1865 (2007).
Römer, G. R. B. E. & Bechtold, P. Electro-optic and acousto-optic laser beam scanners. Phys. Procedia 56, 29–39 (2014).
Botcherby, E. J., Juškaitis, R., Booth, M. J. & Wilson, T. An optical technique for remote focusing in microscopy. Opt. Commun. 281, 880–887 (2008).
Botcherby, E. J., Juskaitis, R., Booth, M. J. & Wilson, T. Aberration-free optical refocusing in high numerical aperture microscopy. Opt. Lett. 32, 2007–2009 (2007).
Botcherby, E. J., Booth, M. J., Juškaitis, R. & Wilson, T. Real-time extended depth of field microscopy. Opt. Express 16, 21843–21848 (2008).
Anselmi, F., Ventalon, C., Bègue, A., Ogden, D. & Emiliani, V. Three-dimensional imaging and photostimulation by remote-focusing and holographic light patterning. Proc. Natl Acad. Sci. 108, 19504–19509 (2011).
Hugonnet, H., Lee, M. & Park, Y. Optimizing illumination in three-dimensional deconvolution microscopy for accurate refractive index tomography. Opt. Express 29, 6293–6301 (2021).
Chang, M., Zhang, P., Sun, J. & Zhang, X. in Advances in Optoelectronics and Micro/nano-optics 1–3 (IEEE, 2010).
Qi, Y. et al. Remote-focusing microscopy with long working distance objective lenses. Appl. Opt. 53, 3473–3478 (2014).
Shaji, A. & Sheeja, M. in 2023 IEEE 15th International Conference on Computational Intelligence and Communication Networks (CICN) 741–745 (IEEE, 2023).
Dogan, H., Dogan, R. O., Ay, I. & Sezen, S. F. DL-EDOF: Novel Multi-Focus Image Data Set and Deep Learning-Based Approach for More Accurate and Specimen-Free Extended Depth of Focus. Journal of Imaging Informatics in Medicine, 1–23 (2024).
Xue, Y., Waller, L., Adesnik, H. & Pégard, N. Three-dimensional multi-site random access photostimulation (3D-MAP). Elife 11, e73266 (2022).
Lutz, C. et al. Holographic photolysis of caged neurotransmitters. Nat. Methods 5, 821–827 (2008).
Mermillod-Blondin, A., McLeod, E. & Arnold, C. B. High-speed varifocal imaging with a tunable acoustic gradient index of refraction lens. Opt. Lett. 33, 2146–2148 (2008).
Deguchi, T. et al. Volumetric Lissajous confocal microscopy with tunable spatiotemporal resolution. Biomed. Opt. Express 11, 6293–6310 (2020).
Kong, L. et al. Continuous volumetric imaging via an optical phase-locked ultrasound lens. Nat. Methods 12, 759–762 (2015).
Duocastella, M., Vicidomini, G. & Diaspro, A. Simultaneous multiplane confocal microscopy using acoustic tunable lenses. Opt. Express 22, 19293–19301 (2014).
Datta, R., Heaster, T. M., Sharick, J. T., Gillette, A. A. & Skala, M. C. Fluorescence lifetime imaging microscopy: fundamentals and advances in instrumentation, analysis, and applications. J. Biomed. Opt. 25, 1–43 (2020).
Levchenko, S. M., Pliss, A., Peng, X., Prasad, P. N. & Qu, J. Fluorescence lifetime imaging for studying DNA compaction and gene activities. Light. Sci. Appl. 10, 224 (2021).
Dufrêne, Y. F. et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol. 12, 295–307 (2017).
Gómez-Varela, A. I. et al. Simultaneous co-localized super-resolution fluorescence microscopy and atomic force microscopy: combined SIM and AFM platform for the life sciences. Sci. Rep. 10, 1122 (2020).
Shepherd, J. W. et al. Correlating fluorescence microscopy, optical and magnetic tweezers to study single chiral biopolymers such as DNA. Nat. Commun. 15, 2748 (2024).
Sirinakis, G., Ren, Y., Gao, Y., Xi, Z. & Zhang, Y. Combined versatile high-resolution optical tweezers and single-molecule fluorescence microscopy. Rev. Sci. Instrum. 83, 093708 (2012).
Yadav, R. & Lu, H. P. Revealing dynamically-organized receptor ion channel clusters in live cells by a correlated electric recording and super-resolution single-molecule imaging approach. Phys. Chem. Chem. Phys. 20, 8088–8098 (2018).
Sasmal, D. K. & Lu, H. P. Single-molecule patch-clamp FRET microscopy studies of NMDA receptor ion channel dynamics in living cells: revealing the multiple conformational states associated with a channel at its electrical off state. J. Am. Chem. Soc. 136, 12998–13005 (2014).
Wu, Y. et al. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. 31, 1032–1038 (2013).
Ning, K. et al. Deep self-learning enables fast, high-fidelity isotropic resolution restoration for volumetric fluorescence microscopy. Light.: Sci. Appl. 12, 204 (2023).
Zhao, F. et al. Deep-learning super-resolution light-sheet add-on microscopy (Deep-SLAM) for easy isotropic volumetric imaging of large biological specimens. Biomed. Opt. Express 11, 7273–7285 (2020).
Park, H. et al. Deep learning enables reference-free isotropic super-resolution for volumetric fluorescence microscopy. Nat. Commun. 13, 3297 (2022).
Fang, C. et al. Minutes-timescale 3D isotropic imaging of entire organs at subcellular resolution by content-aware compressed-sensing light-sheet microscopy. Nat. Commun. 12, 107 (2021).
Acknowledgements
We would like to acknowledge support from the Shenzhen Medical Research Fund (B2301003), the National Natural Science Foundation of China (62065012, 22204106), the Evident & Shenzhen Bay Laboratory Joint Optical Microscopic Imaging Technology Development Program (S234602004-4), the Basic and Applied Basic Research Foundation of Guangdong Province (2021A1515110710), and the Guangdong Provincial Pearl River Talents Program (2021QN02Z631). We thank Feng Zhang at Sino-Galvo Technology for technical support.
Author information
Authors and Affiliations
Contributions
Shangguo Hou conceived the idea and proposed this project; Shangguo Hou, Suhui Deng, and Dongping Zhong supervised this project, including the framework design and improvement. Haoyang Li, Quan Lu, and Wenbo Zhang performed the experiments. Haoyang Li, Quan Lu, Zhong Wang, Yandong Sun, and Yue Hu performed the data analysis. Haoyang Li, Shangguo Hou, Quan Lu, and Zhong Wang wrote the draft of the manuscript. All authors proofread and revised the manuscript. Yu Wu and Lehui Xiao prepared the cell samples.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Engineering thanks Moosung Lee and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Anastasiia Vasylchenkova. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, H., Lu, Q., Wang, Z. et al. Three-dimensional random-access confocal microscopy with 3D remote focusing system. Commun Eng 3, 166 (2024). https://doi.org/10.1038/s44172-024-00320-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44172-024-00320-2