Abstract
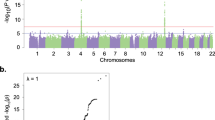
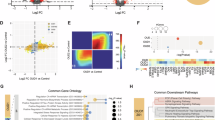
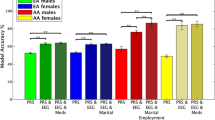
Alcohol use disorder (AUD) is highly heritable and burdensome worldwide. Genome-wide association studies can provide new evidence regarding the etiology of AUD. We report a multi-ancestry genome-wide association study focusing on a narrow AUD phenotype, using novel statistical tools in a total sample of 1,041,450 individuals (102,079 cases; European, 75,583; African, 20,689 (mostly African American); Hispanic American, 3,449; East Asian, 2,254; South Asian, 104; descent). Cross-ancestry functional analyses were performed with European and African samples. Thirty-seven genome-wide significant loci (105 variants) were identified, of which seven were novel for AUD and six for other alcohol phenotypes. Loci were mapped to genes, which show altered expression in brain regions relevant for AUD (striatum, hypothalamus and prefrontal cortex) and encode potential drug targets (GABAergic, dopaminergic and serotonergic neurons). African-specific analysis yielded a unique pattern of immune-related gene sets. Polygenic overlap and positive genetic correlations showed extensive shared genetic architecture between AUD and both mental and general medical phenotypes, suggesting that they are not only complications of alcohol use but also share genetic liability with AUD. Leveraging a cross-ancestry approach allowed identification of novel genetic loci for AUD and underscores the value of multi-ancestry genetic studies. These findings advance our understanding of AUD risk and clinically relevant comorbidities.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
79,00 € per year
only 6,58 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The summary statistics used for the current study were obtained from third party, and are thus not fully available to all authors. Two of them are publicly available (FINNGEN, https://www.finngen.fi/en/access_results, R6 public release; Psychiatric Genomics Consortium (PGC), https://pgc.unc.edu/for-researchers/download-results/). We accessed UK Biobank individual-level genotype to extend publicly available summary statistics for alcohol-related traits GWAS that are otherwise publicly available. Likewise, MVP data can be accessed through the dbGaP website, study phs001672.v9.p1. The summary statistics and data underlying the figures are thus only available to people with registered access to MVP data from the corresponding author or A.S. ([email protected]) upon request.
Code availability
The code for genetic correlation (https://github.com/brielin/Popcorn) and MiXeR (https://github.com/precimed/mixer) is publicly available.
References
Grant, B. F. et al. Epidemiology of DSM-5 alcohol use disorder: results from the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry 72, 757–766 (2015).
Milanzi, E. B. & Ndasauka, Y. in Addiction in South and East Africa: Interdisciplinary Approaches (eds Ndasauka, Y. & Kayange, G. M.) 215–228 (Springer International Publishing, 2019).
Carvalho, A. F., Heilig, M., Perez, A., Probst, C. & Rehm, J. Alcohol use disorders. Lancet 394, 781–792 (2019).
Rehm, J. & Shield, K. D. Global burden of alcohol use disorders and alcohol liver disease. Biomedicines 7, 99 (2019).
Verhulst, B., Neale, M. C. & Kendler, K. S. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol. Med. 45, 1061–1072 (2015).
Kranzler, H. R. et al. Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat. Commun. 10, 1499 (2019).
Walters, R. K. et al. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci. 21, 1656–1669 (2018).
Sanchez-Roige, S. et al. Genome-wide association study meta-analysis of the Alcohol Use Disorder Identification Test (AUDIT) in two population-based cohorts. Am. J. Psychiatry 176, 107–118 (2019).
Zhou, H. et al. Genome-wide meta-analysis of problematic alcohol use in 435,563 individuals yields insights into biology and relationships with other traits. Nat. Neurosci. 23, 809–818 (2020).
Zhou, H. et al. Multi-ancestry study of the genetics of problematic alcohol use in over 1 million individuals. Nat. Med. 21, 1–9 (2023).
Mallard, T. T. et al. Item-level genome-wide association study of the Alcohol Use Disorders Identification Test in three population-based cohorts. Am. J. Psychiatry 179, 58–70 (2022).
Deak, J. D. et al. Genome-wide investigation of maximum habitual alcohol intake in US veterans in relation to alcohol consumption traits and alcohol use disorder. JAMA Netw. Open 5, e2238880 (2022).
Heilig, M. et al. Addiction as a brain disease revised: why it still matters, and the need for consilience. Neuropsychopharmacology 46, 1715–1723 (2021).
Peterson, R. E. et al. Genome-wide association studies in ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 179, 589–603 (2019).
Edenberg, H. J. & McClintick, J. N. Alcohol dehydrogenases, aldehyde dehydrogenases, and alcohol use disorders: a critical review. Alcohol Clin. Exp. Res. 42, 2281–2297 (2018).
Lago, S. G. & Bahn, S. The druggable schizophrenia genome: from repurposing opportunities to unexplored drug targets. npj Genom. Med. 7, 1–13 (2022).
Hindley, G. et al. Charting the landscape of genetic overlap between mental disorders and related traits beyond genetic correlation. Am. J. Psychiatry. 179, 833–843 (2022).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Montemitro, C., Angebrandt, A., Wang, T. Y., Pettorruso, M. & Abulseoud, O. A. Mechanistic insights into the efficacy of memantine in treating certain drug addictions. Prog. Neuropsychopharmacol. Biol. Psychiatry 111, 110409 (2021).
Icick, R. et al. Genetic overlap between mood instability and alcohol-related phenotypes suggests shared biological underpinnings. Neuropsychopharmacology 47, 1883–1891 (2022).
Sun, B. B. et al. Genetic associations of protein-coding variants in human disease. Nature 603, 95–102 (2022).
Gelernter, J. et al. Genome-wide association study of maximum habitual alcohol intake in >140,000 U.S. European and African American veterans yields novel risk loci. Biol. Psychiatry 86, 365–376 (2019).
Müller-Oehring, E. M. et al. Midbrain-driven emotion and reward processing in alcoholism. Neuropsychopharmacology 38, 1844–1853 (2013).
Bogdan, R., Hatoum, A. S., Johnson, E. C. & Agrawal, A. The Genetically Informed Neurobiology of Addiction (GINA) model. Nat. Rev. Neurosci. 24, 40–57 (2023).
Bøhle, K., Otterholt, E. & Bjørkly, S. Is there an association between salivary cortisol and dropping out of inpatient substance addiction treatments? A prospective repeated measures study. Subst. Abuse Res. Treat. 16, 11782218221106797 (2022).
Fish, K. N. & Joffe, M. E. Targeting prefrontal cortex GABAergic microcircuits for the treatment of alcohol use disorder. Front. Synaptic Neurosci. https://www.frontiersin.org/articles/10.3389/fnsyn.2022.936911 (2022).
Trojak, B. et al. Efficacy of transcranial direct current stimulation (tDCS) in reducing consumption in patients with alcohol use disorders: study protocol for a randomized controlled trial. Trials 17, 250 (2016).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Cooper, Y. A. et al. Functional regulatory variants implicate distinct transcriptional networks in dementia. Science 377, eabi8654 (2022).
Adams, C., Conigrave, J. H., Lewohl, J., Haber, P. & Morley, K. C. Alcohol use disorder and circulating cytokines: a systematic review and meta-analysis. Brain Behav. Immun. 89, 501–512 (2020).
de Leeuw, C. A., Mooij, J. M., Heskes, T. & Posthuma, D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol. 11, e1004219 (2015).
Cacabelos, R., Hashimoto, R. & Takeda, M. Pharmacogenomics of antipsychotics efficacy for schizophrenia. Psychiatry Clin. Neurosci. 65, 3–19 (2011).
Falk, D. E., Yi, H. Y. & Hiller-Sturmhöfel, S. An epidemiologic analysis of co-occurring alcohol and tobacco use and disorders: findings from the National Epidemiologic Survey on Alcohol and Related Conditions. Alcohol Res. Health J. Natl Inst. Alcohol Abuse Alcohol 29, 162–171 (2006).
Stinson, F. S. et al. Comorbidity between DSM-IV alcohol and specific drug use disorders in the United States: results from the National Epidemiologic Survey on Alcohol and Related Conditions. Drug Alcohol Depend. 80, 105–116 (2005).
McPheeters, M. et al. Pharmacotherapy for alcohol use disorder: a systematic review and meta-analysis. JAMA 330, 1653–1665 (2023).
Lepreux, G. et al. Recapitulating phenotypes of alcohol dependence via overexpression of Oprk1 in the ventral tegmental area of non-dependent TH::Cre rats. Neuropharmacology 228, 109457 (2023).
Borruto, A. M. et al. NOP receptor antagonism attenuates reinstatement of alcohol-seeking through modulation of the mesolimbic circuitry in male and female alcohol-preferring rats. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 46, 2121–2131 (2021).
Nutt, D. J. The role of the opioid system in alcohol dependence. J. Psychopharmacol. Oxf. Engl. 28, 8–22 (2014).
Hansson, A. C. et al. Dopamine and opioid systems adaptation in alcoholism revisited: convergent evidence from positron emission tomography and postmortem studies. Neurosci. Biobehav. Rev. 106, 141–164 (2019).
Inder, W. J. et al. The effects of alcoholism on the hypothalamic-pituitary-adrenal axis: interaction with endogenous opioid peptides. Clin. Endocrinol. 43, 283–290 (1995).
Hatoum, A. S. et al. Multivariate genome-wide association meta-analysis of over 1 million subjects identifies loci underlying multiple substance use disorders. Nat. Ment. Health 1, 210–223 (2023).
Hatoum, A. S. et al. The addiction risk factor: a unitary genetic vulnerability characterizes substance use disorders and their associations with common correlates. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol. 47, 1739–1745 (2022).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Zhou, H. et al. Genome-wide meta-analysis of alcohol use disorder in East Asians. Neuropsychopharmacology 47, 1791–1797 (2022).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Im, P. K. et al. Alcohol consumption and risks of more than 200 diseases in Chinese men. Nat. Med. 29, 1476–1486 (2023).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Bulik-Sullivan, B. K. et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Saunders, G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612, 720–724 (2022).
Watanabe, K., Umićević Mirkov, M., de Leeuw, C. A., van den Heuvel, M. P. & Posthuma, D. Genetic mapping of cell type specificity for complex traits. Nat. Commun. 10, 3222 (2019).
Holland, D. et al. Beyond SNP heritability: polygenicity and discoverability of phenotypes estimated with a univariate Gaussian mixture model. PLoS Genet. 16, e1008612 (2020).
Frei, O. et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat. Commun. 10, 2417 (2019).
Brown, B. C., Ye, C. J., Price, A. L. & Zaitlen, N. Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 99, 76–88 (2016).
Acknowledgements
This work was partly performed on the TSD (Tjeneste for Sensitive Data) facilities, owned by the University of Oslo and operated and developed by the TSD service group at the University of Oslo, IT Department (USIT). Computations were also performed on resources provided by UNINETT Sigma2, the National Infrastructure for High-Performance Computing and Data Storage in Norway. We gratefully acknowledge support from the NIH (NS057198, EB000790 and 1R01MH124839), the Research Council of Norway (229129, 213837, 223273, 324252, 248980 and 334920), the South-East Norway Regional Health Authority (2017-112 and 2019-108) and KG Jebsen Stiftelsen (SKGJ-MED-021). This project has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement numbers 847776, 964874 and 801133 (Marie Skłodowska-Curie grant agreement). R.I. thanks the INTPART program (principal investigator T.V.L.) for supporting his fellowship at NORMENT. Core funding of third party data acquisition: Million Veteran Program (MVP), United States Department of Veterans Affairs; FINNGEN, Business Finland and the pharmaceutical industry partners (https://www.finngen.fi/en/funding); UK Biobank (UKB), Wellcome Trust medical charity, Medical Research Council, Department of Health, Scottish Government and the Northwest Regional Development Agency (https://www.ukbiobank.ac.uk/learn-more-about-uk-biobank/about-us/our-funding); Psychiatric Genomics Consortium (PGC), US National Institute of Mental Health and the US National Institute of Drug Abuse (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5756100/).
Author information
Authors and Affiliations
Contributions
R.I., A.S., G.H. and O.A.A. were responsible for the study concept and design. R.I., A.S. and G.H. contributed to the acquisition of summary data. A.S. was the main data analyst. B.H., N.K., O.B.S., S.D., H.J.E., H.Z. and O.A.A. assisted with data analysis and interpretation of findings. R.I., A.S., N.K. and B.H. prepared figures. B.H. drafted the paper. O.A.A., O.B.S., W.C., T.V.L., M.C.H., K.S.O., N.K., N.P., O.F., S.B., T.M.S., A.M.D. and J.G. provided critical revision of the paper for important intellectual content. All authors critically reviewed content and approved the final version for publication.
Corresponding author
Ethics declarations
Competing interests
A.M.D. is a founder of and holds equity in Cortechs.ai and serves on its scientific advisory board; he is a member of the scientific advisory boards of HealthLytix and the Mohn Medical Imaging and Visualization Center (Bergen, Norway); and he receives funding through a research agreement between General Electric Healthcare and UCSD. O.A.A. has received speaking honoraria from Lundbeck and has served as a consultant for HealthLytix. The other authors report no financial relationships with commercial interests.
Peer review
Peer review information
Nature Mental Health thanks Colin Hodgkinson, El Cherif Ibrahim and Tianye Jia for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1 and 2.
Supplementary Data 1
Tuning parameters of FUMA SNP2GENE for the main GWAS meta-analysis, by ancestry.
Supplementary Tables
Supplementary Tables 1–6.
Source data
Source Data Table 1
Statistical data for Table 1.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Icick, R., Shadrin, A., Holen, B. et al. Identification of risk variants and cross-disorder pleiotropy through multi-ancestry genome-wide analysis of alcohol use disorder. Nat. Mental Health 3, 253–265 (2025). https://doi.org/10.1038/s44220-024-00353-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44220-024-00353-8