Abstract
Arginine methylation of histones plays a critical role in regulating gene expression. The writers (methyltransferases) and readers of methylarginine marks are well-known, but the erasers–arginine demethylases–remain mysterious. Here we identify Myc-induced nuclear antigen 53 (Mina53), a jumonji C ___domain containing protein, as an arginine demethylase for removing asymmetric di-methylation at arginine 3 of histone H4 (H4R3me2a). Using a photoaffinity capture method, we first identify Mina53 as an interactor of H4R3me2a. Biochemical assays in vitro and in cells characterize the arginine demethylation activity of Mina53. Molecular dynamics simulations provide further atomic-level evidence that Mina53 acts on H4R3me2a. In a transgenic mouse model, specific Mina53 deletion in neural stem/progenitor cells prevents H4R3me2a demethylation at distinct genes clusters, dysregulating genes important for neural stem/progenitor cell proliferation and differentiation, and consequently impairing the cognitive function of mice. Collectively, we identify Mina53 as a bona fide H4R3me2a eraser, expanding the understanding of epigenetic gene regulation.
Similar content being viewed by others
Introduction
Arginine methylation is a prevalent posttranslational modification of intracellular proteins including histones1,2,3,4. Accumulating research has demonstrated a critical role of arginine methylation in regulating a myriad of biological processes, including DNA damage signaling, gene transcription, mRNA translation, and cell fate decisions3,5,6,7,8. Methylation of arginine residues is shown to be catalyzed by at least nine arginine methyltransferases (PRMTs), divided into three subtypes based on the methylation products, namely monomethylation (me1), symmetric dimethylation (me2s), and asymmetric dimethylation (me2a)9,10. These modifications often serve as the recognition motif for the Tudor ___domain-containing effector proteins to translate the methylation-coded information to various functional outputs11,12,13.
Different forms of arginine methylation have been identified on all of the core histones, and have been shown to modulate protein structures and influence protein-DNA interactions to regulate gene expression. The asymmetric dimethylation of histone 4 arginine 3 (H4R3me2a) is a marker of transcriptional activation and is principally catalyzed by the family member PRMT17,14,15,16,17,18. H4R3me2a marker facilitates the recruitment of several histone acetyltransferases to activate gene transcription. Studies also showed that H4R3me2a marker could be recognized by Tudor ___domain-containing proteins including TDRD3, which recruits DNA topoisomerase IIIβ (TOP3B) to facilitate resolution of transcription-mediated R-loops, allowing active gene transcription19,20,21.
Although the writer and readers of H4R3me2a have been well characterized, the eraser of H4R3me2a still remains an enigma. Some members of the JmjC-___domain containing protein family of 2-oxoglutarate-dependent oxygenases have been reported to possess arginine demethylation activity22,23,24,25,26,27. Among them, JMJD6 (JmjC ___domain containing 6) was firstly identified as a putative arginine demethylase of H3R2me2 and H4R3me2, but later biochemical and structural studies instead supported its assignment as a lysine hydroxylase28,29,30. In a recent study, JMJD1B was shown to catalyze demethylation of H4R3me2s to regulate gene expression in hematopoietic stem/progenitor cells24,31. However, no study to date has reported the identification of demethylase for H4R3me2a.
Mina53 (also termed RIOX2) is a direct downstream gene product of c-Myc and belongs to the JmjC-___domain containing protein family. Previous studies suggested its possible lysine demethylase activity towards H3K9me3 in human lung cancer A549 cells, and H3K36me3 in HIV latency cell line J-LAT A232,33. In the present study, we identify Mina53 as an interactor of H4R3me2a mark. Subsequent biochemical, cellular, and structural modeling studies demonstrate that Mina53 acts as an arginine demethylase of H4R3me2a. Using a transgenic mouse model, we further demonstrate that specific deletion of Mina53 in mouse neural stem/progenitor cells (NSPCs) blocks demethylation of H4R3me2a at distinct clusters of genes, and regulates the expression of genes important for NSPC proliferation and differentiation. Consequently, specific deletion of Mina53 in NSPCs impairs learning and memory in mice. Together, our study identifies Mina53 as a bona fide arginine demethylase of H4R3me2a, which is critical for epigenetic programming for neuronal development.
Results
Mina53 is an interactor of H4R3me2a
Identifying H4R3me2a interactors is challenging as the posttranslational modification-mediated protein-protein interactions tend to be weak and transient. To effectively capture these interactions, we designed a modified H4R3me2a peptide (1) containing a minimally perturbative diazirine photoreactive group34 (photoleucine) at position 1 that can covalently capture the interacting proteins upon UV irradiation, and a C-terminal biotin affinity handle to facilitate the subsequent isolation of cross-linked proteins for mass spectrometry analysis (Fig. 1a). As an initial validation of this capturing strategy, we tested whether we could crosslink the well-known H4R3me2a methyltransferase, PRMT1. Flag-tagged PRMT1 was stably expressed in HEK293T cells. Isolated nuclei were then incubated with the modified peptide 1 for 10 min with rotation. After removing the excess peptide 1, photo-crosslinking was performed upon UV irradiation for 10 min. Nuclei lysates were captured with streptavidin beads and analyzed by western blotting. A clear band was shown that is consistent with the crosslinked PRMT1, which was not present in any of the control experiments, thus validating the crosslinking strategy (Supplementary Fig. 1a).
a Schematic for the photoaffinity capture strategy for identification of H4R3me2a interactors (upper), and the structure of H4R3me2a-pLeu-biotin 1 (lower). b Representative immunoblotting analysis of UV-treated crosslinked samples using H4R3me2a-pLeu-biotin 1 or the control peptide. Histone H3 levels serves as a loading control. c Volcano plot of proteins enriched in samples treated with H4R3me2a-pLeu-biotin 1 and samples treated with H4R3me2a peptide without pLeu group on the streptavidin blots is used as the control group. Only genes whose P value is less than 0.05 with |log2 fold change| > 2 are analyzed for further research. d Representative immunoblotting analysis of purified Flag-tagged Mina53 crosslinked with H4R3me2a-pLeu-biotin or H4R3me0-pLeu-biotin. e Microscale thermophoresis analysis of the binding affinity between Mina53 and synthetic FITC-conjugated H4R3me2a peptide (n = 3 independent assays). f Immunoblotting analysis of Flag-Mina53 and H4 upon immunoprecipitation with anti-Flag antibody in HEK293T cells. g Representative immunoblotting analysis of purified His-tagged Mina53 and bulk histones upon immunoprecipitation with anti-His antibody. The experiments related to (b, d) have been repeated three times independently; The experiments related to (f, g) have been repeated twice independently. For statistics in this figure, the two-tail unpaired Student’s-t test was utilized in (c) and (e), and data were shown as means ± SD. Source data are provided as a Source Data file.
Encouraged by the validated result, we next performed a proteome-wide study. Clear differences were observed on streptavidin blots between samples with or without the pLeu group on the streptavidin blots after UV irradiation (Fig. 1b). Isolated proteins were then subjected to on-bead trypsin digestion, and identified by liquid chromatography coupled mass spectrometry (LC-MS). The digested peptides were isotopically derivatized through dimethyl labeling by NaCNBH3 (sodium cyanoborohydride) or NaCNBD3 (deuterated sodium cyanoborohydride) for a high confidence assignment of identified proteins (Supplementary Data 1). Proteomics data were analyzed with the selected cutoff (|log2 fold change| > 2, P < 0.05).
Among the identified proteins, two Tudor ___domain-containing proteins known to bind methylarginine motifs, SND1 and SMNDC1, were enriched in the pLeu-containing peptide sample, along with the expected PRMT1 (Fig. 1c).
Two members of the JmjC ___domain-containing protein family, JMJD1C and RIOX2 (Mina53), were also significantly enriched in the crosslinked sample (Fig. 1c). This is intriguing because this protein family contains enzymes with lysine and/or arginine demethylase activities. Thus, we tested whether these two proteins could be potential arginine demethylases for H4R3me2a. Depletion of Mina53, but not JMJD1C, caused a significant increase in H4R3me2a levels in 293 T cells (Supplementary Figs. 1b, 1c), suggesting that Mina53 may be a demethylase. In addition, we depleted a well-studied JmjC-containing protein−KDM5B in 293 T cells, and analyzed the expression level of H4R3me2a. The result showed that KDM5B depletion had no effect on H4R3me2a levels (Supplementary Fig. 1d). We then further characterized the interaction between Mina53 and H4R3me2a. Flag-tagged Mina53 in the cell lysates was efficiently captured by the pLeu-containing H4R3me2a peptide, whereas much less amount of Mina53 was captured by the pLeu-containing H4R3 peptide without methylation (Fig. 1d), suggesting that me2a modification contributes to the specific interaction between the H4 peptide and Mina53. To further evaluate the effect of R3 modification on the binding between H4 peptides and Mina53, we incubated purified His-tagged Mina53 with H4R3 peptides possessing different R3 modifications. Mina53 could pull down the H4R3me2a peptide in the largest amount, compared to other peptides, with the H4R3me2s peptide in the smallest amount (Supplementary Fig. 1e). The binding affinity of Mina53 with the H4R3me2a peptide was measured to be 27.59 ± 9.81 μM by microscale thermophoresis (MST) (Fig. 1e). In addition, ectopic expression of Flag-tagged Mina53 in 293 T cells followed by immunoprecipitation revealed the association of Mina53 with histone H4 (Fig. 1f). Purified His-tagged Mina53 could also readily pull down H4 when incubated with bulk histones in vitro (Fig. 1g). Together, these data point to the hypothesis that Mina53 may act as an arginine demethylase for H4R3me2a.
Mina53 catalyzes demethylation of H4R3me2a in vitro and in cells
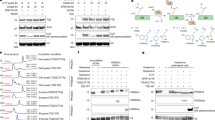
To further explore whether Mina53 catalyzes H4R3me2a demethylation, we bacterially expressed and purified His-tagged WT or the inactive mutant (H179Y/D181G/H240Y) of Mina5335,36, and performed in vitro demethylation reactions with various synthetic methylated peptides using the dot-blotting assay. Blotting signals of H3K9me3 and H3K36me3 peptides were modestly decreased with WT, but not mutant Mina53, consistent with its known lysine demethylation activity (Fig. 2a). Notably, blotting signals of H4R3me2a and H4R3me1 drastically decreased with WT, but not mutant Mina53, while signals of other arginine methylated peptides remained unchanged, indicating the specificity of Mina53 (Fig. 2a, Supplementary Fig. 2a). As expected for a demethylase, omitting key reaction components greatly reduced the H4R3me2a signal, but not the H4R3me2s signal (Fig. 2b). Moreover, high-resolution mass spectrometry analysis revealed WT, but not mutant Mina53, incubation shifted the H4R3me2a peptide peak 14 Da, corresponding to the mono-methylated peptide (Fig. 2c). Incubation with H4R3me1 peptide showed modest non-methylated peptide appearance (Fig. 2d). We further determined the enzyme kinetics of Mina53 using the formaldehyde release assay, which showed a Km value of 9.15 ± 3.71 μM, and a turn-over rate (kcat) of 0.37 ± 0.08 s−1 for the H4R3me2a peptide (Supplementary Fig. 2b). Furthermore, the dot-blotting results with synthetic peptides were recapitulated with isolated bulk histones as substrates (Supplementary Fig. 2c, 2d). Purified Mina53 also catalyzed demethylation of H4R3me2a in bulk histones in a time-dependent manner, with H4R3me1 levels increasing for the first 30 min then gradually decreasing over time (Supplementary Fig. 2e), consistent with the model in which H4R3me1 is the demethylation intermediate of H4R3me2a. Thus, these in vitro results demonstrate that Mina53 serves as an arginine demethylase for H4R3me2a.
a Representative dot-blotting analysis of in vitro demethylation reactions in the presence of wildtype (WT) Mina53 or the catalytically inactive mutant Mina53 and synthetic peptides. b Representative dot-blotting analysis of in vitro demethylation reactions in the presence or absence of various reaction components. Representative mass spectra of demethylation products in reactions with H4R3me2a (c) or H4R3me1 (d) as substrates. Reactions were performed in the presence of WT or the catalytically inactive mutant Mina53 in parallel. Representative immunoblotting analysis of H4R3 methylation status and PRMT1 in HEK293T cells stably expressing WT or the catalytically inactive mutant Mina53 (e), or stably depleting endogenous Mina53 (f). g A representative conformation of the N terminus of H4-Mina53 complex in the H4R3me2a form, obtained from the MD simulation. The electrostatic surface of Mina53 is illustrated, with negatively charged areas in red and positively charged areas in blue. The H4R3 peptide is shown as spheres: R3me2a is colored green, the positively charged residues (K5, K8, K12, K16, and R17) are colored blue, and the other residues are colored white. The experiments related to (e, f) have been performed in duplicate independently.
To determine whether Mina53 mediates demethylation of H4R3me2a and H4R3me1 in cells, we ectopically expressed WT or inactive mutant Mina53 in HEK293T cells or Neuro2A cells, and assessed H4R3me2a and H4R3me1 levels. Expectedly, both H4R3me2a and H4R3me1 levels were significantly decreased upon WT Mina53 expression, while no change was observed upon inactive mutant Mina53 expression, compared with the vector control (Fig. 2e). Interestingly, H3K9me3 and H3K36me3 levels were not affected. Conversely, Mina53 knockdown caused a drastic accumulation of H4R3me2a and H4R3me1 levels, but only a minor change in H3K9me3 and H3K36me3 levels (Fig. 2f). Ectopic expression or knockdown of Mina53 did not impact PRMT1 levels, suggesting that Mina53 directly regulates H4R3me2a and H4R3me1 levels in cells. Collectively, the biochemical and cellular data consistently demonstrate that Mina53 functions as an arginine demethylase for H4R3me2a and H4R3me1.
To reveal the intricate interactions between Mina53 and H4R3me2a, we employed computational modeling. We started with molecular docking to generate a preliminary Mina53-H4R3me2a complex, which was then refined by restrained molecular dynamics (MD) simulations. We confirmed the accuracy of this complex by performing extensive unrestrained MD simulations, which showed that the complex was stable for more than 2 μs (Supplementary Movie 1). This allowed us to examine the atomic-level details of the interaction. We found that H4R3me2a fit snugly on the negatively charged surface of Mina53, spanning the N- and C-terminal domains (Fig. 2g). The R3me2a side chain penetrated deeply into the binding pocket, where it was held firmly by key residues H179, D181, H240, and the Mn2+ and 2-oxoglutarate cofactors (Supplementary Fig. 2f). Moreover, several positively charged H4R3me2a residues (K5, K8, K12, K16 and R17) formed salt bridges with D333, D329, D270, E411, and some other negatively charged residues of Mina53 (Supplementary Fig. 2g), which further stabilized the protein-peptide interaction. To validate the modeling, we generated the double mutant (D333A/D329A) of Mina53, and analyzed the binding affinity with H4R3me2a peptide. The result showed that the mutant displayed a drastic loss of binding with the peptide, as compared with WT Mina53 (Supplementary Fig. 2h). To further explore the specificity of Mina53 towards R3me2a modification, we performed additional simulations with other arginine modification forms (R3me0, R3me1 and R3me2s). We executed simulations lasting 2 μs for each repeat for H4R3me0 and H4R3me1, and 1 μs for each repeat for H4R3me2s, totaling 15 μs of simulation time. These simulations were strategically devised to critically assess how each modified form of arginine is accommodated within the Mina53 binding pocket. The results of these additional simulations (Supplementary Fig. 2i-2j) clearly show that H4R3me2a achieves a notably more favorable position within the binding pocket than both H4R3me2s and the H4R3me0, and exhibits a marginally superior alignment compared to H4R3me1. This is especially evident from the marked difference in the distance between the R3 residue and the pocket (Supplementary Fig. 2j). Thus, our simulations were consistent with experimental data, supporting a model where the Mina53 binding pocket provides exquisite specificity for the dimethylated arginine side chain. Overall, our computational approach revealed the intricate molecular interactions governing H4R3me2a recognition by Mina53, further supporting the assignment of Mina53 as an arginine demethylase.
Mina53-mediated H4R3me2a levels are dynamically regulated in neural stem/progenitor cells
Neural stem/progenitor cells (NSPCs) possess the unique abilities of self-renewal and differentiation into neurons and astrocytes in the brain37,38. The adult neurogenesis process, in which new neurons generated from NSPCs are integrated into neural circuits, is critical for maintaining neuronal activity and cognitive function. Aberrant neurogenesis is closely associated with multiple neurological diseases, including autism, Alzheimer’s diseases, and intellectual disability39,40,41,42,43,44,45.
Accumulating evidence points to the importance of epigenetic regulation in adult neurogenesis43,44,46,47,48,49. Thus, we explored whether Mina53-mediated H4R3me2a demethylation plays a functional role in neurogenesis regulation. We performed Western blotting on proliferating and differentiated NSPCs using isolated NSPCs from mice forebrains (8 weeks old). Upon differentiation, we observed that Mina53 levels were significantly downregulated, with concomitant upregulation of H4R3me2a and H4R3me1 levels as shown in Western blotting and immunostaining (Fig. 3a, Supplementary Fig. 3ab). In contrast, levels of H3K9me3, H3K36me3 and PRMT1 were unaffected. This suggests in NSPCs, H4R3me2a levels are predominantly regulated by Mina53 through its arginine demethylase activity, rather than the lysine demethylase activity. We also found that Mina53 mRNA levels were highest in proliferating NSPCs among differentiated NSPCs, neurons, and astrocytes (Supplementary Fig. 3c). We also analyzed the protein expressions of Mina53 in mice at embryonic days 14 (E14), embryonic days 17 (E17), and at birth (P0). These results showed that Mina53 expression was much higher in embryonic stages than at birth (Supplementary Fig. 3d).
a Representative immunoblotting analysis of histone methylation marks and PRMT1 in proliferating (Proli) and differentiated (Differ) NSPCs. b Representative immunoblotting analysis of H4R3 methylation status and PRMT1 in proliferating NSPCs depleted of Mina53. c Representative images and quantification of BrdU immunofluorescence staining of NSPCs in the presence or absence of Mina53 depletion (n = 3, three different fields of view for each group). d Schematic depiction of mice crossing to generate Mina53 conditional knockout (cKO) mice. e Representative immunoblotting analysis of H4R3 methylation status in NSPCs isolated from control (Ctrl) and Mina53 cKO mice. f Representative images and quantification of Nestin-BrdU immunofluorescence staining of NSPCs isolated from control (Ctrl) and Mina53 cKO mice (n = 3, three mice for each group). The experiments related to (a, b, and e) have been performed in duplicate independently. For statistics in this figure, the two-tail unpaired Student’s-t test was utilized in (c) and (f), and data were shown as means ± SD. Source data are provided as a Source Data file.
To further elucidate the role of Mina53 in regulating NSPCs, we performed acute knockdown of Mina53 using short hairpin RNA (shRNA) and analyzed the effects on NSPC proliferation and differentiation. As expected, Mina53 depletion significantly increased H4R3me2a and H4R3me1 levels in NSPCs (Fig. 3b). Compared with the control group, Mina53 depletion remarkably decreased the percentage of BrdU-positive (BrdU+) cells detected by BrdU incorporation assay (Fig. 3c). Besides, Mina53 depletion also significantly decreased the proportion of Ki67-positive (Ki67+) cells (Supplementary Fig. 3e). In addition, Mina53 knockdown in NSPCs significantly reduced neurosphere size in single-cell clonal assays compared to the control (Supplementary Fig. 3f). When cultured under differentiation conditions, Mina53 knockdown increased the staining intensity and the number of Tuj1-positive (Tuj1+) neurons as well as those of Gfap-positive (Gfap+) astrocytes compared to the control group (Supplementary Fig. 3g, 3h). As Mina53 has been shown to be transcriptionally regulated by c-Myc, we depleted c-Myc in NSPC and detected the expression of genes related to neuronal cell proliferation using real-time PCR (Supplementary Fig. 3i). We observed that c-Myc depletion in NSPCs decreased the expression of Sox2 and Sox10, which are critical markers of stem cell proliferation. Together, these results demonstrate that Mina53 is important for maintaining proliferation and multipotency of NSPCs.
Next, we want to achieve in situ depletion of Mina53 in mice to further explore the effect on NSPCs. Toward this end, we generated a NestinCreERT2:Mina53flox/flox conditional knockout (cKO) mice model by crossing the Mina53flox/flox mice with the NestinCreERT2 mice, which selectively targets Nestin+ NSPCs (Fig. 3d). Multiple doses of tamoxifen were intraperitoneally injected into the eight-week-old cKO mice to induce Mina53 deletion (once per day for 5 consecutive days). The control group of mice was treated with the corn oil. Four weeks after the last tamoxifen injection, NSPCs were isolated for further analysis. Western blotting analysis showed a near complete deletion of Mina53 in cKO NSPCs (Fig. 3e). Consistently, the levels of H4R3me2a and H4R3me1 were significantly increased in cKO cells compared with the control cells (Fig. 3e, Supplementary Fig. 4a). In the BrdU incorporation assay, percentage of BrdU+ cells was much lower in cKO cells compared with that of the control group (Fig. 3f). The cKO cells also displayed a lower percentage of Ki67+ cells (Supplementary Fig. 4b). Under the differentiation condition, cKO cells showed much higher staining intensity and higher proportion of Tuj1+ neurons and Gfap+ astrocytes compared with the control cells (Supplementary Fig. 4c, d). Taken together, these results consistently show that Mina53 regulates H4R3me2a and H4R3me1 levels in NSPCs, and plays an important role in controlling the proliferation and differentiation of NSPCs in vitro.
Mina53 deficiency leads to aberrant neurogenesis and impairs learning and memory in mice
To further investigate the role of Mina53 in neurogenesis, we treated 8-week-old adult mice (NestinCreERT2:Mina53flox/flox v.s Mina53flox/flox) with tamoxifen to induce Mina53 deletion. Four weeks after the first time of tamoxifen injection, BrdU was intraperitoneally injected into mice and mice were sacrificed 4 h after the final injection (Supplementary Fig. 5a). Compared with the control mice, the hippocampus region of cKO mice showed significantly decreased number of BrdU+ cells detected by immunostaining. The percentage of newborn neurons (marked by BrdU+DCX+/BrdU+) was significantly increased in cKO mice (Supplementary Fig. 5b). To explore the relatively long-term effect of Mina53 deletion, we continued to raise the mice for another 4 weeks after inducing Mina53 deletion and BrdU injection (Fig. 4a). Immunostaining of the hippocampus region of the brain showed a significant reduction of BrdU+ cells, but a marked increase of BrdU+NeuN+/BrdU+ cells (indicating mature neurons), in cKO mice compared with control mice (Fig. 4b, c). Together, these in vivo data suggest that loss of Mina53 leads to accelerated differentiation of NSPCs, which likely diminishes the pool of NSPCs for neurogenesis.
a Schematic depiction of inducting Mina53 deletion by tamoxifen, and the subsequent BrdU administration. Representative images (b) and quantification (c) of BrdU-NeuN immunofluorescence staining of the hippocampus region of the brain derived from control (Ctrl) and Mina53 cKO mice (n = 3, three mice for each group). The P value of BrdU+/DAPI, BrdU+ NeuN+/NeuN+ and BrdU+ NeuN-/NeuN- is 0.0012, 0.0086 and 0.0056. d The escape latency during the training period of control (Ctrl) and Mina53 cKO mice (n = 11, eleven mice for each group, P = 6.75E-05). e Representative images of swimming paths of control (Ctrl) and Mina53 cKO mice in Morris Water Maze test (n = 11, eleven mice for each group). f The average swimming speed of control (Ctrl) and Mina53 cKO mice in Morris Water Maze test (n = 11, eleven mice for each group, P = 0.5369). g Time to the platform (left) and the number of platform-crossing during the probe trial of control (Ctrl) and Mina53 cKO mice (n = 11, eleven mice for each group, the P value related to time to platform is 0.0075 and the P value related to the number of platform-crossing during the probe trail is 0.0002). h Relative time spending in the target and other quadrants of control (Ctrl) and Mina53 cKO mice (n = 11, eleven mice for each group, the P value of SE, NW, SW and NE is <0.0001, 0.02189, 0.00117 and 0.09746, respectively). For statistics in this figure, the two-tail unpaired Student’s-t test was used in (c, d, f, g, h), and data were shown as means ± SD. Scale bar, 50 μm. Source data are provided as a Source Data file.
As neurogenesis is critical for the cognitive function of mice, we next examined whether Mina53 deletion in NSPCs affected the learning and memory of mice through several well-established animal behavioral studies (Supplementary Fig. 6a)45,50,51. Adult cKO mice were injected with tamoxifen to induce Mina53 deletion, or injected with corn oil in the control group. Compared with the control group, cKO mice taked more time to reach the visible platform during the training period in the Morris water maze test (Fig. 4d). After training, mice were subjected to probe testing, and the time and route of the mice to get to the invisible platform were recorded (Fig. 4e). Although both groups displayed similar speeds (Fig. 4f), cKO mice had a significant delay in reaching the invisible platform (Fig. 4g). In addition, the less frequency of crossing the platform and time in the target quadrant indicate that the spatial learning and memory abilities of cKO mice are impaired (Fig. 4h).
In the novel object recognition test, no significant difference in the time spent exploring the objects was observed during the training period for both cKO and control mice (Supplementary Fig. 6b). However, during the test period cKO mice spent significantly less time with the novel object compared with control mice (Supplementary Fig. 6c). Moreover, in the Y-maze test, cKO mice showed lower spontaneous alternation without altering total number of entries compared to control mice, indicating that Mina53 is critical for short working memory (Supplementary Fig. 6d and 6e). Collectively, these behavioral studies consistently demonstrate that Mina53 ablation in NSPCs damped the learning and memory of mice, possibly due to the depletion of NSPC pool52.
Mina53-mediated demethylation of H4R3me2a regulates genes involved in NSPC proliferation and differentiation
To further investigate the mechanisms by which Mina53 regulates NSPC proliferation and differentiation, We analyzed gene expression profile in control and Mina53-cKO NSPCs by performing RNA sequencing (RNA-seq). RNA-seq analysis revealed that Mina53 depletion resulted in 1377 differentially expressed genes (DEGs), among which 742 genes were upregulated and 635 genes were downregulated (|log2FC| ≥ 0.5, P < 0.05) (Fig. 5a, Supplementary Data 2). Gene set enrichment analysis (GSEA) of the DEGs showed that they were mainly related to neuronal development, cell growth, cell cycle regulation, and signaling pathways (Fig. 5b). Based on the RNA-seq data, we further validated the expression of several well-known stem cell markers, including Nestin, Sox2, Sox10, and Foxo3, using quantitative reverse transcription-PCR (qRT-PCR) assays, which showed a significant reduction in Mina53-cKO NSPCs compared with control cells (Supplementary Fig. 7a).
a Volcano plot of differentially expressed genes (DEGs) in NSPCs in the presence or absence of Mina53 depletion (n = 3 for each group). b Gene set enrichment analysis of DEGs. c Venn diagram showing the overlapped genes with both increased H4R3me2a occupancy and altered gene expressions in the RNA-seq data. d Enrichment analyses of the upregulated and down-regulated DEGs in control or Mina53 cKO NSPCs. For statistics in this figure, the two-tail unpaired Student’s-t test was used in (a) and (b). Only genes and pathways whose P value are less than 0.05 and absolute value of log2FC are equivalent to or greater than 1.5 are used for further analysis. Scale bar, 50 μm. Source data are provided as a Source Data file and Supplementary Data 2.
As histone methylation plays a critical role in regulating gene expression in cells, we next investigated whether Mina53-mediated demethylation of H4R3me2a directly regulates gene expression involved in proliferation and differentiation of NSPCs. We performed chromatin sequencing using Cleavage Under Targets and Tagmentation (CUT&Tag) in Mina53-cKO and control cells to analyze the genomic distribution of H4R3me2a. Enrichment for occupancy by H4R3me2a was probed with the site-specific antibody in Mina53-cKO and control NSPCs. Analysis of the genome-wide distribution revealed 34.49% of the peaks located in the intron region, 32.69% in the promoter region, and 27.4% in the distal intergenic region of Mina53-cKO cells, compared to 31.48% in the intron region, 38.86% in the promoter region, and 24.98% in the distal intergenic region of control cells (Supplementary Fig. 7b).
Next, we integrated our CUT&TAG data with our RNA-seq data to identify genes directly regulated by Mina53-mediated demethylation of H4R3me2a. We analyzed overlapped genes with both increased H4R3me2a enrichment (log2foldchange > 0.5, P < 0.05) and altered expression. A total of 55 genes were identified (Fig. 5c). As H4R3me2a is known as an active epigenetic marker, it is likely that H4R3me2a induces the expression of inhibitory regulatory factors related to NSPC differentiation thus leading to decreased or increased expression of other genes. GO analysis of the upregulated genes showed that this group of genes are involved in anatomical structure morphogenesis, inflammation, nervous system development and differentiation (Fig. 5d). Among these target genes are Tnc, Gria1, Klf5c, Cbln1 and Mgat5, all of which are known to be important in proliferation and differentiation of stem cells (Supplementary Fig. 7c). Moreover, we depleted Mgat5 in NSPCs and consistently observed a significant reduction of GFAP expression, the marker of astrocyte differentiation (Supplementary Fig. 7d). Together, these results support the notion that Mina53 regulates proliferation and differentiation of NSPCs through demethylation of H4R3me2a epigenetic mark.
Discussion
Our present study demonstrates that Mina53 functions as an arginine demethylase for the H4R3me2a histone mark both in vitro and in vivo. Mina53 deficiency changes the H4R3me2a dynamics at the promoter region of distinct sets of genes involved in proliferation and differentiation of NSPCs, thereby altering gene expressions and consequently impairing adult neurogenesis in cKO mice. Consistent with previous reports that the appearance of the H4R3me2a mark specifies NSPC differentiation, our results show that H4R3me2a levels were significantly upregulated in differentiated NSPCs. NSPCs with Mina53 depletion showed a higher tendency to differentiate into neurons, indicating that Mina53-regulated H4R3me2a is involved in neuronal development. We propose that Mina53 is critical for maintaining H4R3me2a demethylation state, which inhibits expression of differentiation genes and inhibitory transcription factors, thereby enabling NSPCs to maintain multipotency. As PRMT1 is known to generate the H4R3me2a mark, it is likely that these two enzymes of opposing functions work together to control the dynamic level of H4R3me2a to govern the on/off states of certain genes under different conditions. Deficiency in either PRMT1 or Mina53 would disrupt the homeostasis of the histone mark, leading to aberrant neuronal development. On the other hand, PRMT1 is capable of catalyzing arginine methylation on both histones and non-histone proteins. We also envision that Mina53 may catalyze arginine demethylation on other protein substrates in addition to H4R3. Thus, PRMT1 and Mina53 may have distinct sets of substrates in cells. It is intriguing to explore the cellular substrates of Mina53, which may provide important insight to further understand dynamic arginine methylation in cells.
Recent studies indicate that members of the JmjC protein family are potential arginine demethylases. However, the activities of most of these proteins have not been conclusively characterized. It is possible that there exist multiple demethylases targeting the same methylarginine sites on a given protein. Indeed, Mina53 depletion in our study only increased the H4R3me2a level in a group of genes. Further exploration of JmjC ___domain-containing proteins likely provides new opportunities to advancing the field of arginine demethylation.
Methods
All research procedures related to mice described in this study were performed in compliance with the ethical regulations by the Research Ethical Committees of Zhejiang University with the permission number 25115.
Cell culture
293 T cells and Neuro2A cells were purchased from ATCC. Cells and cultured in DMEM (HyClone) supplemented with 10% FBS (Excell), penicillin and streptomycin in a cell culture incubator containing 5% carbon dioxide at 37 °C. Cells are changed every month and only cells free from mycoplasma contamination were used in our experiments. As for extracted NSPCs from mice, cells were cultured in DMDM/F12 medium (Gibco) supplemented with 2% B27 (Thermo Fisher Scientific), penicillin, streptomycin, EGF and FGF, (20 ng/mL, Peprotech) to keep proliferation. As to induce the differentiation of NSPCs, poly-L-Ornithine and laminin were added to cell culture dishes at a concentration at 1 μg/μL and 5 μg/μL, separately. Then NSPCs cells were added to the dishes in DMEM/F-12 medium supplemented with 2% B27, and RA (retinoic acid, Sigma) with forskolin (Sigma) at the final concentration of 1 μM and 5 μM, separately.
Immunostaining
The cells or brain sections were washed three times with cold PBS and blocked by adding 3% FBS and 0.1% Triton-X-100 in PBS for 1 h followed by adding primary antibody in 3% FBS diluted with PBS for 2 hours at room temperature or overnight at 4 °C. Then cells were washed 3 times with PBS and the fluorophore-conjugated secondary antibody and DAPI were added to cells for 1 h. In order to stain BrdU, 1 M HCl was used to treat samples for 30 min and then immunostaining process was conducted. The antibodies were all listed in Supplementary Table 1.
Co-immunoprecipitation (co-IP) and immunoblotting analysis
Cells were lysed using the Co-IP lysis buffer (30 mM HEPES, 85 mM KCl, 0.5% NP-40 and EDTA-free protease inhibitors, pH 7.4) on ice for half an hour and then sonicated. After concentration at 12,000 g, the supernatant was collected and incubated with anti-Flag-M2 beads (Thermo) at 4 °C overnight. After washed three times with Co-IP lysis buffer, flag peptide (GeneScript) was added to anti-Flag-M2 beads for 6 hours and the elution was detected by Western blotting.
Dot blotting assay
Synthetic peptides were incubated with full length His-tagged Mina53 purified from E. coli, and cofactors for 2 h at 37 °C. Reactions were quenched using 2 mM EDTA and then desalted with Sep-Pak C18 column (Waters). Peptides were eluted from the column using 30% acetonitrile containing 0.01% TFA. After lyophilization, peptides were resolved in water and further detected with indicated histone methylation antibodies. Synthetic peptides were listed in the Supplementary Table 2.
Quantitative PCR (qPCR)
Total RNA was extracted using TRIzol reagent (Sigma) and then the concentration of mRNA was quantified by Thermo Scientific NanoDrop 2000c. After reverse transcription using HiScript IV 1st Strand cDNA Synthesis Kit (Vazyme), the relative expression level of related genes was detected by RT-PCR. Sequence of relative primers used in this article were provided in Supplementary Table 3.
Synthesis of Fmoc-pLeu-OH
The commercial ketone ((S)-2-((tert-butoxycarbonyl)amino)-4-oxopentanoic acid) (4.99 g, 21.6 mmol) was dissolved in anhydrous methanol (100 mL), and then ammonia (7 M in methanol, 24.1 mL, 168.5 mmol) was added slowly with syringe at −30 °C, the reaction mixture was stirred at −30 °C and 0 °C for another 2 h, respectively. After finish of this step, the reaction mixture was cooled to −15 °C, followed by slow addition of hydroxylamine-O-sulfonic acid (2.93 g, 25.9 mmol) in anhydrous methanol (10 mL) with syringe. The reaction mixture warmed up to room temperature and stirred for 14 hours. After completion of this reaction, precipitate was removed by filtration and the solvent was removed under vacuum to obtain the crude. Anhydrous methanol (50 mL) was added to redissolve the crude, followed by addition of triethylamine (6.01 mL, 43.2 mmol). Iodine in anhydrous methanol was added to the reaction mixture at 0 °C until the brown color persisted, stirring at room temperature for 2 hours. After finished of this reaction, it was concentrated under vacuum, extracted with DCM, reduced with Na2S2O3, washed with brine, and dried with Na2SO4 to obtain the intermediate (Boc-protected diazirine). The intermediate (6.2 g) was dissolved in TFA/DCM (1:1, 40 mL) and stirred at room temperature for 2 h. The solvent was removed under vacuum and the residue was redissolved in water/dioxane (1:2, 60 mL), followed by adding NaHCO3 (496 mg, 5.90 mmol). Fmoc-OSu (1.99 g, 5.90 mmol) was dissolved in dioxane (8 mL) and added slowly to the reaction mixture with syringe over 30 min, the mixture was stirred at room temperature for 24 h. After completion of this reaction, the solvent was removed under vacuum and redissolved in water (50 mL). The mixture was adjusted to be pH 3-4 with 4 M HCl, extracted with ethyl acetate, washed with saturated brine, dried with Na2SO4, and purified with flash column chromatography (DCM/methanol, 10:1) to obtain the white solid (915 mg) at 10.2% yield.
1H-NMR (500 MHz, MeOD): δ 7.79 (d, J = 7.5 Hz, 2H), 7.71 (dd, J = 7.0, 5.0 Hz, 2H), 7.38 (t, J = 7.5 Hz, 2H), 7.29 (t, 7.5, 2H), 4.40 (dd, J = 10.5, 7.0 Hz, 1H), 4.33 (dd, J = 10.5, 7.0 Hz, 1H), 4.25 (t, J = 7.0 Hz, 1H), 4.16 – 4.06 (m, 1H), 2.00 (dd, J = 15.0, 4.5 Hz, 1H), 1.63 (dd, J = 15.0, 11.0 Hz, 1H), 1.04 (s, 3H).
13C-NMR (126 MHz, MeOD): δ 157.03, 143.91, 143.82, 141.19, 127.38, 126.77, 124.95, 124.93, 119.51, 66.70, 50.30, 36.31, 23.64, 18.52.
HR-MS: [M + H]+ m/z 366.1435 (Calculated:366.1454), [M+Na]+ m/z 388.1277 (Calculated: 388.1273).
Peptide synthesis
Synthesis of peptides was performed based on standard fluorenyl methoxycarbonyl (Fmoc)-solid-phase peptide synthesis (SPPS) by using a Liberty BlueTM Automated microwave peptide synthesizer (CEM Corp., U.S.A.). Firstly, the Rink Amide-AM resin (0.1 mmol) was bubbled with N2 in DMF for 10 min at room temperature. Secondly, Fmoc- group was removed with a solution of 4-methylpiperidine (3 mL, vol/vol, 20% in DMF) for 1 min at 90 °C. Thirdly, Fmoc-AA-OH was coupled to the resin at 80 °C for 4 min in the presence of 5 equivalents of DIC and 10 equivalents of Oxyma. After completion of the peptide, the resin was washed with DMF, DCM and MeOH in sequence and air-dried under room temperature.
The air-dried peptides were cleaved from resin with cocktail (4 mL, vol/vol, TFA: TIPS: phenol: water, 88:2:5:5) for 3 h at room temperature. The resin was removed and cold diethyl ether (36 mL) was added to the filtrate to precipitate the peptide, followed by centrifuge. The sediment was washed with cold diethyl ether (40 mL) again and dried under room temperature to obtain the crude. The crudes were purified by HPLC, lyophilized to powder and stored at −20 °C until usage.
Biotin-pLeu-H4R3me2a, sequence: pLeuGRme2aGKGGKGLGKGGAKKbio-NH2,
[M + H]+ calculated for 1835.15, found 1835.93; [M + H-N2]+ calculated for 1807.15, found 1807.92.
Biotin-H4R3me2a, sequence: SGRme2aGKGGKGLGKGGAKKbio-NH2,
[M + H]+ calculated for 1796.15, found 1795.90.
Formaldehyde releasing assay
To examine the conversion of the methyl group into formaldehyde, assays were carried out in the abovementioned reaction buffer, and analyzed by the Hantzsch reaction using AAPT, a modified NASH method. Briefly, after TCA precipitation and the addition of 1% Tris buffer, equal volume of AAPT buffer was added to the supernatant and the mixtures were incubated at room temperature for 30 min. Signals were detected using a PHERAstar FS (BMG Labtech) plate reader under 355 nm excitation and 470 nm emission. As to calculate the concentration of formaldehyde, 37% formaldehyde were used as the standard sample and the absorbances of a series of formaldehyde buffer with different concentrations were used to generate the standard curve. The Km and Kcat values were calculated according to the Michaelis-Menten equation.
Analysis of in vitro demethylation reaction products
C18 ZipTip (Waters) was used to desalt the demethylation reaction mixture. The C18 ZipTips were activated using 80% acetonitrile with 0.1% TFA (trifluoroacetic acid), then equilibrated with 0.1% TFA. TFA and acetonitrile were separately added to the mixture to a final concentration of 0.2% and 2%, and then the reaction mixture was added to the C18 ZipTips. The C8 ZipTips were washed with 0.1% for three times and the bound peptides were eluted from the C18 ZipTips with 30% acetonitrile. After lyophilization, the eluted peptides were detected by LC-MS. The peptides were separated by a 10 min gradient elution at a flow rate 0.300 µL/min with the Shimadzu LC-40 UPLC System during the during the HPLC process. Then separated peptides is injected into the SCIEX ZenoTOF 7600 mass spectrometer with an ACQUITY UPLC HSS T3 (2.1 × 100 mm, 1.8 μm particle size)-type analytical column. 0.1% formic acid in water was used as mobile phase A and 100% acetonitrile and 0.1% formic acid was used as mobile phase B. The peptides were ionized with Turbo V ion source (ESI) in positive ion polarity mode. Ionization source conditions: Spray voltage +5500 V; source temperature 500 °C, ion source gas 1 pressure 45 psi, curtain gas pressure 35 psi, ion source gas 2 pressure 45 psi, CAD gas pressure 7 psi. The mass spectrometer was operated in the TOF MS mode and the scan range was 400-1500 m/z. The relative abundance of peptides was analyzed and output using the SCIEX OS 3.1 software.
Lentivirus production and infection
For lentiviral infection of NSPCs, lentiviruses containing scramble or shRNAs targeting Mina53 (Mutiplicity of Infection, MOI = 10) were obtained from the OBiO Tech Corp. of Shanghai. NSPCs were plated in a 6-well plate at a density of 5 × 105 cells, to which lentiviruses were added 6 h later. After 24 h, the medium was half-replaced and then the lentiviruses were supplemented one more time. Cells were collected at the indicated time points for subsequent assays.
For lentiviral infection of HEK293T and Neuro2A cells, cells were plated into the 6 cm culture dish prior to infection. Lentiviruses targeting Mina53 and JMJD1C were added when the cell density reached 30% confluency. The medium was replaced 6-8 h post infection. Cells were collected 72 h after the infection. Sequences of shRNAs have been provided in Supplementary Table 4.
Neurosphere formation assay
A 70 μm cell strainer (FALCON, Durham, USA) was used to filtered out the NSPCs to dissociate to single cells. Single-cell suspension was added into a 96-well plate. Images were captured by using a Nikon inverted microscope on days 7 and 14. The fresh medium was replaced every 3 days. ImageJ was used to analyze the cross-sectional area of the neurospheres.
In vitro and in vivo neurogenesis assays
NSPCs were cultured on pre-coated coverslips in the proliferation medium containing 5 μM BrdU for 8 h for the in vitro proliferation assay and then fixed by adding 4% paraformaldehyde. Then the immunostaining process was performed as described below.
As for the evaluation of the proportion of in vivo proliferation and differentiation of NSPCs, BrdU was injected to mice every 4 h for 6 times (50 mg/kg, i.p.) and then mice were sacrificed for detect the differentiation of NSPCs,.
As for the detection of NSPCs, BrdU (50 mg/kg, i.p.) was injected to mice every 8 h for 6 times. 4 weeks later, mice were sacrificed and the brains wee collected and dehydrated by using 30% sucrose. Cryostat (Leica, CM1950) was used to prepare cryo-section in the coronal plane of these brain samples. 4–6 sections per brain was analyzed for immunostaining related to BrdU, DCX and NeuN staining.
Recombinant expression of Mina53 in E. coli
The expression construct pET28a-His6-Mina53 was transfected into E. coli BL21 (DE3), and cells were cultured at 37 °C to an OD 600nm ≈ 0.6 in LB broth. IPTG (0.25 mM final concentration) was added to LB to induce the expression of Mina53 at 14 °C for 16 h. Then E. coli BL21 was collected and centrifuged. After these processes, PBS buffer containing 20 mM imidazole was used to resuspended these cells. After sonication (30% amplitude, 5 s on/10 s off, 99 cycles), the soluble fraction was collected and incubated with Ni2+-NTA agarose column. After washed 3 times with the binding buffer (30 mM Hepes, 150 mM NaCl, 30 mM imidazole, pH7.5), proteins were eluted from the Ni2+-NTA agarose column using the elution buffer (30 mM Hepes, 150 mM NaCl, 300 mM imidazole, pH7.5). Microsep was used to concentrate purified proteins.
Crosslinking of PRMT1 and Mina53 with the pLeu containing peptide
The pLeu containing peptides (final concentration 0.6 μM) were resuspended in 100 μL cross-linking buffer (20 mM HEPES, 1.5 mM MgCl2, 150 mM KCl, EDTA-free protease inhibitors, pH 7.5) containing 6 μM recombinant PRMT1 or Mina53. The mixtures were incubated at 37 °C for 10 min before irradiation with 365 nm UV light for 10 min on ice. Then, the binding buffer (25 mM Hepes, 150 mM NaCl, 0.5% v/v NP-40, 1 mM DTT, pH 7.6) and Streptavidin Agarose Resin (Thermo scientific) were added into the mixture for immunoprecipitation. The IP was performed at 4 °C overnight. Three times of binding buffer, two times of 0.5% SDS in PBS, two times of 1 M NaCl in PBS, two times of tris-buffered saline was successively used to wash the beads. Finally, beads were boiled for 20 min in the SDS loading buffer. Samples were analyzed by Western blotting.
Analysis of crosslinking products by LC-MS
HEK293T cells were lysed in RSB buffer (10 mM HEPES, 150 mM NaCl, 0.2% NP-40, Protease Inhibitor Cocktail, pH 7.5) for 10 min 4 °C. After centrifugation, the crude nuclei were isolated and resuspended with cross-linking buffer (100 mM HEPES, 1.5 mM MgCl2, 150 mM KCl, EDTA-free protease inhibitors, pH 7.5). Then pLeu containing peptides were resuspended in the nuclei at a concentration of 100 μM. The mixtures were then incubated at 4 °C for 20 min protected from light, and followed by irradiating using UV at 365 nm for 15 min on ice. After centrifugation at 200 g for 5 min, the nuclei were washed with cross-linking buffer and resuspended in 500 μL binding buffer (25 mM tris, 300 mM KCl, 0.1% v/v NP-40 alternative, 0.5 mM DTT, pH 7.5). After sonication (30 W amplitude, 5 s on/10 s off, 12 cycles), Streptavidin Agarose Resin were added. The immunoprecipitation was performed at 4 °C for 2 h. Next, beads were washed using 3x binding buffer, 2 × 1 M NaCl in 50 mM Tris (pH7.5), 1 × 2 M urea in 50 mM Tris (pH7.5), 1 × 1 mM EDTA in 50 mM Tris (pH7.5), 1 × 50 mM Tris (pH7.5), sequentially. The bound proteins were subjected to on-bead tryptic digestion, and protein identification by LC-MS.
Peptides were separated with a 120 min gradient elution at a flow rate 0.300 µL/min with the Thermo Vanquish Neo integrated nano-HPLC system followed by Thermo Exploris 480 mass spectrometer detection. There is an analytical column in the spectrometer (75 µm ID, 150 mm length; Upchurch, Oak Harbor, WA, C-18 resin packed in it). 0.1% formic acid in water was used as mobile phase A and 80% acetonitrile with 0.1% formic acid was used as mobile phase B. Xcalibur 4.1 software was inset to operate the mass spectrometer. The AGC target was set as 5e4, and the maximum injection time was 50 ms. Thermo Xcalibur Qual Browser and Proteome Discoverer were used to analyze collected mass spectrum data for the database searching against UniProtKB (UP000005640). The search parameters of Sequest were set as follows: a 10 ppm precursor mass tolerance and 0.02 Da fragment ion tolerance, and up to 2 internal cleavage sites. For protein quantification, re-quantification was enabled and the minimum ratio count was set to one. Only proteins quantified in both experimental replicates were used for next analyze and protein ratios were calculated with values from experimental replicates. Two-tailed unpaired Student’s t-test was sued to calculate the P values.
Only proteins with a ratio greater than 5.0 and a P-value less than 0.05 were defined as significant. And the volcano plot was created by GraphPad prism software.
Microscale thermophoresis (MST)
Monolith NT.115 instrument (Nano Temper Technologies) was used to perform MST assay. His-tagged Mina53 was diluted to indicated concentrations (from 3 nM to 0.1 mM), and incubated with 25 nM of synthetic FITC-H4R3me2a peptide for 15 min in MST buffer (30 mM Hepes, 150 mM NaCl, pH7.5). Then, relative mixtures were loaded into standard glass capillaries (Monolith NT Capillaries, Nano Temper Technologies) to perform the MST assay. NanoTemper software was used to calculate the KD values.
CUT&Tag assay
HyperactiveTM In-Situ ChIP Library Prep Kit from Illumina (Vazyme Biotech, China) was used to perform CUT&TAG assay. First, collected cells were incubated with ConA beads. Then digitonin was added to the mixture to permeate cell membrane, which was followed by incubation with primary H4R3me2a antibody (Active motif) for 2 h at room temperature. After washed with binding buffer, the secondary IgG antibody was added for 1 h. Next, the hyperactive pA-Tn5 transposase was added to the mixture and then Tn5 transposase was used to linking DNA fragments with P5 and P7 adapters for further amplified by PCR using P5 and P7 primers. After this process, libraries were sequenced on the Illumina NovaSeq6000 platform, and 150 bp paired-end reads were generated. The sequence data was analyzed by fatsp software to produce the clean reads. Then Bowtie2 was used to aligned the clean reads to the Mus musculus reference genome mm39. Then the data was further analyzed using SEACR software. Finally, Genomics Viewer (IGV) was used to visualize the peak distribution along genomic regions and ChIPseeker was used to find peak distribution and annotation.
As for further analyze, Manorm software was used to find different peaks between case and control groups. The significantly different peaks were identified as p < 0.05 and |log2(fold change)|å 1.
RNA-seq
RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA) was used to assed the amounts and integrity of RNA. Extracted RNA from each sample was used at same amount (3 μg) which was determined as input. AMPure XP beads was used to purify PCR product derived from the input sample to generate library. Then the library preparations were sequenced by using an Illumina Hiseq platform (Illumina NovaSeq6000). Raw data (raw reads) of fastq format were processed and Hisat2 v2.0.5 was used to align clean data to the Mus musculus reference genome mm10. FPKM was used to find genes with different expression levels, p < 0.05 and |log2(fold change)|å 0.5 were set as the threshold for significantly differential expression.
Animals
All animal procedures used in this study were performed according to the protocols approved by the Institutional Animal Care and Use Committee of Zhejiang University with the permission number 25115. NestinCreERT2 mice (C57BL/6 J) and Mina53flox/+ mice (C57BL/6 J) were purchased from the Jackson Laboratory and the Cyagen Laboratory, separately. Mina53flox/flox mice were crossed with NestinCreERT2 mice to generate NestinCreERT2;Mina53flox/flox transgenic mice. The genotypes of mice were confirmed before use and related primers was listed in Supplementary Table 5. Only male mice were used for this study.
Behavioral test
Eleven adult (8-week-old) Mina53flox/flox mice or NestinCreERT2;Mina53flox/flox mice were injected with tamoxifen (SIGMA) (100 mg/kg, i.p. once/day, for 5 consecutive days), respectively. Four weeks post the first tamoxifen injection, the behavioral tests were performed.
The Morris water maze (MWM) test was conducted in a circular pool (with a diameter of 120 cm) filled with water to make the platform invisible. MWM test consisted of two stages: training and testing. During the training stage, mice were trained in the water maze to find the hidden platform (with a diameter of 10 cm) from four different starting positions (northeast, northwest, southeast, southwest). If mice failed to find the platform, experimenters will place them on the platform. This process remained for 10 s. As for each mouse, this test was performed four trials every day and the duration were four days. One day after training, the platform was removed to perform the probe trial. There are 2 min for the mice to search in the pool and MazeScan software (Acimetrics) was used to analyze the time to platform, quadrant time, number of platform crossings, traveled total distance, and average speed of each mouse which had been videotaped.
In the New object recognition test (ORT), mice were individually placed in the testing box (with a length, width and height of 40 cm, 40 cm, 40 cm, separately) for habituation before the test. On the training day, mice received 10 min to explore two objects of the same shape which were placed in the center of the chamber. On the testing day, the memory of each mice was tested by placing them back into the chamber again with two objects (a familiar one from the previous test and a novel one with a different shape). During the testing, the condition including the spatial ___location for the objects remained the same. 70 % ethanol was used to clean the box after each test to avoid odor cues.
The Y-maze has three equal-length arms (40 cm long and 9 cm wide with 16 cm-high walls) which are symmetrically disposed at 120° angle from each other. There were 8 min for mice to explore three arms freely.
To evaluate the locomotion of animals in the Y-maze, the number of arm entries of each mice was recorded. Spontaneous alternation was defined as total alternations divided by the number of maximum alternations (number of arms entered-2).
Molecular modeling, docking and molecular dynamics simulations
The Mina53 complex was constructed by docking a histone 4 arginine 3 (H4R3) peptide containing an asymmetrically di-methylated arginine (Rme2a) onto the crystal structure of Mina53 (PDB ID: 4BXF) using AutoDock Vina34,53. Missing residues were modeled based on the predicted structure from AlphaFold254. Mn(II) and 2-oxoglutarate (2OG) were retained in the active site throughout the simulations. The complex was placed in a periodic cubic box with dimensions of 10.0 nm, solvated with TIP3P water molecules, and ions (Na+ and Cl−) were added to a concentration of 0.15 M, resulting in ~90,000 atoms in total. All simulations were conducted using the CHARMM36m force field55. After energy minimization, the system underwent equilibration through 1 ns NVT and 1 ns NPT simulations. A production simulation followed, maintaining the temperature at 300 K using the v-rescale thermostat. Neighbor searching was performed every 5 steps, and electrostatic interactions were calculated using the PME algorithm with a 1 nm cut-off, using a reciprocal grid of 80 × 80 × 80 cells and 4th-order B-spline interpolation. A 1 nm cut-off was applied for Van der Waals interactions, and hydrogen mass repartitioning, with a single LINCS iteration (expansion order 4), allowed for a 4 fs integration time step. A 500 ns MD simulation was first conducted to assess the stability of the docked structure. This was followed by a 250 ns restrained MD simulation, where a harmonic potential was applied to the sidechain of the peptide di-methylated arginine to hold its position within the binding site, allowing the rest of the peptide to freely explore the conformational space. The optimized structure obtained from this simulation was found to be highly stable in a subsequent 2000 ns unbiased MD simulation. The final frame of this simulation was used to analyze interactions between Mina53 and the peptides. To explore Mina53’s specificity for the R3me2a modification, models were generated for other arginine modifications (monomethylated H4R3me1, symmetrically dimethylated H4R3me2s, and unmodified H4R3me0) using CHARMMGUI’s non-standard amino acid substitution module56. Two replicate simulations were performed for each system, with 2 μs per repeat for H4R3me0 and H4R3me1, and 1 μs per repeat for H4R3me2s, totaling 15 μs of simulation time.
All MD simulations were run using Gromacs 2020.657. The PLUMED driver was used to analyze the distance between the center of mass of the R3 sidechain and the p53 binding pocket for each peptide (H4R3me0, H4R3me1, and H4R3me2s)58. Structural illustrations were prepared using PyMol (The PyMOL Molecular Graphics System, Version 2.5 Schrödinger, LLC).
Statistics and reproducibility
Statistical analyses and representations were performed using GraphPad Prism 8.0.2. Detailed information regarding the nature of entity for “n”, exact n values for each group and experiments, test statistics method and displayed error bars are described in related figure legends to determine the significance between group comparisons. N value of each experiments presented in each provided graph is equal or greater than 3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD045247. The RNA-seq and CUT&Tag data generated in this study have been deposited to the NCBI Gene Expression Omnibus (GEO) datasets with the accession number GSE275270 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi). Imaging files have been deposited to the Zenodo database with following links: https://doi.org/10.5281/zenodo.14176224. All the other data which support the findings of this study are available with in the article, supplementary information and source data. Source data are provided with this paper.
References
Blanc, R. S. & Richard, S. Arginine methylation: the coming of age. Mol. Cell 65, 8–24 (2017).
Chang, K. et al. Critical roles of protein arginine methylation in the central nervous system. Mol. Neurobiol. 60, 6060–6091 (2023).
Di Lorenzo, A. & Bedford, M. T. Histone arginine methylation. FEBS Lett. 85, 2024–2031 (2011).
Bedford, M. T. & Clarke, S. G. Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 (2009).
Guccione, E. & Richard, S. The regulation, functions and clinical relevance of arginine methylation. Nat. Rev. Mol. Cell Biol. 20, 642–657 (2019).
Wang, W. et al. Asymmetrical arginine dimethylation of histone H4 by 8-oxog/OGG1/PRMT1 is essential for oxidative stress-induced transcription activation. Free Radic. Biol. Med. 164, 175–186 (2021).
Yu, W. et al. Histone tyrosine sulfation by SULT1B1 regulates H4R3me2a and gene transcription. Nat. Chem. Biol. 19, 855–864 (2023).
Liu, X. et al. Arginine methylation of METTL14 promotes RNA N(6)-methyladenosine modification and endoderm differentiation of mouse embryonic stem cells. Nat. Commun. 12, 3780 (2021).
Wu, Q. et al. Protein arginine methylation: from enigmatic functions to therapeutic targeting. Nat. Rev. Drug Discov. 20, 509–530 (2021).
Dai, W. et al. Protein arginine methylation: an emerging modification in cancer immunity and immunotherapy. Front. Immunol. 13, 865–964 (2022).
Gayatri, S. & Bedford, M. T. Readers of histone methylarginine marks. Biochim. Biophys. Acta 1839, 702–710 (2014).
Chen, C. et al. Deciphering arginine methylation: Tudor tells the tale. Nat. Rev. Mol. Cell Biol. 12, 629–642 (2011).
Simcikova, D., Gelles-Watnick, S. & Neugebauer, K. M. Tudor-dimethylarginine interactions: the condensed version. Trends Biochem Sci. 48, 689–698 (2023).
Wang, H. et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science 293, 853–857 (2001).
Huang, S., Litt, M. & Felsenfeld, G. Methylation of histone H4 by arginine methyltransferase PRMT1 is essential in vivo for many subsequent histone modifications. Genes Dev. 19, 1885–1939 (2005).
Li, X. et al. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 115, 2028–2037 (2010).
Beacon, T. H., Xu, W. & Davie, J. R. Genomic landscape of transcriptionally active histone arginine methylation marks, H3R2me2s and H4R3me2a, relative to nucleosome depleted regions. Gene 742, 1445–1493 (2020).
Yao, B. et al. PRMT1-mediated H4R3me2a recruits SMARCA4 to promote colorectal cancer progression by enhancing EGFR signaling. Genome Med. 13, 58 (2021).
Yang, Y. et al. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell 40, 1016–1023 (2010).
Yang, Y. et al. Arginine methylation facilitates the recruitment of TOP3B to chromatin to prevent R loop accumulation. Mol. Cell 53, 484–497 (2014).
Yuan, W. et al. TDRD3 promotes DHX9 chromatin recruitment and R-loop resolution. Nucleic Acids Res. 49, 8573–8591 (2021).
Wang, L., Wen, M. & Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science, 365, eaav0758 (2019).
Chang, B. et al. JMJD6 is a histone arginine demethylase. Science 318, 444–447 (2007).
Li, S. et al. JMJD1B demethylates H4R3me2s and H3K9me2 to facilitate gene expression for development of hematopoietic stem and progenitor cells. Cell Rep. 23, 389–403 (2018).
Walport, L. J. et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat. Commun. 7, 11974 (2016).
Bonnici, J. et al. The catalytic domains of all human KDM5 JmjC demethylases catalyse N-methyl arginine demethylation. FEBS Lett. 597, 933–946 (2023).
Liu, H. et al. Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proc. Natl Acad. Sci. USA 114, 7717–7726 (2017).
Webby, C. J. et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science 325, 90–93 (2009).
Wang, F. et al. JMJD6 promotes colon carcinogenesis through negative regulation of p53 by hydroxylation. PLoS Biol. 12, 1001819 (2014).
Cockman, M. E. et al. Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6. Proc. Natl Acad. Sci. USA 119, 2201483119 (2022).
Burton, A. J. et al. In situ chromatin interactomics using a chemical bait and trap approach. Nat. Chem. 12, 520–527 (2020).
Chen, B. et al. Mdig de-represses H19 large intergenic non-coding RNA (lincRNA) by down-regulating H3K9me3 and heterochromatin. Oncotarget 4, 1427–1437 (2013).
Huang, H. et al. A CRISPR/Cas9 screen identifies the histone demethylase MINA53 as a novel HIV-1 latency-promoting gene (LPG). Nucleic Acids Res. 47, 333–347 (2019).
Zheng, L. et al. JMJD1B mediates H4R3me2s reprogramming to maintain DNA demethylation status in neural progenitor cells during embryonic development. Cell Insight 2, 100114 (2023).
Lu, Y. et al. Lung cancer-associated JmjC ___domain protein mdig suppresses formation of tri-methyl lysine 9 of histone H3. Cell Cycle 8, 2101–2109 (2009).
Chowdhury, R. et al. Ribosomal oxygenases are structurally conserved from prokaryotes to humans. Nature 510, 422–426 (2014).
Liu, D. et al. Coating materials for neural stem/progenitor cell culture and differentiation. Stem Cells Dev. 29, 463–474 (2020).
Zhang, Z. et al. Histone methylations define neural stem/progenitor cell subtypes in the mouse subventricular zone. Mol. Neurobiol. 57, 997–1008 (2020).
Brennand, K. J. & Talkowski, M. E. Xenopus models suggest convergence of gene signatures on neurogenesis in autism. Neuron 109, 743–745 (2021).
Packer, A. Neocortical neurogenesis and the etiology of autism spectrum disorder. Neurosci. Biobehav. Rev. 64, 185–195 (2016).
Ribeiro, F. F. & Xapelli, S. An overview of adult neurogenesis. Adv. Exp. Med Biol. 1331, 77–94 (2021).
Kempermann, G. et al. Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23, 25–30 (2018).
Szulwach, K. E. et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189, 127–141 (2010).
Wakabayashi, T. et al. MicroRNAs and epigenetics in adult neurogenesis. Adv. Genet 86, 27–44 (2014).
Anacker, C. & Hen, R. Adult hippocampal neurogenesis and cognitive flexibility—linking memory and mood. Nat. Rev. Neurosci. 18, 335–346 (2017).
Guo, J. U. et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351 (2011).
Yao, B. et al. Epigenetic mechanisms in neurogenesis. Nat. Rev. Neurosci. 17, 537–549 (2016).
Berg, D. A. et al. A common embryonic origin of stem cells drives developmental and adult neurogenesis. Cell 177, 654–668 (2019).
Ziffra, R. S. et al. Single-cell epigenomics reveals mechanisms of human cortical development. Nature 598, 205–213 (2021).
Lithopoulos, M. A. et al. Neonatal hyperoxia in mice triggers long-term cognitive deficits via impairments in cerebrovascular function and neurogenesis. J. Clin. Investig. 132, e146095 (2022).
Yoshikawa, Y. et al. Nox4 promotes neural stem/precursor cell proliferation and neurogenesis in the hippocampus and restores memory function following trimethyltin-induced injury. Neuroscience 398, 193–205 (2019).
Moreno-Jimenez, E. P. et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 25, 554–560 (2019).
Eberhardt, J. et al. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model 61, 3891–3898 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Jo, S. et al. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput Chem. 29, 1859–1865 (2008).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2, 19–25 (2015).
The PLUMED consortium. Promoting transparency and reproducibility in enhanced molecular simulations. Nat. Methods, 16, 670–673 (2019).
Acknowledgements
We thank Dr. Qingyun Yang and the Mass Spectrometry & Metabolomics Core Facility at Westlake University for the proteomic study. We thank the excellent technical support from the core facility of Central Laboratory at the First Affiliated Hospital of Zhejiang University. This work was supported by the Fundamental Research Funds for Central Universities (K20220228, 226-2024-00055 to W.Y. and Y.W.), the National Natural Science Foundation of China (NSFC, grant nos. 22325704, 92353303, 32271331 to W.Y.; 92049108, 82371182 to X.L., 32371300 to Y.W.), the National Key R&D Program of China (2021YFF1200404 to Y.W.).
Author information
Authors and Affiliations
Contributions
W.Y. conceived the project, and designed cell biology and biochemistry experiments; X.L. designed experiments related to transgenic mice; L.Z., J.S., X.Z., Y.L., X.H., and Y.Y. performed cell biology, biochemistry and animal behavioral experiments; K.Z., and M.W. performed mass spectrometry analysis; Y.W. performed molecular dynamic simulations experiments; W.Y., M.W., Y.W., and X.L. analyzed the data; W.Y. wrote the paper with inputs from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Netty Santoso and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, L., Zhao, X., Sun, J. et al. Mina53 demethylates histone H4 arginine 3 asymmetric dimethylation to regulate neural stem/progenitor cell identity. Nat Commun 15, 10227 (2024). https://doi.org/10.1038/s41467-024-54680-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54680-6