Abstract
Single-cell RNA sequencing (scRNA-seq) has advanced our understanding of cell types and their heterogeneity within the human liver, but the spatial organization at single-cell resolution has not yet been described. Here we apply multiplexed error robust fluorescent in situ hybridization (MERFISH) to map the zonal distribution of hepatocytes, spatially resolve subsets of macrophage and mesenchymal populations, and investigate the relationship between hepatocyte ploidy and gene expression within the healthy human liver. Integrating spatial information from MERFISH with the more complete transcriptome produced by single-nucleus RNA sequencing (snRNA-seq), also reveals zonally enriched receptor-ligand interactions. Finally, MERFISH and snRNA-seq analysis of fibrotic liver samples identify two hepatocyte populations that expand with injury and do not have clear zonal distributions. Together these spatial maps of the healthy and fibrotic liver provide a deeper understanding of the cellular and spatial remodeling that drives disease which, in turn, could provide new avenues for intervention and further study.
Similar content being viewed by others
Introduction
The liver is composed of parenchymal, non-parenchymal, and immune cells that are organized into anatomic structures called lobules, which are 0.5–1 mm in diameter and defined by sites of portal venous and arterial inflow and central venous outflow1,2. Within the lobule, hepatocytes are classically organized into three zones, with zone 1 proximal to the portal vein and hepatic artery, zone 3 proximal to the central vein, and zone 2 in the intermediate region3,4. Hepatocyte zonation matches function to the physiologic environment in both space and time5. For example, hepatocytes in zone 1 are more involved in gluconeogenesis and beta-oxidation, reflecting the relatively oxygen- and nutrient-rich environment near the portal vein and hepatic artery, whereas hepatocytes in zone 3 are more active in glycolysis and lipogenesis, reflecting the depletion of oxygen and nutrients near the central vein3,4,6. Beyond metabolism, morphogen expression, such as that of Wnt-family members, is also shaped by this zonation and can vary in time7. Thus, this zonal organization is critical for our understanding of hepatocyte activity. However, many details of this zonation, including how hepatocytes adjust their gene expression along the portal-central vein axis and the function of this potential spatial fine tuning remain unclear in the human liver, as current single-cell analyses of the human liver have either measured gene expression without spatial data or evaluated spatial data without single-cell resolution.
The liver is also composed of non-parenchymal cells, which include hepatic stellate cells (HSCs), resident macrophages, endothelial cells (ECs), and cholangiocytes8,9. While sub-populations of macrophages and HSCs in the healthy human liver have been described in single-cell data10,11,12, these subpopulations, and potential gene expression variations within, have not been resolved within the lobule organization at single-cell resolution in the human liver. As such, their role within the spatial context of the liver also remains unclear.
In addition to spatial ___location, another feature of hepatocytes that may drive functional heterogeneity is their ploidy13. Specifically, hepatocytes in the adult liver can either be single or multinucleated, and nuclei themselves can vary in ploidy. Early in life, most human hepatocytes are diploid and contain a single nucleus; ploidy increases with age such that in adults, about a third of hepatocytes are multinucleated, or their nuclei contain more than two copies of each chromosome14,15,16. Yet the spatial organization of multinucleated hepatocytes and the impact of nuclear content on gene expression remains poorly understood17.
Here, we apply image-based spatial transcriptomics (multiplexed error robust fluorescence in situ hybridization; MERFISH) and single-nucleus RNA sequencing (snRNA-seq) to the same samples of healthy human liver in order to construct spatial maps of hepatocytes and non-parenchymal cells at single-cell and transcriptome-wide resolution. This analysis allowed us to define gradients of gene expression across human hepatocyte zones, determine the cell types where receptor-ligand co-expression are in spatial proximity to promote crosstalk, and evaluate the relationship between ploidy and gene expression within hepatocytes. We also performed similar measurements in fibrotic liver samples to understand the changes in hepatocytes and non-parenchymal cells that occur with chronic injury. These analyses provide a single-cell spatial transcriptomic map of the human liver, annotating the gradient in hepatocyte gene expression from the portal area to central vein and show that hepatocyte ploidy is equally distributed across zones within the lobule and does not affect differential gene expression. In addition, we define subsets of spatially distinct macrophages and HSC populations, and identify the expansion of two hepatocyte populations with chronic injury, which together provide an approach and framework to understand changes that occur with human liver disease at single-cell resolution in space. More broadly, the framework we provide for the joint spatial and transcriptomic analysis of single cells within the healthy and diseased human liver may prove useful for the further study of the molecular, cellular, and tissue remodeling that occurs in human liver disease.
Results
Spatial organization of hepatocytes in the human liver
To explore the spatial and cellular organization of the human liver, we collected human liver tissue from the margins of surgical resections of three adult patients (two female and one male; Supplementary Table 1). While few patients undergo liver resection without suspicion of disease, the absence of clear signs of acute or chronic injury in H&E images supported our assignment of these samples as ‘Healthy’ (Supplementary Fig. 1 and Supplementary Table 1). We designed a MERFISH panel targeting 317 genes with a focus on hepatocytes (Methods; Supplementary Data 1), and then characterized the expression of these genes within cryosections taken from these samples using MERFISH18,19,20.
The distribution of key individual genes revealed the rich spatial architecture of the liver and provided an initial validation of our measurements. We observed sheets of cells expressing ALDOB, consistent with hepatocytes21 (Supplementary Fig. 2), while CD5L expression was clustered in spaces between hepatocytes consistent with resident macrophages located in sinusoids10 (Supplementary Fig. 2a–f). PDGFRB expression was concentrated along the edges of ALDOB-expressing hepatocytes in a distribution expected for HSCs, which are located in the subendothelial space11 (Supplementary Fig. 2a–f). KRT7 and SOX9 were enriched in periportal areas consistent with cholangiocytes and include clusters suggestive of longitudinal sections of small bile ducts (Supplementary Fig. 2g–l), while DNASE13 and INMT expression were scattered through the parenchyma consistent with the distribution of ECs10 (Supplementary Fig. 2g–l).
To define individual cells within our data, we included in our MERFISH measurements an immunofluorescence stain against a pan-cell surface marker, the Na+/K+-ATPase22. This antibody stain was incorporated into our MERFISH readout through the use of an oligonucleotide-tagged secondary antibody22 (Methods). We then leveraged Cellpose23 to define cell boundaries in three dimensions from these co-stains, assigned RNAs within these boundaries, and applied an RNA-based segmentation routine, Baysor22, to adjust and improve these boundaries and recover cells for which the co-stains did not provide clear boundaries (Supplementary Fig. 3). Following this analysis and removal of cells with small numbers of transcripts, we compiled a dataset of ~90,000 putative cells from the healthy human liver (Supplementary Fig. 4).
The expression of all 317 genes was quantified in each cell to generate a count matrix for the human liver samples. The count matrix for the pooled data was then visualized by Uniform Manifold Approximation and Projection (UMAP) after the application of batch correction (Methods; Fig. 1a). Three clusters of hepatocytes were defined along with two HSC clusters, two macrophage clusters, one EC cluster, and one cholangiocyte cluster. Each cluster was defined by established markers, supporting our cluster assignment (Fig. 1b; Supplementary Data 2), and limited evidence of marker gene expression outside of the cell types they define was observed, supporting our segmentation (Supplementary Fig. 3c). Reflecting both the hepatocyte focus of our MERFISH panel as well as the larger size of hepatocytes, hepatocyte clusters contained more RNA counts and more expressed genes than other clusters (Supplementary Fig. 4b–d). Nonetheless, with the exception of lymphocyte and neutrophil populations, which did not contain sufficient marker genes in our panel, the lower counts observed per cell for non-hepatocyte clusters still allowed us to clearly define all other major cell classes expected within the liver (Fig. 1a, b).
a Uniform manifold approximation and projection (UMAP) of all cells measured with MERFISH in the healthy human liver. b Heatmap displaying differentially expressed genes that identify each cluster. Normalized gene expression is indicated by color. Each column represents a single cell, and cells are grouped by cluster as indicated by color at the left of the heatmap. Genes are indicated on the top of the heatmap, and groups of genes characteristic of individual clusters are indicated on the bottom. Hierarchical clustering is shown at the right. Genes enriched in each cluster are shown. Genes may appear more than once if enriched in more than one cluster. c–e Distribution of cells expressing SDS and CYP2A6 (enriched in zone 1, c), ALDOB and ADH1B (most enriched in zone 2, d), and CYP2E1 and CYP1A2 (enriched in zone 3, e) in a section of liver tissue. Scale bars: 1000 μm. Expression level is measured in normalized gene expression. f Dot plot quantifying expression of genes mapped in c–e where the size of the dot represents the percentage of cells expressing a specific gene, and the color intensity indicates mean expression. g mRNA distributions across a single lobule. Each dot represents an individual mRNA transcript for the indicated gene oriented from a portal region at the top to a central region at the bottom. The relative abundance of each transcript across the lobule was then plotted (lower panel) for the same field of view. Scale bars: 50 μm. h Spatial distribution of cells assigned to each cluster of hepatocytes (Hep 1, Hep 2, or Hep 3). Hep 1 cells (purple) map to periportal areas (zone 1), Hep 3 cells (pink) map to pericentral areas (zone 3), and Hep 2 cells (brown) map between Hep 1 and Hep 3 cells (zone 2). Scale bar: 1000 μm.
We next explored the spatial distribution of hepatocytes by evaluating the expression of individual genes (Fig. 1c–f, Supplementary Fig. 5a). SDS was enriched in zone 1 (periportal), while CYP2E1 expression increased in zone 3 (pericentral), as previously described in mouse24,25,26,27 (Fig. 1c, e, f; Supplementary Fig. 5a). In addition, we found that CYP2A6 and ASS1 were enriched in portal regions similar to SDS, while CYP1A2 and GLUL were enriched in central regions similar to CYP2E1 (Fig. 1c, e, f–g). ALDOB and ADH1B were expressed in all hepatocytes but were enriched in zone 2, with ALDOB expression shifted towards zones 1 and 2 and ADH1B expression shifted towards zones 2 and 3 (Fig. 1d, f). These results showed that MERFISH captures differences in gene expression across zones within the hepatic lobule, and we next asked how effectively we could define zonation with the full MERFISH probe set. Mapping hepatocyte clusters 1, 2, and 3 (Fig. 1a) back to tissue sections showed clear zonal distribution, with hepatocyte cluster 1 localizing to zone 1, hepatocyte cluster 3 to zone 3, and hepatocyte cluster 2 to zone 2 (between zone 1 and 3; Fig. 1h, Supplementary Fig. 5b, c). Collectively, these observations are consistent with the classic zonation described in human and mouse liver4,24,25,26, supporting our MERFISH measurements.
While our analysis revealed that hepatocytes can be separated into three populations, we noted that these populations are contained within a single group of cells in the UMAP representation of the data (Fig. 1a). This representation suggests that the classic liver zones represent approximations of a continuous variable in gene expression in hepatocytes determined by the spatial relationship between portal and central areas of the lobule. To explore this possibility further, we performed a pseudotime analysis on gene expression of hepatocytes and found that the pseudotime mapped to a continuous distribution in space (Supplementary Fig. 5d–f). This gradient of gene expression could also be observed across individual lobules (Fig. 1g), where many genes show a gradient in gene expression from periportal to pericentral regions (Supplementary Fig. 5g). Collectively, these observations suggest that spatial gene expression in human hepatocytes is better described by a continuous variable determined by the relative distance between the portal and central areas of the lobule, similar to observations in mouse7,26,27.
Spatial organization of non-parenchymal cells in the human liver
We next expanded our analysis to include the spatial organization of non-parenchymal cell types (Fig. 2). Two macrophage populations were visualized in the healthy liver and showed different patterns of distribution (Fig. 2a, b, c, Supplementary Fig. 6a). Macrophage (Mac) cluster 1 was enriched in the periportal area and also dispersed through the lobules, while cells from Mac 2 were scattered more diffusely through the lobules. Both macrophage populations were identified by expression of CD74, while Mac 2 cells also expressed CD5L and MARCO (Fig. 2d, i; Supplementary Fig. 6b), most consistent with non-inflammatory macrophages or Kupffer cells10,11,21.
a, b Spatial distribution of all cells defined by MERFISH within the architecture of the healthy liver for tissue from two different donors, donor 1 (a) and donor 2 (b). Scale bars: 1000 μm. c, e, g Spatial distribution of macrophage (c), HSC (e), and cholangiocyte and EC (g) populations mapped in the same tissue section as a. Scale bars: 1000 μm. d, f, h Spatial distribution of cells in the same tissue section as (a) colored by the normalized expression of the indicated marker genes. CD74 and CD5L identify macrophages (d), PDGFRB and LAMB1 identify HSCs (f), KRT7 identifies cholangiocytes, and DNAS1L3 identifies ECs (h). Scale bars: 1000 μm. i Dot plot quantifying expression of genes that identify non-parenchymal cells in the liver. The size of the dot represents the percentage of cells expressing a specific gene, and the color intensity indicates mean expression.
Two clusters were also identified demonstrating patterns of gene expression consistent with HSCs (Fig. 2a, b, e; Supplementary Fig. 6c). These cells also displayed different spatial distributions, with cells from HSC 1 enriched in periportal regions and spread through the lobules, while cells from HSC 2 were scattered more diffusely through the lobules without periportal enrichment. These two populations differed primarily in the levels of CD74 expression, while both populations expressed PDGFRB and LAMB1 (Fig. 2f, i; Supplementary Fig. 6d).
Cholangiocytes and ECs were also identified by MERFISH and followed predicted distributions (Fig. 2a, b, g; Supplementary Fig. 6e). Cholangiocytes were localized to portal regions and were identified by expression of KRT7 (Fig. 2h, left; Supplementary Fig. 6f, left), while ECs, many of which line the sinusoids, were identified more diffusely through the liver, and expressed DNASE1L3 (Fig. 2h, right; Supplementary Fig. 6f, right). We did not note clear patterns of zonation in gene expression within these non-parenchymal populations28,29,30, suggesting that, if such gradients exist in the human liver, our MERFISH library does not contain the proper genes to capture them.
Integration of MERFISH with snRNA-seq data
MERFISH identifies parenchymal and non-parenchymal cells in human liver tissue at single-cell resolution and provides expression data for hundreds of genes, but it does not cover the full transcriptome. To address this limitation, we next performed snRNA-seq analysis for the same human liver samples analyzed by MERFISH, profiling ~15,000 nuclei after filtering (Supplementary Fig. 7a). UMAP visualization and Leiden clustering revealed a similar diversity of cell populations in the snRNA-seq data to what we observed in the MERFISH data (Fig. 3a, b, Supplementary Fig. 7b, c). We then leveraged standard data integration tools to jointly cluster hepatocytes in the snRNA-seq data with those measured in MERFISH (Fig. 3a, b; Supplementary Fig. 8a). Visualization of the snRNA-seq data after this integration showed that we could identify hepatocytes zones 1, 2, and 3 as seen in the MERFISH data, but the axis of zonation within hepatocytes was not as clear in the snRNA-seq data as it was in the MERFISH measurements (Fig. 1a; Fig. 3a).
a UMAP of snRNA-seq data for healthy human liver. Annotations for non-parenchymal cells were determined from the snRNA-seq alone while annotations for the hepatocytes were determined by merging data from both MERFISH and snRNA-seq and leveraging both marker gene expression and label-transfer of MERFISH labels. b Heatmap displaying differentially expressed genes that identify each cluster of snRNA-seq nuclei. Normalized expression level is indicated by the color bar. Each column represents a single cell, and cells are grouped by cluster as indicated at the top of the heatmap. Genes are indicated to the left of the heatmap, and groups of genes characteristic of individual cell clusters are indicated on the right. Hierarchical clustering is shown at the bottom. c Putative receptor-ligand interactions identified between zonal hepatocytes and notable non-parenchymal cells. Receptor-ligand interactions were mapped between each zone of hepatocytes (zone (z) 1, z2, z3). Only interactions that are uniquely enriched by hepatocyte zonation are shown. Gray circles indicate no interaction, and colored circles (zone 1 hepatocytes - purple; zone 2 hepatocytes - brown; zone 3 hepatocytes - pink) indicate receptor ligand co-expression.
Leveraging the snRNA-seq alone, we also defined two populations of HSCs, two populations of macrophages, a population of cholangiocytes, and a population of ECs (Fig. 3a, b; Supplementary Data 2). The gene expression profiles of the snRNA-seq clusters identified with this analysis had expression profiles consistent with the corresponding MERFISH clusters, supporting their assignment, cross validating the expression profiles measured with both techniques, and providing further support for the MERFISH segmentation (Supplementary Fig. 8b-d). However, we did note some evidence of lower HSC and macrophage marker expression in the cholangiocytes seen with snRNA-seq relative to those seen with MERFISH, consistent with modest segmentation challenges with these cells (Supplementary Fig. 8d). In addition, the snRNA-seq data also identified a population of non-macrophage immune cells, which were not defined in the MERFISH data (Fig. 3a).
Hepatocyte zonation shapes interactions with non-parenchymal cells
While we did not observe clear zonal distributions in non-parenchymal cells, we noted that the variation in gene expression in hepatocytes could nonetheless create a zonality to the interactions between hepatocytes and these other cell populations. To explore this possibility, we leveraged CellPhoneDB31 with our snRNA-seq data to explore potential modes of cell-cell interactions that could be shaped by liver zonation (Methods; Supplementary Data 3). Specifically, we focused this analysis only on receptor-ligand interactions that are uniquely enriched between hepatocytes of specific zones and the cell types in close proximity to these zonated hepatocytes (Fig. 3c, Supplementary Data 3). For example, in the periportal region hepatocytes (zone 1) produced TGFB3, which can interact with TGFBR1 expressed in Mac 1 and Mac 2 cells. PLA2G2A, a secretory phospholipase32, can interact with integrins a4b1 and a5b1, allowing periportal hepatocytes to signal to HSC 1, HSC 2, Mac 1, Mac 2, and ECs. IL1RN produced by periportal hepatocytes can also antagonize IL1R expressed by Mac 1 cells. TF produced by Mac 1 cells can signal to TFRC on periportal hepatocytes, while HSC 1 and ECs expressed EFNA5 and EFNB2, respectively to signal to portal hepatocytes through EPHA2 and EPHB1.
In the pericentral region, hepatocytes could signal to HSC 1 and HSC 2 cells through expression of GDF7, which can be recognized by multiple BMP receptors in HSC 1 and HSC 2 cells. Expression of TENM3 or TENM2 by pericentral hepatocytes is recognized by ADGRL2 and/or ADGRL3, which are expressed by HSC 1, HSC 2, and Mac 1 cells. VEGFA, expressed by pericentral hepatocytes is recognized by NRP2, expressed by HSC 1 cells and NRP1 expressed by Mac 2 cells. HEBP1 from pericentral hepatocytes could also signal to Mac 1 and Mac 2 cells expressing FPR3, while pericentral expression of CCL16 could signal to CCR1 expressed on Mac 2 cells. THBS1 and THBS2 produced by HSCs can signal to CD36 expressed in pericentral hepatocytes. THBS1/2 is detected at the highest level in HSCs and can signal to CD36 expressed by pericentral hepatocytes (Supplementary Data 3), while the THBS1-CD36 receptor-ligand pair is also enriched between pericentral hepatocytes and macrophages. GAS6 produced by HSC 1 and ECs can signal to pericentral hepatocytes through MERTK, while SEMA4D expressed by Mac 1 and Mac 2 cells, and VEGFB produced by Mac 2 cells can be recognized by PLXNB2 and NRP1, respectively, both of which are expressed by pericentral hepatocytes. ECs and Mac 2 cells also produce WNT2B, which can be recognized by FZD6-LRP5/6 in periportal hepatocytes, with the highest interaction scores observed between ECs and zone 3 hepatocytes (Supplementary Data 3). These findings highlight receptor-ligand interactions that may regulate cross-talk between hepatocytes and non-parenchymal cells in zone specific patterns.
Effect of nuclear content on gene expression in hepatocytes
Hepatocytes have the capacity to replicate their nuclei without dividing, generating polyploid and multinucleated cells14,15,16. It is not clear if additional nuclear content leads to defined gene expression changes that could modulate function, or to what degree zonation influences nuclear content in the human liver. As an image-based approach to transcriptomics, MERFISH naturally provides a measure of the nuclear content of each cell. To leverage this capability, we counted the number of nuclei observed in individual hepatocytes and correlated this property with aspects of gene expression and spatial ___location. Indeed, we observed both single and multinucleated hepatocytes in the MERFISH data (Fig. 4a) and found that approximately a third of hepatocytes were multinucleated (Fig. 4b). The distribution of single and multinucleated hepatocytes did not change across hepatocyte zones (Fig. 4c, d), and we did not observe any significant changes in expression of genes between hepatocytes with one or two nuclei as quantified by MERFISH (Fig. 4e). Nonetheless, the total RNA counts per cell increased with nuclei number (Fig. 4f, left) as did the cell area (Fig. 4f, center), such that multinucleated hepatocytes tended to be larger and contain proportionally more RNA transcripts, but RNA density was not dependent on the number of nuclei (Fig. 4f, right). These results suggest that multinucleated hepatocytes may contain more RNA transcripts within proportionally larger cells, which may underscore an increased metabolic capacity. However, this increase does not appear to be associated with differential gene expression or ___location of hepatocytes within the lobule, suggesting that these cells do not have distinct functional roles in the healthy liver, at least within the pathways we explored with MERFISH.
a Example MERFISH DAPI images showing hepatocytes containing single and multiple nuclei. Examples of multinucleated cells are indicated by white arrowheads. DAPI staining marks nuclei (white), and cell boundaries (light blue) were determined by Baysor with input from Cellpose. Scale bars: 20 μm. b Distribution of number of nuclei per hepatocyte. c Hepatocyte UMAP, as in Fig. 1, colored by the number of nuclei found within each hepatocyte. Source data are provided as a Source Data file. d Spatial distribution of hepatocytes with 1 or 2 detected nuclei in healthy human liver (donor 1, left and donor 2, right). Scale bar: 1000 μm. e Dot plots of hepatocyte genes for hepatocytes with 1 or 2 nuclei. The size of circles represents the percentage of cells expressing a gene in each group and the color intensity indicates mean expression. f Distribution of the transcripts per cell (left), area per cell, (middle) and RNA density per cell (right) for hepatocytes with the indicated nuclear content. Source data are provided as a Source Data file.
Applying spatial transcriptomics in liver fibrosis
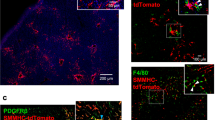
In order to explore the spatial and cellular remodeling that occurs in the context of disease, we next performed MERFISH on human liver samples from patients with liver injury and fibrosis (Supplementary Table 1 and Supplementary Fig. 1). We leveraged the same protocols as described above to image and segment cells and, in total, imaged ~220,000 cells (Supplementary Fig. 9). In order to compare hepatocytes between normal and fibrotic liver, we applied batch correction methods to jointly integrate MERFISH data across all samples (Fig. 5a, b; Supplementary Fig. 10). Instead of three zones of hepatocytes, this joint analysis produced two: a periportal cluster of hepatocytes (Portal Hep) and a pericentral cluster of hepatocytes (Central Hep), which collectively contained zone 1, 2 and 3 hepatocytes from healthy liver (Fig. 5a, Supplementary Fig. 9b-d, Supplementary Fig. 10, Supplementary Data 2). In addition, two clusters were also identified that were almost entirely composed of hepatocytes from fibrotic injury (Fibrotic Hep 1 and 2) (Fig. 5a, Supplementary Fig. 9b-d, Supplementary Fig. 10, Supplementary Data 2). Periportal hepatocytes and pericentral hepatocytes retained their expected localization in the setting of chronic liver injury (Fig. 5c), while the two clusters of hepatocytes that emerged with fibrotic injury were distributed throughout the lobules (Fig. 5d). A single cluster of HSCs and a single cluster of macrophages were also identified (Fig. 5a, b; Supplementary Data 2). HSCs displayed bands of cells across lobules, while macrophages were spread more diffusely through the lobules (Fig. 5e). Thus, we find that hepatocyte zonation is at least partially maintained with fibrotic injury, while two injury-related hepatocyte states are enriched that do not appear to retain zonation.
a UMAP of fibrotic tissue measured with MERFISH. Clusters are indicated by color. Portal Hep and Central Hep show similarities to hepatocytes identified as zone 1, 2, and 3 cells from healthy liver, and Fibrotic Hep 1 and Fibrotic Hep 2 contain hepatocyte clusters that emerge with fibrotic injury. The HSC cluster shows similarities to both HSC 1 and HSC 2 cells from healthy liver, and the Mac cluster has similarities to both Mac 1 and Mac 2 clusters from healthy liver. b Relative abundance of each cell type identified in each sample (left to right: healthy donors 1–3 followed by fibrotic donors 4–6). Cell types are colored as in a. Source data are provided as a Source Data file. c–e Spatial distribution of cell populations in the fibrotic liver (donor 4), highlight hepatocytes that are similar to those seen in healthy samples (c), hepatocyte populations that are identified only when including fibrotic liver samples (d), and key non-parenchymal cells (e). Scale bar: 1000 μm. f–h Dot plot quantifying expression of genes differentially expressed in clusters seen in healthy measurements (Healthy zone 1, 2, or 3) or in fibrotic samples (Portal Hep, Central Hep, Fibrotic Hep 1, or Fibrotic Hep 2) for hepatocytes (f), macrophages (g), and HSCs (h). Macs Healthy 1 and Healthy 2 refer to Mac 1 and Mac 2 cells in healthy liver, and HSCs Healthy 1 and Healthy 2 refer to HSC 1 and HSC 2 cells in healthy liver.
We next evaluated changes in gene expression between hepatocytes in healthy livers and those with fibrotic injury using MERFISH (Fig. 5f; Supplementary Data 4). For this analysis, we first separated healthy hepatocytes into their original three zones (Healthy zone 1, 2, 3) and compared expression to portal and central hepatocytes from fibrotic liver (Portal Hep and Central Hep; Supplementary Data 4). Portal and central hepatocytes from the fibrotic liver retain the same zonal expression of marker genes as healthy zone 1, 2, and 3 and are characterized primarily by a reduction in expression of these genes. We then evaluated gene expression of the expanded fibrotic populations (Fibrotic Hep 1 and 2) (Fig. 5f, bottom; Supplementary Data 4). The Fibrotic Hep 1 cluster showed a general reduction in expression of genes enriched in hepatocytes, while the Fibrotic Hep 2 cluster showed an increase in expression of several notable genes, including SLC7A2, CPS1, ATP9A, DTX1, POR and EXT1 compared to all other hepatocyte populations.
Non-hepatocyte clusters also showed disease-associated gene expression changes. For example, fibrotic macrophages showed a loss of MARCO, CD5L, CD68 and retained an expression pattern that more closely matched Mac 1 cells in normal liver (Fig. 5g), while fibrotic HSCs maintained high levels of COL1A1 expression without a clear positive signature from the other genes included in the MERFISH panel (Fig. 5h).
Transcriptome-wide expression changes in fibrosis
To expand our analysis transcriptome-wide, we next performed snRNA-seq on the same samples characterized with MERFISH, collecting ~13,500 nuclei across these three samples (Fig. 6; Supplementary Fig. 11). Analysis of the snRNA-seq data alone produced the same non-parenchymal clusters as seen with MERFISH; however, to better integrate the labeling of the hepatocytes, we jointly integrated and clustered hepatocytes between these two measurements (Fig. 6a, b; Supplementary Fig. 12a, b). Importantly, this analysis revealed nuclei in the snRNA-seq data that were similar to the two fibrotic hepatocyte clusters (Fibrotic Hep 1 and Fibrotic Hep 2) seen in the MERFISH measurements, supporting the emergence of these two disease-associated populations (Supplementary Fig. 12a, b).
a UMAP of snRNA-seq data for injured human liver. Annotations for the hepatocytes were determined by merging data from both MERFISH and snRNA-seq and leveraging both marker gene expression and label-transfer of MERFISH labels. b Heatmap displaying differentially expressed genes that identify each snRNA-seq cluster. Normalized expression level is indicated by the color bar on the right. Each column represents a single nucleus, and nuclei are grouped by cluster as indicated at the top of the heatmap. Genes are indicated at the bottom of the heatmap, groups of genes characteristics of individual cell clusters are indicated on the top, and cells are grouped by cluster as indicated by color at the left. c Dot plot quantifying expression of genes differentially expressed in clusters seen in healthy measurements (Healthy zone 1, 2, or 3) or in fibrotic samples (Portal Hep, Central Hep, Fibrotic Hep 1, or Fibrotic Hep 2) for hepatocytes. Dot plots quantifying expression of genes differentially expressed in macrophages (d) and HSCs (e) are shown. Macs Healthy 1 and Healthy 2 refer to Mac 1 and Mac 2 cells in healthy liver, and HSCs Healthy 1 and Healthy 2 refer to HSC 1 and HSC 2 cells in healthy liver.
We then used the more complete transcriptomic data from snRNA-seq to evaluate differences in gene expression between hepatocytes in healthy liver and livers with fibrotic injury. Indeed, a large functional diversity of genes were differentially expressed between hepatocytes within these conditions (Supplementary Data 4). To organize this functionality, we performed gene ontology (GO) analysis, which showed that genes induced with injury were enriched across many metabolic pathways (Supplementary Data 5). We thus focused on the more general category of metabolic processes (GO:0008152) to capture these changes in gene expression (Fig. 6c, Supplementary Data 6). When comparing all healthy to all injured hepatocytes, the most highly enriched group of metabolic genes include those with dehydrogenase, oxygenase, and hydrolase activity along with nuclear receptors THRB, RORA, and ESR1 (Supplementary Data 4 and Supplementary Data 6). Increased expression was also observed in the ligases CPS1 and GLUL and the transporter SLC25A15, which are involved in the urea pathway (Supplementary Data 6).
With injury, hepatocytes that maintain portal and central zonation (Portal Hep and Central Hep) both continued to show increased expression of hydrolases, oxygenases, and the nuclear receptors THRB, RORA, and ESR1 in comparison to healthy hepatocytes from zones 1, 2, and 3, and induction of dehydrogenases was most pronounced in Central Heps with injury (Fig. 6c, Supplementary Data 4 and Supplementary Data 6). While oxygenases increase in both Portal and Central Heps with injury, CYP2E1 shows the highest expression in Central Heps with injury (Fig. 6c, Supplementary Data 4 and Supplementary Data 6). The ligase CPS1 is increased in both populations, and GLUL remains more restricted to Central Heps and ASS1 to Portal Heps with injury. Within the hepatic populations that emerge with fibrotic injury and do not show zonal distribution (Fibrotic Hep 1 and 2, Fig. 5d), we again observe an enrichment in metabolic pathways, primarily those involving fatty acids and amino acids, while pathways involving RNA processing and ribosome biosynthesis are reduced (Supplementary Fig 13a, b). Both Fibrotic Hep 1 and Fibrotic Hep 2 continue to show enriched expression of the nuclear receptors THRB, RORA, and ESR1 (Fig. 6c, Supplementary Data 4 and Supplementary Data 6) and maintain increased expression of CPS1 and SLC25A15. Fibrotic Hep 1 cells are relatively more enriched in oxygenases compared to Fibrotic Hep 2 cells and are notable for increased expression of the phospholipase PNPLA333, along with ASPG, CPEB4, and PRKAG2. Fibrotic Hep 2 cells show relative enrichment in expression of hydrolases, heat shock proteins (HSPH1 and HSP90AA1, while HSPH1 is enriched in both Fibrotic Hep 1 and 2), and PLCB1 compared to Fibrotic Hep 1 cells. The transmembrane receptor EGFR is increased in Fibrotic Hep 1 and Fibrotic Hep 2 cells, while the transmembrane receptor GHR is increased in all injured hepatocyte populations, and LEPR is enriched in injured Portal Heps and Fibrotic Hep 1 and Fibrotic Hep 2 populations (Fig. 6c, Supplementary Data 4 and Supplementary Data 6). These results show the metabolic impact of chronic injury on hepatocytes, including common activation of nuclear hormone receptors and genes that regulate the urea pathway, and more restricted induction of transmembrane receptors, heat shock proteins, and the phospholipase PNPLA3.
Gene expression was also evaluated in non-parenchymal cells. With injury, macrophages showed changes in gene expression in pathways including cell signaling, cell migration, and immune response when compared to Mac 1 and Mac 2 from healthy livers (Fig. 6d, Supplementary Data 2). Change in gene expression in HSCs with injury included those involved with the extracellular matrix, cell migration, and cell adhesion when compared to HSC 1 and HSC 2 in healthy livers (Fig. 6e, Supplementary Data 2). ECs and cholangiocytes also showed changes in gene expression across categories including cell adhesion and cell surface receptor signaling with injury (Supplementary Fig. 12c, d; Supplementary Data 2). These results show that the macrophage population defined with injury is distinct from Mac 1 and Mac 2 identified in healthy donors. Similarly, the HSC population defined in injury is distinct from HSC 1 and HSC 2 in healthy donors.
Discussion
Significant advances in mapping the human liver at single-cell resolution have been achieved recently with scRNA-seq and snRNA-seq10,11,21,34. In parallel, platforms such as Visium12 now provide spatial information, but it has remained challenging to assign transcripts to individual cells and cell types located in close proximity. Here we complement these previous efforts by developing protocols for the application of MERFISH in human liver samples and using these methods to achieve spatial transcriptomic analysis of >300 genes at single-cell resolution in the human liver, profiling ~310,000 cells in both healthy and diseased liver. Combining MERFISH with snRNA-seq then allowed us to analyze the broader transcriptome within spatially resolved cell types and cell sub-types.
From these measurements, we were able to resolve three clusters of hepatocytes that are spatially distinct across the lobule in healthy human liver, characteristic of separation of hepatocytes into zones 1, 2, and 3 moving from periportal to pericentral regions. While MERFISH allows to us to define these zones, it is also evident through pseudotime analysis, measurement of gene expression across individual lobules, and transcriptional analysis performed across lobules, that zonation reflects a gradient from periportal to pericentral regions rather than distinct zones. Thus, the classic three zones are, in reality, an approximation for the continuous gene expression and functional changes that occur in space with human hepatocytes—a proposal consistent with recent single-cell analysis in the murine liver26,27.
With the development of fibrotic injury, many hepatocytes maintained a zonal phenotype, but we also observed the emergence of two additional hepatocyte populations with fibrotic injury that show diffuse distribution through the lobules without a portal or central bias (Fibrotic Hep 1 and 2 populations; Fig. 5a, b, d). Specific genes identified as differentially expressed can shift between MERFISH and snRNA-seq and may reflect differences between nuclear and cytoplasmic distribution of some transcripts, but a common theme from both analyses was enrichment of genes in the urea pathway with fibrotic injury. CPS1 (Carbamoyl phosphate synthetase I) is responsible for converting ammonia to carbamoyl phosphate to enter the urea cycle35, SCL7A2 encodes an arginine/ornithine/lysine transporter and provides ornithine (either directly or through conversion of arginine) to interact with carbamoyl phosphate to form citrulline36,37, and SLC25A15, is an ornithine transporter in the urea cycle38 (Figs. 5f and 6c). In addition, Fibrotic Hep 1 cells showed enrichment in genes including PNPLA3, CPEB4, and ASPG, which are linked to hepatic steatosis, steatohepatitis, and/or fibrosis33,39,40,41, and THRB, which encodes the protein target of resmetirom, a medication shown to be effective in treating nonalcoholic steatohepatitis42, together highlighting additional pathways that may be altered in disease.
Two populations of macrophages were identified by MERFISH and show different distribution patterns through the lobule. Mac 2 cells were identified as MARCO+, CD5L+, and CD68+ with increased expression of VISG4 compared to Mac 1 cells and were located more diffusely through the lobule, most consistent with Kupffer cells. Mac 1 cells were MARCO- and CD5L- with lower levels of CD68. While Mac 1 cells were also found through the lobule, they did exhibit a distinct enrichment in portal areas in contrast to Mac 2 cells. Further comparisons of gene expression between Mac1 and Mac 2 with snRNA-seq confirmed these expression patterns along with increased CD163, VCAM1, and TMEM26 in Mac 2 cells (Supplementary Data 2), all of which are evolutionarily conserved markers of Kupffer cells43. While Mac 2 cells expressed higher levels of IL18 compared to Mac 1 cells, we do not observe the increased expression of GPNMB, SPP1, or LGALS1 in Mac 1 cells necessary to link them to lipid associated macrophages (LAMs), identified in the livers of healthy mice and expanded with steatosis43,44. With the development of fibrotic injury, the macrophage population lost expression of MARCO, CD5L, and CD68 in the MERFISH data, and snRNA-seq analysis also showed reduction in MARCO and CD5L as well as VCAM1 and CD163. While these features suggest movement away from Kupffer cell markers, IL18 expression was maintained, and gene expression remained distinct from the Mac 1 population (Fig. 6d).
MERFISH also identified two populations of HSCs. HSC 1 was enriched in the portal areas compared to HSC 2, and both populations were found distributed through the lobules. Within the genes evaluated by the MERFISH probes, the MHC class II chaperone CD7445 showed the greatest differential expression between the two HSC populations, with highest expression observed in HSC 1 cells. A similar effect was also observed in the snRNA-seq data. CD74 is expressed by HSCs, can be induced by inflammatory signals such as IFN-γ and can promote immune activation through MHC class II-mediated interactions46,47. It is currently unclear if these CD74 positive and negative populations represent HSCs with distinct antigen presentation abilities or possibly different histories of activation. Overall, the injured HSC expression profile was distinct from HSC 1 and HSC 2 populations (Fig. 6e).
With single-cell spatial resolution, we evaluated receptor-ligand coexpression in cells across hepatocyte zonation. Our MERFISH panel was not designed to explore a diverse set of functional pathways in HSCs, macrophages, or ECs across zones, but the current data do allow us to ask about specific interactions driven by differential expression of receptors or ligands in hepatocytes across zones. Indeed, we observe that the zonal expression of receptors and ligands in hepatocytes can shape the potential interactions with non-parenchymal cells. In particular, we identified signaling interactions including a thrombospondin signal (THBS1/2) from HSCs to CD36 in zone 3 hepatocytes, which may influence metabolic dysfunction associated steatotic liver disease (MASLD)48, and the interaction of WNT2B, expressed by ECs and Mac 2 cells, with FZD6 and LPR5/6 expressed by zone 3 hepatocytes49,50 helps validate this analysis. More broadly, this analysis, like all single-cell receptor-ligand analyses, produced only hypotheses of potential interactions. Nonetheless, this rich set of hypotheses for spatially organized interactions could prove useful in directing future functional studies.
MERFISH also provides data about nuclei and cell boundaries in addition to gene expression, allowing us to link nuclei content to gene expression within individual cells. We found that approximately a third of adult hepatocytes are multinucleated14,16, in agreement with previous descriptions of human liver. We did not quantify ploidy separately, but based on these studies, we would predict that >80% of these multinucleated hepatocytes are 2N14,15,16. While we did not observe a change in relative gene expression with increase in nuclear content, we did observe an increase in RNA transcripts. In addition, multinucleated hepatocytes also tend to be larger in size, such that mRNA levels were proportional to cell size. These findings are consistent with the interpretation that cells with more nuclei are both larger15 and express higher levels of RNA. Most of the transcripts identified in hepatocytes are cytoplasmic, but transcripts are also present in the nucleus. It is possible that MERFISH data could be used to evaluate transcriptional activity between different nuclei in the same hepatocyte, but this will require additional tools to reconstruct nuclei from the stacked images and control for the fraction of each nuclei visualized in an individual cell.
Taken together, our study applied MERFISH to create spatial transcriptomic maps of the healthy and fibrotic liver at single-cell resolution to define spatially and transcriptionally distinct sub-populations of hepatocytes, macrophages, and HSCs within hepatic lobules and distinct hepatocyte sub-populations that expand with fibrotic injury. By combining MERFISH with snRNA-seq, we extended the transcriptional profiles of cell types in these spatial maps to understand unique receptor-ligand interactions involving hepatocytes and non-parenchymal cells across hepatocyte zones and defined changes in gene expression in spatially-resolved hepatocyte populations with injury. Finally, by evaluating nuclear content, we found that multinucleated hepatocytes do not show differences in relative gene expression compared to mono-nucleated hepatocytes, but tend to be larger and produce more total transcripts. Future studies will now be able to extend these approaches, expanding probe set libraries to encompass additional RNA species to evaluate all cell types in the liver and their broader transcriptional profiles at single-cell spatial resolution in health and disease.
Methods
Tissue collection
All liver samples were collected from excess surgical tissue at Massachusetts General Hospital in accordance with protocols approved by the Mass General Brigham (MGB) or Dana Farber Cancer Institute (DFCI) Institutional Review Boards (IRBs) including approval to provide data on age by decade and sex. All MERFISH and snRNA-seq data presented within are from matched samples. Sex was not considered as a variable due to the small sample size. Patients were consented under MGB protocol 2019P001245 (AM-061) or DFCI protocol 02-240 (all other samples). Patients consented under 2019P001245 received compensation of $10 USD and a parking pass. Patients consented under 02-240 did not receive compensation. Tissue collected was used for analysis, and additional tissue is not available for distribution.
MERFISH sample preparation
Liver tissue sections (3–5 mm thickness) were fixed by submerging in fresh 4% v/v paraformaldehyde (PFA; EMS 15710) in 1X Phosphate Buffered Saline (PBS; Ambion AM9625) for 3–5 hours at 4 °C with gentle rocking. After fixation, tissue samples were then transferred to a 30% w/v sucrose (VWR, 0335) solution in 1X PBS supplemented with 4% v/v PFA and incubated overnight at 4 °C with gentle rocking. After washing in 1X PBS, samples were placed in cryomolds on dry ice, covered with Optimal Cutting Temperature media (OCT; Tissue-Tek 4583), wrapped in aluminum foil, and stored at −80 °C until use.
MERFISH library preparation
A panel of 317 genes were selected with a focus on those differentially expressed between hepatocyte clusters12 supplemented with common markers of non-parenchymal liver cells10. The MERFISH encoding probes targeting these genes were designed using a previous pipeline19,51. Briefly, each probe contained a 30-nt-long region specific to the targeted RNA, concatenated with a series of readout sequences that defined the binary barcode assigned to that RNA. Each mRNA was targeted with 72 such probes. To encode these 317 genes, we selected a 22-bit-long, constant Hamming weight and Hamming distance code with a Hamming weight of 4 and a minimum Hamming distance of 4. Each of the 72 encoding probes contained two readout sequences and the complement of all encoding probes to a given gene contained instances of the four readouts associated with the 4 bits in which they had a ‘1’. This barcoding scheme contains 332 barcodes and we leveraged the 15 not used to encode RNAs as ‘blank’ barcode controls, not assigned to an RNA (Supplementary Data 1).
Templates for these probes were designed by concatenating a primer and a T7 promoter to the sequences (Supplementary Data 1), as described previously, and the template pool was ordered from Twist Biosciences. These templates were amplified into encoding probes in a protocol that involved PCR, in vitro transcription, reverse transcription, alkaline hydrolysis, and SPRI purification, as described previously19,20,51,52.
MERFISH staining
The general workflow for MERFISH sample preparation includes sectioning, permeabilization, cell boundary staining, RNA probe hybridization, acrylamide gel embedding, digestion, and photobleaching of samples. These protocols have been described previously19,20,51,52. Briefly, OCT-embedded tissues were cryosectioned at a thickness of 7 μm, and slices were transferred onto silanized, poly-lysine coated 40-mm circular coverslips with fiducial beads, prepared as described previously19,20,51,52. Following sectioning, tissue slices were briefly fixed with 4% v/v PFA in 1X PBS at room temperature for 10 minutes, washed thrice with 1X PBS for 5 minutes, and then permeabilized overnight at 4 °C in 70% ethanol. Samples were rehydrated in 2X saline sodium citrate (SSC; Ambion AM9763).
To prepare samples for antibody staining of cell boundaries, tissue samples were treated with 0.05% (v/v) proteinase K (New England Biolabs [NEB]; P8107S) in pre-warmed (37 °C) 2X SSC for 10 minutes at 37 °C. Samples were then rinsed with 2X SSC before incubating with a blocking buffer (10% BSA, 3% v/v 6% v/v murine RNase inhibitor [NEB, M0314L] in 2X SSC) for 30 minutes. The sample was then stained with a primary antibody against the Na+/K+-ATPase (2 µg/ml, Abcam; Ab76020) in the same blocking buffer for 30 minutes at room temperature. Excess primary antibody was removed from the samples with three, 10-minute, 2X SSC washes at room temperature. The sample was stained with a secondary antibody using the same protocol as the primary antibody (3.75 µg/ml; ThermoFisher; A16112). The secondary antibody was labeled with an oligonucleotide using previous protocols53.
Probes were hybridized to the sample at a concentration of 6–10 µM in a 30% v/v formamide (Fisher Scientific, AM9342), 10% w/v dextran sulfate (VWR, 97062-828), 1 mg/mL yeast tRNA (ThermoFisher, 15401029) solution in 2X SSC for 48 hours at 37 °C in a humidified oven. Samples were rinsed by letting the samples sit in 30% v/v formamide in 2X SSC before hybridization for at least 4 hours and twice after hybridization for 30 minutes each time at 37 °C.
To clear samples, they were embedded in a polyacrylamide gel and then digested for two days with proteinase K, as described previously19,20,51. Briefly, samples were embedded in a thin polyacrylamide film by inverting them onto a GelSlick-coated microscope slide with a droplet of 4% acrylamide solution (4% v/v 20:1 acrylamide:bis-acrylamide [BioRad, 1610144] with 0.15% v/v TEMED [Sigma, T7024] and 0.30% v/v ammonium persulfate [Sigma, 215589]). After the gel polymerized for 2 hours, samples were covered with a digestion solution (1% proteinase K, 20% v/v SDS [ThermoFisher, AM9823], 0.25% v/v triton-X [Sigma, T8787] in 2X SSC) and incubated at 37 °C for 24 hours. Samples were then rinsed multiple times with 2X SSC. In order to remove autofluorescence, samples were photobleached under a blue LED light at 4 °C for 24 hours. Samples were stored at 4 °C until imaging.
snRNA-seq
Single nucleus extraction was performed based on previously-described protocols54. A 2X stock of salt-Tris solution (ST buffer) containing 292 mM NaCl (Thermo Fisher Scientific, AM9759), 20 mM Tris-HCl pH 7.5 (Thermo Fisher Scientific, 15567027), 2 mM CaCl2 (VWR International Ltd, 97062-820), and 42 mM MgCl2 (Sigma Aldrich, M1028) was prepared in ultrapure water. The day of each experiment, for each sample, Tween with salts and Tris (TST) buffer was made from 1 ml of 2X ST buffer, 60 µl of 1% Tween-20 (Sigma Aldrich, P-7949, 0.03% final), 10 µl of 2% BSA (NEB, B9000S, 0.01% final) and 930 µl of nuclease-free water and supplemented with 1U/ml Protector RNase inhibitor (Millipore Sigma, 3335402001). 1X ST buffer was prepared by dilution 2X ST with ultrapure water (Thermo Fisher Scientific, 10977023) and supplemented with 0.5U/ml Protector RNase Inhibitor (Millipore Sigma, 3335402001).
On dry ice, a section of frozen tissue was placed into a gentleMACS C Tube (Miltenyi Biotec, 130-093-237) with 2 ml of TST buffer. Tissue was immediately dissociated using a gentleMACS Dissociator (Miltenyi Biotec, 130-096-427) using the m_Spleen_01_01 program twice and incubated on ice for 5 min to complete a 10 min incubation in Tween with salts and Tris (TST) buffer. C tubes were centrifuged at 4 °C for 2 min at 500 g. The pellet was resuspended in TST buffer, filtered through a 40 µm Falcon cell strainer (VWR, 43-50040-51) into a 50 ml conical tube. The strainer was washed with 1 ml 1X ST buffer before use. An additional 1 ml of 1X ST buffer was used to wash the gentleMACS C Tubes and filter, and another 1 ml was added to the filter for a final wash. The sample was transferred to a 15 ml conical tube and centrifuged at 4 °C for 10 min at 500 g. The pellet was resuspended in 1X PBS (-Mg/-Ca, Gibco, 10010023), 1% BSA (NEB, B9000S), and 1U/ml Protector RNase Inhibitor (between 100-200 μl depending on pellet volume). The nucleus solution was filtered through a 35 µm Falcon cell strainer (Corning, 352235). Nuclei were counted using a INCYTO C-chip disposable hemocytometer (VWR, 22-600-100).
Sample processing
MERFISH measurements
MERFISH measurements were performed as described previously using a custom-microscope system52. The entire slice was first imaged using a 10X objective and the DAPI stain, and this mosaic was used to select the region of interest for MERFISH measurements.
snRNA-seq
8000–12,000 nuclei of the single nucleus suspension were loaded onto the Chromium Chips for the Chromium Single Cell 3′ Library (Chromium Next GEM Single Cell 3’ Kit v3.1; PN-1000268, PN-1000120, PN-1000215) according to the manufacturer’s recommendations (10x Genomics). Gene expression libraries were constructed and indexed according to manufacturer’s instructions and pooled for sequencing on a NovaSeq 6000 sequencer (Illumina). All libraries were sequenced to a targeted depth of 400 million reads in the following configuration: R1: 28 bp; R2: 90 bp; I1, I2: 10 bp.
Analysis
MERFISH decoding
MERFISH decoding was performed using a previously described pipeline19,20,51. Briefly, images from a single field-of-view (FOV) were stacked into movement-corrected stacks containing all z-planes and imaging channels, an optimal weighting was determined for the relative intensity of different image frames, and pixels were assigned to individual barcodes using a soft decoding approach based on nearest neighbors with a Euclidean distance metric. Adjacent pixels assigned to the same barcode were aggregated to form a single RNA.
RNAs were assigned to cells using a combination of Cellpose23 (version 0.7.2) and Baysor22 (version 0.5.0). Briefly, we trained a cellpose model based on the Na+/K+-ATPase immunofluorescent stain and created a label matrix using cellpose and this model for each FOV. Cellpose was applied to each z-plane separately, and then overlapping boundaries were combined across z-planes to create a 3D segmentation. We leveraged our previous pipeline19,20,51 to identify cell boundaries, connect boundaries across FOV boundaries, and parse RNAs into these boundaries. This initial segmentation result was then refined and improved with Baysor using the following parameters: scale 10, scale standard deviation 50%, molecules/cell 3, and prior confidence 0.50.
MERFISH single-cell analysis
The output of Baysor was analyzed using Scanpy55 (version 1.8.1). Initial processing concatenated the MERFISH data into a single AnnData structure, filtered cells with less than 3 genes and 15 RNA counts, normalized the remaining cells, and applied a logarithmic scaling. Harmony was used as our data integration method in order to account for sample variation56. For the analysis of the healthy MERFISH data, we used 100 PCA components and a final Leiden resolution of 1.1 For the analysis of the MERFISH fibrotic data, we used 150 PCA components and a final Leiden resolution of 1.2
snRNA-seq
Samples were demultiplexed using Illumina’s bcl2fastq conversion tool and the 10x Genomics pipeline Cell Ranger (version 7.0.1) to perform alignment against the 10x Genomics pre-built Cell Ranger reference GRCh38-2020-A (introns included). Cells with mitochondrial or ribosomal counts >10% and cells with <500 genes or UMI < 500 were removed. Ambient RNA was removed using the SoupX57 package (version 1.6.2). We focused on ambient RNA from genes enriched in hepatocytes, as hepatocytes constituted almost 70% of cells in the dataset. We included hepatocyte genes previously used for ambient RNA removal (CYP2E1, ASGR1, CYP3A4, SCD, SLP1, CYP2B6, CPS1, CRP, A2M, and ALB)12 but included ALB in a separate filter from the other genes due to ts high detection across clusters. We added a third filter (CYP27A1, CYP4A11, CYP4V2, CYP2B7P, CYP2A6, CYP2A7, CYP3A5, CYP2C9, CYP2C8, CYP4F3, CYP3A43) based high expression in hepatocytes in the current dataset. Next, doublets and multiplets were filtered out using DoubletFinder v2.0.458 for each individual sample. Each sample’s count was normalized by the SCTransform method in Seurat v4.3.0 with mitochondrial reads regressed out.
Healthy and diseased samples were each merged into individual Seurat objects, and genes were projected into principal component space using principal component analysis (RunPCA). Integration and batch correction across subjects were performed with the Harmony50 R package. The first 20 principal components processed with Harmony were used as inputs for the FindNeighbours and FindClusters functions. Clustering for healthy samples was performed at a resolution of 1, chosen from tested resolutions (0.1, 0.2, 0.3, 0.4 0.5, 0.6, 0.7, 0.8, 0.9 and 1.0). The same resolution was used for diseased samples.
A shared-nearest-neighbor graph was constructed based on the Euclidean distance metric in principal component space, and cells were clustered using the Louvain algorithm. The RunUMAP function with default settings was used to calculate 2D UMAP coordinates to identify distinct cell populations. Cell types were assigned to clusters using ScType59 with liver as the tissue annotation. We identified hepatocyte, HSC, cholangiocyte, endothelial cell (EC), macrophage, and an additional immune system cell, populations. A few sub-clusters within the hepatocyte populations were labeled as unknown by ScType and were manually assigned the hepatocyte label based on gene expression. All reported cluster labels for snRNA-seq data from healthy or diseased samples reflects the assignments made from the analysis of the snRNA-seq data alone, with the exception of the hepatocyte sub-divisions, which were jointly labeled by co-integrating with MERFISH (see below). We performed joint integration of hepatocytes with MERFISH as the zonal gene expression axis was not as apparent in snRNA-seq analysis, and initial analysis of these data alone did not recapitulate clusters that defined these gene expression gradients as clearly as MERFISH.
Cluster-level quality control was then performed using MALAT1, a long noncoding RNA transcript retained in the nucleus52, as an indicator of high-quality snRNA-seq clusters60,61. We removed two clusters from the healthy dataset and two from the fibrotic injury dataset where greater than 15% of cells contained fewer than three MALAT1 reads. In contrast, fewer than 2.7% of cells in any other cluster showed fewer than three MALAT1 reads.
Integration of MERFISH and snRNA-seq data
To co-integrate the healthy MERFISH and snRNA-seq measurements to identify the zonal gradients within hepatocytes within the snRNA-seq data, we concatenated the two separate AnnData structures containing just hepatocyte cells identified from the Leiden clustering described above. Expression was logarithmically transformed, z-scored, and the genes were trimmed to just those measured with MERFISH before concatenation. These cells were then integrated using Harmony in order to compare gene expression markers and labels across cell types, leveraging the inbuilt algorithms and defaults associated with scanpy. This co-integrated data was clustered using Leiden clustering in order to assess and group the hepatocytes between the two modalities. The individual Leiden clusters were then annotated by leveraging a combination of marker expression and the existing annotations derived from the clustering analysis of the MERFISH data alone, as described above. Importantly, this approach was used only to apply labels to the snRNA-seq cells, and all expression analysis derived from this co-integration was taken from the snRNA-seq cells.
Ligand-receptor analysis
Receptor-ligand interactions were evaluated using snRNA-seq data using CellPhoneDB31,62. We applied 1000 iterations and considered ligands and receptors for analysis if expressed by at least 10% of cells (threshold = 0.1). Significant interactions between hepatocytes and non-parenchymal cells (p-value of 0.05 or less) were only selected if the interactions involved hepatocytes from one or two zones. Predicted interactions were excluded if 1) the interaction score for a significant interaction was less than the interaction score for non-significant interaction between hepatocytes and the indicated cell type and 2) the ligand was expressed at a higher level in the receptor cell than the ligand cell. Only interactions defined as Ligand-Receptor are displayed.
Analysis of gene expression between hepatocytes in healthy liver and livers with fibrotic injury
Differential expression analysis was performed for the indicated comparisons using scanpy’s ranked genes function using the t-test model. Genes were ranked by scores to identify the 100 most induced genes for each comparison within hepatocytes. GO analysis (biologic process; Panther release 19.0) was performed as indicated for all healthy hepatocytes compared to all hepatocytes in livers with fibrotic injury (Supplementary Data 5). Results showed enrichment of metabolic pathways, and the broader category of metabolic process (GO:0008152) was selected as the broadest category to evaluate changes in metabolic genes. Genes within this pathway were then evaluated for each subsequent comparison, and were further characterized by Panther protein class.
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) was performed using GSEApy (version 1.1.3)63 to compare Fibrotic Hep 1 and Fibrotic Hep 2 with both healthy hepatocytes and all identified hepatocytes. The terms were derived from the “GO_Biological_Process_2021” list.
Statistics and reproducibility
Statistical analyses were performed as described. No statistical method was used to predetermine sample size for MERFISH or snRNA-seq. No data that met the described criteria were excluded from the analyses. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The MERFISH raw and processed data generated in this study have been deposited in datadryad under the doi: 10.5061/dryad.37pvmcvsg [https://datadryad.org/stash/dataset/doi:10.5061/dryad.37pvmcvsg]. The raw snRNA-seq data used in this study are available in the GEO database under accession code: GSE210077. Processed data for snRNA-seq have been deposited in datadryad under the doi: 10.5061/dryad.37pvmcvsg [https://datadryad.org/stash/dataset/doi:10.5061/dryad.37pvmcvsg]. To aid in the visualization of these data, a web viewer has been created for the MERFISH and snRNA-seq presented in this paper. This viewer can be accessed at: https://moffittlab.github.io/visualization/2024_Human_Liver/index.html. Source data are provided with this paper.
References
Rezania, V., Coombe, D. & Tuszynski, J. A. A physiologically-based flow network model for hepatic drug elimination III: 2D/3D DLA lobule models. Theor. Biol. Med. Model 13, 9 (2016).
Teutsch, H. F. The modular microarchitecture of human liver. Hepatology 42, 317–325 (2005).
Gebhardt, R. Metabolic zonation of the liver: Regulation and implications for liver function. Pharm. Therapeut 53, 275–354 (1992).
Jungermann, K. & Keitzmann, T. Zonation of Parenchymal and Nonparenchymal Metabolism in Liver. Annu Rev. Nutr. 16, 179–203 (1996).
Martini, T., Naef, F. & Tchorz, J. S. Spatiotemporal Metabolic Liver Zonation and Consequences on Pathophysiology. Annu. Rev. Pathol.: Mech. Dis. 18, 439–466 (2023).
Kietzmann, T. Metabolic zonation of the liver: The oxygen gradient revisited. Redox Biol. 11, 622–630 (2017).
Droin, C. et al. Space-time logic of liver gene expression at sub-lobular scale. Nat. Metab. 3, 43–58 (2021).
Damm, G. et al. Human parenchymal and non-parenchymal liver cell isolation, culture and characterization. Hepatol. Int. 7, 951–958 (2013).
Roberts, S., Ludwig, J. & Larusso, N. The pathobiology of biliary epithelia. Gastroenterology 112, 269–279 (1997).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Ramachandran, P. et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 38, 1–29 (2019).
Andrews, T. S. et al. Single‐Cell, Single‐Nucleus, and Spatial RNA Sequencing of the Human Liver Identifies Cholangiocyte and Mesenchymal Heterogeneity. Hepatol. Commun. 6, 821–840 (2021).
Tanami, S. et al. Dynamic zonation of liver polyploidy. Cell Tissue Res. 368, 405–410 (2017).
Kudryavtsev, B. N., Kudryavtseva, M. V., Sakuta, G. A. & Stein, G. I. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch. B 64, 387–393 (1993).
Watanabe, T. & Tanaka, Y. Age-related alterations in the size of human hepatocytes. Virchows Arch. B 39, 9–20 (1982).
Saeter, G. et al. Changes in ploidy distributions in human liver carcinogenesis. J. Natl Cancer Inst. 80, 1480–1485 (1988).
Matsumoto, T., Wakefield, L. & Grompe, M. The Significance of Polyploid Hepatocytes During Aging Process. Cell Mol. Gastroenterol. Hepatol. 11, 1347–1349 (2021).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. 113, 11046–11051 (2016).
Moffitt, J. R. et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl Acad. Sci. 113, 14456–14461 (2016).
Aizarani, N. et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature 472, 199–204 (2019).
Petukhov, V. et al. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 40, 345–354 (2022).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2020).
Braeuning, A. et al. Differential gene expression in periportal and perivenous mouse hepatocytes. Febs J. 273, 5051–5061 (2006).
Saito, K., Negishi, M. & Squires, E. J. Sexual dimorphisms in zonal gene expression in mouse liver. Biochem. Biophys. Res. Commun. 436, 730–735 (2013).
Halpern, K. B. et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017).
González-Blas, C. B. et al. Single-cell spatial multi-omics and deep learning dissect enhancer-driven gene regulatory networks in liver zonation. Nat. Cell Biol. 26, 153–167 (2018).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
Dobie, R. et al. Single-Cell Transcriptomics Uncovers Zonation of Function in the Mesenchyme during Liver Fibrosis. Cell Rep. 29, 1832–1847.e8 (2019).
Inverso, D. et al. A spatial vascular transcriptomic, proteomic, and phosphoproteomic atlas unveils an angiocrine Tie–Wnt signaling axis in the liver. Dev. Cell 56, 1677–1693.e10 (2021).
Vento-Tormo, R. et al. Single-cell reconstruction of the early maternal–fetal interface in humans. Nature 563, 347–353 (2018).
Lambeau, G. & Gelb, M. H. Biochemistry and Physiology of Mammalian Secreted Phospholipases A2. Annu. Rev. Biochem. 77, 495–520 (2008).
Romeo, S. et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 40, 1461–1465 (2008).
Sharma, A. et al. Onco-fetal Reprogramming of Endothelial Cells Drives Immunosuppressive Macrophages in Hepatocellular Carcinoma. Cell 183, 377–394.e21 (2020).
Martínez, A. I., Pérez-Arellano, I., Pekkala, S., Barcelona, B. & Cervera, J. Genetic, structural and biochemical basis of carbamoyl phosphate synthetase 1 deficiency. Mol. Genet. Metab. 101, 311–323 (2010).
Matsumoto, S. et al. Urea cycle disorders—update. J. Hum. Genet. 64, 833–847 (2019).
Yahyaoui, R. et al. A new metabolic disorder in human cationic amino acid transporter‐2 that mimics arginase 1 deficiency in newborn screening. J. Inherit. Metab. Dis. 42, 407–413 (2019).
Fiermonte, G. et al. The Mitochondrial Ornithine Transporter Bacterial Expression, Reconstitution, Functional Characterization, And Tissue Distribution Of Two Human Isoforms*. J. Biol. Chem. 278, 32778–32783 (2003).
Valenti, L. et al. Homozygosity for the patatin‐like phospholipase‐3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology 51, 1209–1217 (2010).
Maillo, C. et al. Circadian- and UPR-dependent control of CPEB4 mediates a translational response to counteract hepatic steatosis under ER stress. Nat. Cell Biol. 19, 94–105 (2017).
Xanthakos, S. A. et al. High Prevalence of Nonalcoholic Fatty Liver Disease in Adolescents Undergoing Bariatric Surgery. Gastroenterology 149, 623–634.e8 (2015).
Harrison, S. A. et al. A Phase 3, Randomized, Controlled Trial of Resmetirom in NASH with Liver Fibrosis. N. Engl. J. Med. 390, 497–509 (2024).
Guilliams, M. et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022).
Remmerie, A. et al. Osteopontin Expression Identifies a Subset of Recruited Macrophages Distinct from Kupffer Cells in the Fatty Liver. Immunity 53, 641–657.e14 (2020).
Stockinger, B. et al. A role of la-associated invariant chains in antigen processing and presentation. Cell 56, 683–689 (1989).
Maubach, G., Lim, M. C. C., Kumar, S. & Zhuo, L. Expression and upregulation of cathepsin S and other early molecules required for antigen presentation in activated hepatic stellate cells upon IFN-γ treatment. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 1773, 219–231 (2007).
Viñas, O. et al. Human hepatic stellate cells show features of antigen‐presenting cells and stimulate lymphocyte proliferation. Hepatology 38, 919–929 (2003).
Zeng, H. et al. CD36 promotes de novo lipogenesis in hepatocytes through INSIG2-dependent SREBP1 processing. Mol. Metab. 57, 101428 (2022).
Wang, B., Zhao, L., Fish, M., Logan, C. Y. & Nusse, R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 524, 180–185 (2015).
Preziosi, M., Okabe, H., Poddar, M., Singh, S. & Monga, S. P. Endothelial Wnts regulate β‐catenin signaling in murine liver zonation and regeneration: A sequel to the Wnt–Wnt situation. Hepatol. Commun. 2, 845–860 (2018).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Cadinu, P. et al. Charting the cellular biogeography in colitis reveals fibroblast trajectories and coordinated spatial remodeling. Cell 187, 2010–2028.e30 (2024).
Wang, G., Moffitt, J. R. & Zhuang, X. Multiplexed imaging of high-density libraries of RNAs with MERFISH and expansion microscopy. Sci. Rep. 8, 4847 (2018).
Slyper, M. et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat. Med. 26, 792–802 (2020).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 9, giaa151 (2020).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 8, 329–337.e4 (2019).
Ianevski, A., Giri, A. K. & Aittokallio, T. Fully-automated and ultra-fast cell-type identification using specific marker combinations from single-cell transcriptomic data. Nat. Commun. 13, 1246 (2022).
Clarke, Z. A. & Bader, G. D. MALAT1 expression indicates cell quality in single-cell RNA sequencing data. bioRxiv 2024.07.14.603469. https://doi.org/10.1101/2024.07.14.603469 (2024).
Montserrat-Ayuso, T. & Esteve-Codina, A. Revealing the Prevalence of Suboptimal Cells and Organs in Reference Cell Atlases: An Imperative for Enhanced Quality Control. bioRxiv 2024.04.18.590104. https://doi.org/10.1101/2024.04.18.590104 (2024).
Garcia-Alonso, L. et al. Single-cell roadmap of human gonadal development. Nature 607, 540–547 (2022).
Fang, Z., Liu, X. & Peltz, G. GSEApy: a comprehensive package for performing gene set enrichment analysis in Python. Bioinformatics 39, btac757 (2022).
Acknowledgements
The authors thank Jasneet Aneja for assistance in organizing the collection of liver tissue, Kenneth Tanabe for assistance with sample collection, and Nicole Brousaides and the MGH Histopathology Research Core for help sectioning tissue. We thank Sonya MacParland, Gary Bader, Tallulah Andrews, and Jawairia Atif for helpful discussions and assistance in selecting genes for the MERFISH probe set. We thank Cristin McCabe for assistance with data management, and Adam Slamin, Dan Dubinsky, and the Broad Genomics Platform for help with generation of single-nucleus RNA sequencing data. A.C.M. was supported by internal funds from UMass Chan, and R.J.X., J.R.M., and A.C.M were supported by grants from the Chan Zuckerberg Initiative.
Author information
Authors and Affiliations
Contributions
B.P., B.R.W., J.R.M, and A.C.M. conceived the study and designed the experiments. Samples were prepared for MERFISH by B.P. and imaged by B.R.W. with support from A.S. and E.T.E. snRNA-seq was performed by L.A.-Z. and S.M. with support from Å.S., E.T.E, J.D., and R.J.X. Computational analyses were performed by B.R.W., B.P., and R.U.R. Visualization tools were built by L.G. and T.L. The manuscript was written by B.R.W., B.P., J.R.M, and A.C.M. with input from all other authors.
Corresponding authors
Ethics declarations
Competing interests
R.J.X. is a co-founder of Celsius Therapeutics and Jnana Therapeutics, board director at MoonLake Immunotherapeutics, and consultant to Nestlé. J.R.M is a co-founder of, stake-holder in, and advisor for Vizgen. J.R.M. is an inventor on patents associated with MERFISH applied for on his behalf by Harvard University and Boston Children’s Hospital. J.R.M.‘s interests were reviewed and are managed by Boston Children’s Hospital in accordance with their conflict-of-interest policies. J.R.M. has received research funding from Merck for unrelated projects. A.C.M. has received research funding from Boehringer Ingelheim and GlaxoSmithKline for unrelated projects. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Bruce Wang, and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Watson, B.R., Paul, B., Rahman, R.U. et al. Spatial transcriptomics of healthy and fibrotic human liver at single-cell resolution. Nat Commun 16, 319 (2025). https://doi.org/10.1038/s41467-024-55325-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-55325-4
This article is cited by
-
Hepatic stellate cells: balancing homeostasis, hepatoprotection and fibrogenesis in health and disease
Nature Reviews Gastroenterology & Hepatology (2025)
-
The spatial and temporal activation of macrophages during fibrosis
Nature Reviews Immunology (2025)