Abstract
The search for simple principles that underlie the spatial structure and dynamics of plant communities is a long-standing challenge in ecology1,2,3,4,5,6. In particular, the relationship between species coexistence and the spatial distribution of plants is challenging to resolve in species-rich communities7,8,9. Here we present a comprehensive analysis of the spatial patterns of 720 tree species in 21 large forest plots and their consequences for species coexistence. We show that species with low abundance tend to be more spatially aggregated than more abundant species. Moreover, there is a latitudinal gradient in the strength of this negative aggregation–abundance relationship that increases from tropical to temperate forests. We suggest, in line with recent work10, that latitudinal gradients in animal seed dispersal11 and mycorrhizal associations12,13,14 may jointly generate this pattern. By integrating the observed spatial patterns into population models8, we derive the conditions under which species can invade from low abundance in terms of spatial patterns, demography, niche overlap and immigration. Evaluation of the spatial-invasion condition for the 720 tree species analysed suggests that temperate and tropical forests both meet the invasion criterion to a similar extent but through contrasting strategies conditioned by their spatial patterns. Our approach opens up new avenues for the integration of observed spatial patterns into ecological theory and underscores the need to understand the interaction among spatial patterns at the neighbourhood scale and multiple ecological processes in greater detail.
Similar content being viewed by others
Main
Species-rich plant communities such as tropical forests have been investigated by ecologists for decades, but explaining their high species richness remains a challenge for ecological theory1,2,3,4,5,6,15,16. Although numerous studies have been devoted to this issue, mechanistic connections among features of plant communities and species coexistence are incompletely understood7,8,9. For example, a key feature in forests is the spatial aggregation of tree species, which has long been used to infer mechanisms that contribute to coexistence17,18,19,20,21. This is because aggregation is related to ecological processes such as negative conspecific density dependence17,18,22, dispersal limitation2,23, mycorrhizal associations12,13,14 and habitat association24,25. Conspecific spatial aggregation Ω is usually defined as the average density D of conspecific trees in neighbourhoods around individual trees of the same species divided by the mean tree density λ of the species in the forest plot20,25. Hence, Ω describes the extent to which trees of the same species tend to occur in spatial clusters. Several theoretical and observational studies suggest that conspecific aggregation is related to species abundance, whereby species with lower abundance show higher levels of aggregation8,19,20,21,26,27,28. However, other studies have reported only weak relationships between aggregation and species abundance29 (Supplementary Text). Nevertheless, a relationship between aggregation and abundance could have important consequences for the maintenance of high species richness. This is because recent theoretical work7,8,9 has indicated a connection between conspecific aggregation and the rare species advantage required for coexistence.
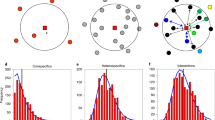
Different aggregation–abundance relationships are possible. At one extreme, when conspecific clusters form mostly near adults (for example, owing to short-distance dispersal7,8,26 and/or mycorrhizal associations12,13,14), the density D of conspecific trees in the neighbourhood of individual trees will be similar for species with low and high abundance, but species with lower abundance will have fewer clusters (compare Fig. 1a and Fig. 1b). Thus, aggregation (that is, Ω = D/λ)20,25 increases if abundance (and therefore the mean tree density λ) decreases. At the other extreme, when local clusters are created away from conspecific adults8 (for example, owing to clumped animal seed dispersal (zoochory) or canopy gaps24,30,31,32,33), fewer seeds will reach the cluster locations if the species has lower abundance. Consequently, the neighbourhood density D will become proportional to abundance and therefore aggregation Ω becomes independent of abundance (compare Fig. 1a with Fig. 1c).
a, Illustration of a simulated pattern of a species with abundance N = 500 individuals in a 25-ha area with mean neighbourhood density D = 0.0116 trees per m2, mean tree density λ = 0.002 trees per m2 and aggregation Ω = D/λ = 5.8 (colours represent different clusters of individual trees of the same species). b, Entire clusters of the pattern in a were removed (N = 100). Through this step, the neighbourhood density was approximately maintained (D = 0.0107), but because λ was reduced by a factor of 1/5, aggregation increased approximately 5 times (Ω = 26.7). c, Individuals of the pattern in a were randomly removed (N = 100). Through this step, D and λ were reduced by a factor of 4.93 and 5.0, respectively, which approximately maintained aggregation (Ω = 5.9). We estimated D and Ω for 10-m neighbourhoods around the focal individuals.
Given the links between conspecific aggregation, negative density dependence and coexistence7,8,9,26, the aggregation–abundance relationship may be related to the latitudinal diversity gradient, which is proposed to be driven by ecological, evolutionary, regional or historical effects34. Here we conduct a comprehensive analysis of both how spatial neighbourhood patterns of trees derived from large forest inventories6 and the relationship between aggregation and abundance change with latitude. We propose underlying ecological mechanisms and integrate our results into mathematical theory to investigate how the aggregation–abundance relationship may affect the rare species advantage and thereby species coexistence (Box 1).
Although aggregation can be defined in various ways19,20,25,29,35 (Extended Data Fig. 1), we derive measures of spatial patterns from established approaches8,36,37,38 that model the effect of neighbours on the performance of individual plants (Box 1). This links the competition of individual trees with the dynamics of species at the community scale. We illustrate our new theory using the example of neighbourhood competition, in which tree survival is reduced in areas of high tree density by competition for space, light or nutrients39, or through predators or pathogens17,18.
A latitudinal trend in aggregation
Using data on 720 focal species in 21 temperate, subtropical and tropical forest plots with sizes of 20–50 ha from a global network of forest research plots (CTFS-ForestGEO)6 (Extended Data Table 1), we found that species with lower abundance tended to be more aggregated than species with higher abundance (Fig. 2). Notably, when describing the relationship between observed aggregation kff* and abundance Nf* for each forest plot by a power law \({k}_{{ff}}^{* }=a\,{N}_{{f}}^{* e}\) (refs. 21,27,28) (Fig. 2a,b), the exponent e followed a marked latitudinal gradient (Fig. 2c and Extended Data Fig. 2). Tropical forests showed a weak negative relationship between aggregation and abundance (that is, exponent values close to zero; Fig. 2a and Extended Data Fig. 3a–f). By contrast, in temperate forests, species with low abundance showed generally high aggregation, and aggregation strongly decreased with increasing abundance (that is, exponent values mostly below –0.58; Fig. 2b and Extended Data Fig. 3n–u). In contrast to conspecifics, heterospecific associations were not related to abundance (except for some weak correlations in temperate forests; Extended Data Fig. 4).
a, Aggregation values for species in a tropical forest (Mo Singto (MST) plot) plotted over their respective abundances per ha (dots) and fitted linear regression between ln(kff) and ln(Nf) (line). b, Same as a, but for a temperate forest (Donglingshan (DLS) plot). c, Latitudinal gradient in the exponent of the aggregation–abundance relationship for the 21 forest plots. To show the overall trend in the data, we fit a linear regression. Aggregation is defined in Box 1 equation (3) and was estimated for neighbourhoods of 15 m. We used in our analyses 720 species with at least 50 large trees20 (diameter at breast height ≥ 10 cm). For plot characteristics, sample sizes and plot acronyms see Extended Data Table 1. Circles of subtropical plots are marked with a red edge.
The latitudinal gradient in the relationship between conspecific aggregation and abundance (Fig. 2c) and the absence of such a relationship for heterospecifics (Extended Data Fig. 4) suggested that simple principles may drive the complex spatial structure and dynamics of plant communities across latitudinal gradients. We also observed similar latitudinal gradients in the proportion of species that show mostly animal seed dispersal11 and in the proportion of species with an arbuscular mycorrhizal (AM) association14 (Extended Data Table 1). Temperate forests are usually dominated by ectomycorrhizal (EM) tree species, whereas tropical forests are dominated by AM tree species14.
For the combination of the two traits (that is, zoochory and AM association), there was an even stronger latitudinal gradient (Fig. 3a), which suggested an explanation of the observed latitudinal gradient in the aggregation–abundance relationship (Fig. 3b). Species-specific EM fungi facilitate conspecific recruitment by forming a physical sheath around young feeder roots14,40 and counteracting negative competitor-driven or pathogen-driven effects12,13,14, thereby leading to increased aggregation. Therefore, seed dispersal close to conspecific adults should be advantageous in temperate forests, where many species show an EM association. By contrast, seed dispersal farther away from conspecific adults should be more advantageous for AM trees, given that an AM association provides less protection against competitors or pathogens that accumulate near conspecifics than an EM association13,40. In summary, we propose that the key mechanism that leads to the different responses of aggregation to abundance is the way that local clusters emerge with respect to conspecific adults. That is, in tropical forests, mechanisms such as animal seed dispersal lead to the emergence of clusters away from adults, whereas in temperate forests, clusters form close to adults8 (Fig. 1).
a, Latitudinal gradient in the proportion of species per plot that show both mostly animal seed dispersal and AM association. b, Relationship between the exponent of the aggregation–abundance relationship and the proportion of species per plot that show mostly animal seed dispersal and AM association for the 21 forest plots. For plot characteristics, sample sizes, raw data, plot acronyms and relationships with animal seed dispersal and AM association, see Extended Data Table 1. To outline the overall trend in the data, we fit a polynomial regression of order 2 in a and we fit a linear regression in b. Most species showed either AM or EM associations. Coloured discs indicate the example plots presented in Fig. 2a,b.
Regardless of the ultimate mechanisms that generate it, the systematic change in the relationship between aggregation and abundance with latitude (Fig. 2c) has important implications for coexistence dynamics and theory. Stable coexistence requires that the abundance of a newly invading (or an almost extinct) species increases1,5,8,9 (that is, a rare species advantage). Common non-spatial models that feature this invasion criterion ignore the possibility of a negative aggregation–abundance relationship by assuming that the invading species does not suffer from conspecific competition9. This assumption was met in our data for tropical forests, where aggregation was weakly related to abundance (Fig. 2a,c). In this case, mean conspecific neighbourhood densities \(\bar{{C}_{f}}\) were almost linearly related to species abundance Nf (as constant aggregation \({k}_{{ff}}^{* }\) led to \(\bar{{C}_{f}}=c{k}_{{ff}}^{* }{N}_{f}^{* }\); Box 1 equation (3); Fig. 1c). However, an increase in aggregation with decreasing abundance, as observed for temperate forests (that is, higher latitudes; Fig. 2b,c), challenges the common assumption of invasion analysis. This is because local conspecific neighbourhood densities were almost independent of abundance, given that the exponent of the aggregation–abundance relationship approached values of −1 (from \(\bar{{C}_{f}}=c{k}_{{ff}}^{* }{N}_{f}^{* }\) and \({k}_{{ff}}^{* }=a/{N}_{f}^{* }\) follows \(\bar{{C}_{f}}={ca}\); Box 1 equation (3); Fig. 1b). Thus, individuals of species with low and high abundance experience similar degrees of conspecific competition. Consequently, existing invasion analysis can break down9. Spatial aggregation of trees is therefore closely linked to species coexistence, and we need new theories to determine whether and under which circumstances species with low abundance are likely to increase9.
Including aggregation into the theory
A theory that describes how the response of conspecific aggregation to abundance influences species coexistence requires a dynamic population model that relates competition at the population level to spatial patterns in the neighbourhood of individual trees. Here we derived such an approach8, which incorporated information on spatial patterns at the scale of individual trees provided by the ForestGEO datasets6. We exemplified our theory using a simple model, which uses the following assumptions: (1) reproduction is density-independent7; (2) survival of individual trees is reduced in areas of high local tree density, as described by neighbourhood crowding indices36,37,38 (Box 1); and (3) immigration may occur at a constant rate:
Here Nf,t is the abundance of the focal species f at time t, the time interval Δt is the 5-year census interval, \({\widetilde{\lambda }}_{f}({N}_{f,t})\) is the per capita population growth rate of species f as a function of species abundance Nf,t, sf is a density-independent per capita background survival rate, rf is the per capita recruitment rate, βff is the conspecific neighbourhood-scale competition coefficient and vf is a parameter that governs the magnitude of a constant immigration rate rf vf (ref. 41). Wf(Nf,t), the fitness factor42, is the average of the sum Cof + Iof of conspecific and heterospecific neighbourhood crowding taken over all individuals o of a focal species f (Box 1, equation (6)) and incorporates information on spatial patterns, abundance and the aggregation–abundance relationship into our model (equations (11b) and (11f)). To keep our example model simple, we did not consider tree size.
We used a spatially explicit and individual-based implementation of our model8, in which spatial patterns (measured by \({k}_{{ff}}^{* },{k}_{{fh}}^{* }\)) emerged as a consequence of spatially explicit recruitment of offspring and negative density dependence (Methods). Changing only the way offspring are placed relative to conspecific adults led to the range of observed exponents of the aggregation–abundance relationship (Extended Data Fig. 5). When most offspring were placed close to their parents, locally high adult densities were maintained through the continuous placement of new individuals into these clumps, which was controlled through subsequent thinning due to density-dependent mortality. This mechanism led to an aggregation–abundance relationship similar to that of temperate forests8, and we found that the community cannot be invaded by a species at low abundance (Extended Data Fig. 5a,f). However, if fewer recruits were placed close to their parents, then the dependence of aggregation on abundance was weaker (Extended Data Fig. 5b–e). Consequently, a species at low abundance can invade because it experiences reduced competition (that is, lower values of total crowding \(\bar{{C}_{f}}+\bar{{H}_{f}}\)) (Extended Data Fig. 5j,o). This rare species advantage emerges as a consequence of a positive fitness-density covariance8,42 if a species at a lower abundance is surrounded by a lower number of conspecific neighbours (that is, lower values of \(\bar{{C}_{f}}\); Extended Data Fig. 5o,t).
By taking a mean-field approach43 (that is, diffuse competition at the community scale8) and assuming zero-sum dynamics2 (that is, the total number of trees is fixed; Extended Data Fig. 5f–j), we decoupled the multispecies model (equation (1a)) by means of the fitness functions
where \({k}_{ff}^{* }\), \({k}_{{fh}}^{* }\), \({N}_{f}^{* }\) and J* are the observed aggregation, heterospecific association, abundance and total community size, respectively, bf is the exponent of a corrected power-law aggregation–abundance relationship (equation (12)), Bf is the average competitive strength of one heterospecific neighbour of species f relative to that of one conspecific and c is a scaling factor (Box 1). We parameterized our mathematical model (equations (1a) and (1b)) by using information from our large forest plots. All parameters of the model, and the measures of spatial patterns, were species-specific. However, given that we had only limited information, we used the same parameter value for all species for several parameters (that is, rf, sf and bf). For the estimation of βff, we assumed that the observed abundance values were close to equilibrium (see Methods for details).
An expanded spatial-invasion criterion
The effect of aggregation on coexistence can be analysed by using an invasion criterion1,9. An invading (or almost extinct) species will be able to increase from a low abundance if its per capita population growth rate (equation (1a) and Fig. 4a) is sufficiently positive (that is, a rare species advantage). Our spatial-invasion criterion therefore requires that the (scaled) per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{s})/{r}_{f}\) should have at a low abundance Ns at least a value δ larger than zero (equation (17a)). For the case in which spatial patterns are the only mechanism to facilitate coexistence (that is, no niche overlap or immigration; Bf = 1, vf = 0), we obtained the simple invasion criterion
where Ns is the small invasion abundance (here Ns = 5 or 10) and Nf* the observed abundance, bf is the exponent of the corrected aggregation–abundance relationship (equation (12)), and the quantity ρf = Bf (kfh*/(kff* − kfh*))(J*/Nf*) (equation (17d)) is a risk factor, as larger values of ρf make it more difficult to meet the invasion criterion. The risk factor combines the main influence of niche differences (Bf), spatial pattern (kff* and kfh*) and relative abundance (Nf*/J) on the invasion criterion into a single compound index.
a, The scaled per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{f})/{r}_{f}\) for five example species of different forests plotted over abundance Nf. We scaled \({\widetilde{\lambda }}_{f}({N}_{f})\) with the reproduction rate rf to obtain a quantity that is comparable among forest plots. The species include Castanopsis acuminatissima of the MST plot (bf = −0.3), Ficus langkokensis of the Xishuangbanna plot (bf = −0.52), Carya tomentosa of the Tyson Research Center plot (bf = −0.75), Ostrya virginiana of the Wabikon plot (bf = −0.92) and Maackia amurensis of the Changbaishan plot (bf = −1.08). We also show the theoretical values of \({\widetilde{\lambda }}_{f}({N}_{f})/{r}_{f}\) for bf = 0 (black) and −1 (grey). b, Same as a, but for the averages over all focal species in tropical, subtropical and temperate forests. c, Trade-off between the exponent bf of the aggregation–abundance relationship (x axis) and the maximal risk factor ρf,max (equation (19)) that just satisfies the spatial invasion criterion (equations (2) and (18)): temperate forests (smaller bf) show smaller risk factors ρf than tropical forests (larger value of bf). The lines show ρf,max for a small scaled growth rate δ = 0.0035 (see equation (19)) and example ratios of Ns/Nf* = 5/50 (red) and Ns/Nf* = 5/5,000 (black), where Ns (=5) is the small invasion abundance and Nf* (=50 and 5,000). The circles show for each species the risk factor ρf and the cyan discs mark the 33 out of 720 species that do not satisfy the criterion (19 species with ρf > 350 are not visible). The data are from scenario 1 (that is, no niche differences, no immigration and observed equilibrium abundances) (Extended Data Table 1).
Equation (2) suggests that temperate and tropical forests use contrasting strategies to fulfil the invasion criterion (Fig. 4c). Tropical forests showed smaller negative values of the exponent bf that allow them to have larger values of the risk factor, whereas temperate forests showed smaller values of the risk factor but larger negative values of the exponent bf. Moreover, less abundant species (that is, larger values of \({N}_{s}/{N}_{f}^{* }\) in equation (2)) must show, across all latitudes, larger aggregation kff* than more abundant species to produce smaller values of the risk factor (the red versus black line in Fig. 4c). Moreover, larger niche differences (that is, Bf decreases) in combination with larger aggregation kff* enhance coexistence (equations (18) and (19)). We anticipate that this effect would be particularly important for the persistence of low-density species.
To assess the order of magnitude of the effects of spatial structure, niche differences and immigration on the per capita population growth rate, we investigated four scenarios (Table 1). Coexistence was generally facilitated to a similar extent in the analysed forest plots by the observed spatial patterns (Table 1, Fig. 4b and Extended Data Fig. 6a,e). For example, model results for scenario 1 (without niche differences and without immigration) produced at a small abundance of 10 individuals mean-scaled per capita growth rates of 0.036, 0.043 and 0.05 for tropical, subtropical and temperate forests, respectively (Table 1). Note that these values produced stable dynamics in our simulations (Extended Data Fig. 5i,j). Species in temperate forests tended to show slightly higher values of the scaled per capita growth rates on average compared with tropical forests (Table 1 and Fig. 4b).
Using expanded versions of the model, adding niche differences (Bf < 1) and/or a small constant immigration rate (vf = 0.1) (Table 1) led to further increases in the scaled per capita growth rates of similar magnitude. Although the effect of a small immigration rate on the per capita population growth rate rapidly declined with increasing species abundance (Table 1 and Extended Data Fig. 6g,h), the effect of spatial patterns and niche differences persisted for larger abundance values (Extended Data Fig. 7).
Discussion
Developing an approach that integrates the observed spatial patterns of individual trees in forests with ecological processes into mathematical theory is a considerable challenge. Here we presented a unified framework that combines spatial point process theory with population models to derive expectations about how interactions between different spatial patterns and processes affect the ability of invading species to expand. The framework relies on spatial patterns, such as conspecific aggregation and heterospecific association, which link neighbourhood-scale competition of individual trees with species dynamics at the community scale. Our theory led to a closed-form expression for the per capita population growth rate of species as a function of spatial patterns, demography, niche overlap and immigration (equations (1a) and (1b) and (13)), which facilitated a general understanding of spatial coexistence in forests (equations (2) and (18)).
Several important ecological insights resulted from this new theory. First, we showed that the spatial patterns found in forests have in general a stabilizing effect under neighbourhood competition7,8, as they substantially increased the per capita population growth rate of an invading species compared with a value of zero for the non-spatial case (Table 1). This result challenges the prevalent perspective that spatial patterns alone cannot promote coexistence44,45. However, this assertion arises from the assumption of previous models7,26,35,44,46 that place recruits close to their parents, which leads to destabilizing negative aggregation–abundance relationships (with exponent values close to −1) (Fig. 1 and Extended Data Fig. 5). Our results therefore highlight the crucial role that spatial patterns and the aggregation–abundance relationship have in shaping coexistence outcomes.
Second, our spatial theory suggests that temperate and tropical forests both satisfy the spatial invasion criterion required for coexistence to a similar extent (Table 1), but they do so in contrasting ways. Tropical forests are not subject to destabilizing negative aggregation–abundance relationships (that is, they show low negative values of the exponent bf) owing to the combined effect of animal seed dispersal and an AM association, but show higher values of the risk factor ρf (Fig. 4c and Extended Data Fig. 8b,c). By contrast, temperate forests show a stronger negative dependency of aggregation on abundance, but this disadvantage is outweighed by lower values of the risk factor through higher relative species abundance and/or stronger aggregation facilitated by an EM association12 (Fig. 4c and Extended Data Fig. 8a,c,d).
Third, the negative aggregation–abundance relationship found here for temperate forests challenges the implicit assumption of common non-spatial implementations of the invasion criterion1,9 that an invading species does not suffer from conspecific competition9 (such as shown in Extended Data Fig. 5t). In violation of this assumption, if aggregation is caused by short-distance dispersal, most individuals will be close to a conspecific competitor, even if the species abundance is low (Extended Data Fig. 5p). This mechanism leads to conspecific population-level competition coefficients that are not constant as commonly assumed47, but increase with decreasing abundance (equation (11c)). We developed a new theory that overcame this issue and enabled us to determine whether and under what circumstances the abundance of invading species are likely to increase in a spatial context (equation (18)). We propose that similar criteria can also be derived for other population models (Supplementary Text).
Our spatial analysis of 21 large forest plots revealed a latitudinal gradient in the strength of the relationship between conspecific aggregation and abundance (Extended Data Fig. 2d). Our model simulations (Extended Data Fig. 5) provided evidence that this latitudinal gradient is related to seed dispersal, and on the basis of our correlation analysis (Fig. 3), we propose that it arises as an interaction between animal seed dispersal11 and mycorrhizal associations12,13,14. Nonetheless, given that multiple ecological variables correlate with latitude, other mechanisms could contribute to this pattern. However, a recent global study10 outlined the joint evolution of mycorrhizal symbiosis, seed dispersal and pollination in tree species because they each interact in ways that are mediated by the spatial structure of tree populations. Their results support our dispersal–mycorrhiza hypothesis, whereby most AM-associated trees have biotic seed dispersal and biotic pollination, with long dispersal distances, whereas most EM-associated trees have abiotic seed dispersal and wind (abiotic) pollination mode, with shorter dispersal distances10. The dispersal–mycorrhiza hypothesis therefore provides an extra dimension to the study of negative conspecific density dependence. An integrated understanding of the interacting effects of animal seed dispersal and mycorrhizal associations will be fundamental to our understanding of the forces that structure forest diversity and composition.
The simple spatial invasion criterion corroborated our hypothesis that simple principles may drive the complex spatial structure and dynamics of plant communities across latitudinal gradients, but this structural simplicity hides substantial biological complexity. Moving forward, we suggest that methods that model the forces that structure forest diversity and composition should incorporate mechanistic representations of the processes that can potentially drive the aggregation–abundance relationship. Here we showed that they may include seed dispersal11,48, habitat association24 and its interactions with mycorrhizal associations10,12,13,14 as main processes. A priority would be to consider species-specific aggregation–abundance relationships and to study how the exponent bf depends on the traits and the dispersal syndrome of the species. More detailed versions of our spatial theory may also consider variation in tree size, as included in classical neighbourhood crowding indices36,37, to describe asymmetric competition. Preliminary analyses suggest that our key results hold when including the size of trees and their growth.
Overcoming limitations of non-spatial models, as often used in contemporary coexistence theory1,4,15,16,47,49, requires approaches that explicitly consider the lower-level processes that generate the phenomenon of interest50. Our results demonstrated the utility of functions that describe the parameters of the population models, here the population-level competition coefficients αfi (between species i and f; equation 11d), as a function of neighbourhood-scale competition coefficients βfi and measures of spatial patterns (that is, kff, kfh and Bf; equations (11c) and (11d))8. Our scaling approach had the advantage that the parameters of these functions can be determined from field measurements, such as the ForestGEO plots6 in our case. Taking this approach, we demonstrated that spatial patterns that emerge from neighbourhood-scale processes have a key role in species coexistence, which underscores the need to understand the mechanisms that underpin the spatial heterogeneity of forests in greater detail.
Methods
Study areas
Twenty-one large forest dynamic plots of areas between 20 and 50 ha with similar numbers of tropical, subtropical and temperate forests were used in the current study (Extended Data Table 1 and Supplementary Table 1). The forest plots are part of the ForestGEO network6 and are located in Asia and the Americas ranging in latitudes from 6° 40′ N to 48° 08′ N. Tree species richness among these plots ranges from 36 to 468. All free-standing individuals with diameter at breast height (d.b.h.) ≥ 1 cm were mapped, their size measured and identified. We focused our analysis here on individuals with d.b.h. ≥ 10 cm (resulting in 313,434 individuals) and tree species with more than 50 individuals (resulting in initially 737 species). The 10-cm size threshold excluded most of the saplings and enabled comparisons with previous spatial analyses. Shrub species were excluded. We also excluded 15 species with low aggregation (that is, kff* < kfh*; Box 1), which would lead to negative growth rates at small abundance values: ten of them from BCI, two from MST, two from NBH and one from FS (for definitions of plot acronyms, see Extended Data Table 1). These (generally less abundant) species are probable relicts of an earlier successional episode when they were more abundant19,51. We also excluded the two species Picea mariana and Thuja occidentalis of the Wabikon forest that are restricted to a patch of successional forest that was logged approximately 40 years ago52.
Most forest plots (18 out of our 21 plots, those with more than 1 census) enabled the estimation of the average mortality risk of individuals with d.b.h. ≥ 10 cm within one census period. We estimated mortality across all species and obtained for each forest plot one average mortality rate for trees with d.b.h. ≥ 10 cm (Extended Data Table 1). We also determined for all species used in our analyses the mycorrhizal association types based on available global datasets53,54,55 and website sources (https://www.mycorrhizas.info/). To determine whether a species is mainly dispersed by animals (zoochory), we used the Seed Information Database (https://ser-sid.org/) of the Society for Ecological Restoration and the Royal Botanic Gardens Kew and available literature56. Species without descriptions of mycorrhizal associations and dispersal modes were assigned according to their congeneric species. The proportion of focal species with zoochory, with AM association and with both are shown in Extended Data Table 1.
Proxy for pairwise competition strength between species
Some of our analyses required the ratio βfj/βff (Box 1) that describes the relative competitive effect of individuals of species i on an individual of the focal species f at the neighbourhood scale36,57. In general, it is challenging to derive estimates for the pairwise competition coefficients37 because this would require unfeasibly large datasets to obtain a sufficient number of neighboured f–j species pairs for less abundant species. We therefore compared two scenarios. In scenario 1, we assumed that conspecific and heterospecific individuals compete equally, thus βfj/βff = 1. In scenario 2, we assumed that individuals that are close relatives compete more strongly or share more natural competitors or pathogens than distant relatives58 (that is, βfj/βff < 1). As proxy for this effect, we used phylogenetic distances59, given in millions of years (Myr), as a surrogate for the relative competition strength because they are available for the species in our plots based on molecular data or the Phylomatic informatics tool60. To obtain consistent measurements for the ratio βfj/βff among forest plots, phylogenetic similarities βfj/βff were scaled between 0 and 1, with conspecifics set to 1 and a similarity of 0 assumed for a phylogenetic distance of 1,200 Myr, which was larger than the maximal observed distance (1,059 Myr). This was necessary to avoid discounting crowding effects from the most distantly related neighbours58.
For plots without molecular data, we used the V.PhyloMaker2 package (v.0.1.0)61 to generate a phylogenetic tree for each plot using GBOTB.extented.WP.tre updated from the dated megaphylogeny GBOTB62 as a backbone. For the other eight plots with molecular data, we followed a previously reported method63 to build the phylogenetic tree based on DNA barcode data. We then used the cophenetic function in the picante package (v.1.8.2)64 to calculate phylogenetic distance for each plot. For this, we assumed that functional traits are phylogenetically conserved36,58,65. The analyses to generate phylogenetic trees and to calculate phylogenetic distances were performed using R (v.4.3.2)66.
Crowding indices describing competition of individual trees at the neighbourhood scale
We assumed in our example model that survival of a focal tree k is reduced in areas of high local density of conspecifics and heterospecifics (that is, neighbourhood crowding), for example, through competition for space, light or nutrients, or predators or pathogens17,18,39,67, whereas reproduction is density-independent with per capita rate rf. However, our approach was also able to deal with alternative assumptions on the processes driven by neighbourhood crowding. For example, analogous models were derived for crowding effects on the reproductive rate and/or the establishment of offspring (Supplementary Text).
We describe the neighbourhood crowding around a given tree o of a focal species f by commonly used neighbourhood crowding indices36,37,58,68,69,70 (Box 1), but used separate indices for conspecific and heterospecific trees. The conspecific crowding index Cof of a given individual o of a given focal species f counts the number nf of conspecific neighbours j that have a distances doj smaller than a given neighbourhood radius r, but weights each neighbour o by its inverse distance 1/doj, assuming that farther away neighbours compete less (equation (7a)). The heterospecific crowding index Hof does the same with all heterospecifics (equation (7b)), and the heterospecific interaction crowding index Iof weights heterospecifics additionally by their relative competitive strength βfj/βff (equation (7c); see above ‘Proxy for pairwise competition strength between species’). Thus, we estimated for each individual o three crowding indices:
conspecific crowding:
heterospecific crowding:
with niche differences:
where ni is the number of neighbours of species i within distance r of the focal individual, doj is the distance between the focal individual o and its jth neighbour of species i, and βfi/βff is the competitive effect of one individual of species i relative to that of the focal species f (refs. 36,37,68,69,70).
Survival probability of individual trees
To link the survival of an individual o to its crowding indices Cof and Iof, we followed earlier work on individual neighbourhood models36,37,58,69,70 and assumed that the survival probability sof of a tree o of species f is given by
where sf is a density-independent background survival rate of species f and βff is the neighbourhood-scale conspecific competition coefficients of species f (ref. 36). Statistical analyses with neighbourhood crowding indices have shown that the growth and survival of trees depend on their neighbours mostly within distances r of up to 10 or 15 m (ref. 70). We therefore estimated all measures of spatial neighbourhood patterns with a neighbourhood radius of r = 15 m.
Average survival rate of species
We used scale transition theory42 and spatial point process theory25 to transfer the individual-based microscale information on the number and distance of conspecific and heterospecific neighbours of focal individuals, which are provided by the ForestGEO census maps, into macroscale models of community dynamics8. To this end, we averaged the survival probabilities sof of all individuals o of the focal species f (equation (8)), to obtain the average population-level survival rate \({\bar{s}}_{f}\), for which we derived a closed-form expression for gamma-distributed crowding indices8:
where \(\bar{{C}_{f}}\) and \(\bar{{I}_{f}}\) are the average crowding indices, and γfC = ln(1 + DfC βff)/(DfC βff) and γfI = ln(1 + DfI βff)/(DfI βff) arise through the averaging step because of the nonlinearity in equation (8)8, and are driven by the dispersion (that is, the variance-to-mean ratio) DfC and DfI of the distribution of the crowding indices \(\bar{{C}_{f}}\) and \(\bar{{I}_{f}}\), respectively. In our case of high survival, where DfC βff and DfI βff are both small, γfC and γfI are near one and can be neglected (that is, γfC ≈ 1 and γfI ≈ 1).
Link between average crowding indices and spatial patterns
To incorporate the population-level survival rate (equation (9)) into our population model, we decomposed the average crowding indices into species abundance and measures of spatial patterns (Box 1). In brief, we did this by expressing the crowding indices in terms of the pair correlation function, a basic summary function of spatial statistics25, and the mean density λf = Nf/A of the species f across the whole plot of area A (see equations (S1)–(S8) in the Supplementary Text). The resulting measures kff and kfh of spatial patterns quantify the increase or decrease in average conspecific and heterospecific neighbourhood crowding, respectively, relative to the reference case without spatial patterns. For conspecifics, we expect under a random distribution of the focal species a mean crowding index of \(\bar{{C}_{f}}={c}{N}_{f}\) (where c = 2π r/A is a scaling factor, with A being the area of the plot and r the radius of the neighbourhood; see equation (S7) in the Supplementary Text), and for heterospecifics, we expect under independent placement (of the focal species with respect to the heterospecifics) a mean crowding index of \(\bar{{H}_{f}}=c\sum _{i\ne f}{N}_{i}\) (ref. 25). We therefore obtained
where we define Bf in equation (10c) as \({B}_{f}=\bar{{I}_{f}}/\bar{{H}_{f}}\) to be the average competitive strength of one heterospecific neighbour relative to that of one conspecific. In a subsequent step, we assumed that Bf is approximately constant in time (that is, our mean-field approximation).
The quantity kff in equation (10a) measures spatial patterns in conspecific crowding of species f (kff > 1 indicates aggregation, and kff < 1 regularity), and the quantity kfh in equation (10b) measures patterns of heterospecific association around the focal species f (kfh < 1 indicates segregation, and kfh > 1 attraction). Note that our measure of conspecific aggregation, which weights neighbours by distance, is highly correlated to Condit’s omega measure of aggregation20 that counts the number of neighbours without weighting by distance (Extended Data Fig. 1). We also found that the strength of the latitudinal gradient in the exponent of the aggregation–abundance relationship (expressed as the R2) was for a radius of, for example, r > 10 m basically independent of the neighbourhood area over which conspecific aggregation was measured (Extended Data Fig. 2c,f). This was expected because of the distance-weighting (Box 1), whereby distant neighbours contribute little to total neighbourhood crowding.
Mean-field assumption
A crucial insight used in our approach8 is that crowding competition of individual trees, as described by equation (8), leads to diffuse competition at the population level in species-rich communities. That is, when taking a mean-field approximation43,71, the species-specific competition strengths of heterospecifics can be replaced in the macroscale model by a temporally constant average heterospecific competition strength Bf (ref. 8) (equation (10c)), which summarizes the emerging effects of the pairwise neighbourhood-scale competition coefficients βfi/βff at the population level. For species-rich forests at or near a stationary state, Bf is a good approximation of a species-specific constant (see the supplementary text in ref. 8). As we will see, a constant Bf simplifies the matrix of the community-level competition coefficients (equation (11d)) and enables analytical expressions of the invasion condition and the equilibria Nf* of our multispecies model (equations (11a) and (11b)) for the case that aggregation is independent of abundance.
Zero-sum assumption
Local density dependence on survival as assumed here (equation (8)) controls local tree densities and causes approximate zero-sum dynamics2, in which the total number J of individuals remains approximately a constant J*. For example, zero-sum dynamics emerged in our individual-based simulations of the extended multispecies model (blue lines in Extended Data Fig. 5f–j). The number of heterospecifics is therefore given in good approximation by \(\sum _{i\ne f}{N}_{i}={J}^{* }-{N}_{f,t}\). Using the zero-sum approximation together with the mean-field approximation (equation (10c)) in equation (11f) decouples the multispecies dynamics and enabled us to investigate the dynamics of individual species in good approximation (equation (13)).
The corrected aggregation–abundance relationship
In Fig. 2 and Extended Data Fig. 3, we fitted a phenomenological power-law for the species of a given forest plot, where the x value was the logarithm of abundance Nf and the y value the corresponding logarithm of kff (refs. 21,27,28). However, this led to values of kff close to zero for large abundance values, which would indicate strongly regular patterns25 not found in the data. Instead, in the extreme case without an aggregation mechanism (that is, random placement of offspring), crowding competition will lead to the repulsion of conspecifics comparable to heterospecific association kfh of heterospecifics. Thus, to avoid a bias, we used the quantity kff – kfh as the y value in our fit (Extended Data Fig. 9). This required the assumption that kff > kff, which was satisfied for 98% of our species (722 out of a total of 737 species; see the section ‘Study areas’). We leave investigation of the specific cases where kff < kfh to future studies.
Basic multispecies model M1
Our basic multispecies model (M1) for the per capita growth rate of species f is given by
where Nf,t is the abundance of species f at time step t, sf is a density-independent per capita background survival rate, rf is the per capita recruitment rate and βff is the neighbourhood-scale conspecific competition coefficients of species f. The biological information on neighbourhood crowding competition was incorporated into the fitness factor42 Wf, which results from combining equations (9) and (10):
The community-level competition coefficients αfi of this model are therefore given by
Thus, even if the conspecific neighbourhood-scale competition coefficients βff were constant, the corresponding community-level coefficients αff were not necessarily constant. Instead, they depended on abundance if aggregation kff depended on abundance8, as observed in many of our forest dynamics plots (Fig. 2). Not considering the effect that crowding can have on the competition coefficients αff and αfi is a common (implicit) assumption of models used in coexistence theory4,15,42,47,49. Note that equation (11a) together with the fitness factor of equation (11b) enabled us to construct a reference model for the constants kff and kfh, for which the equilibria Nf* and the conditions for feasibility and invasiveness can be analytically derived8,72, and the corresponding non-spatial model (that is, \({k}_{{ff}}^{* }={k}_{{fh}}^{* }=1\)) without niche differences (that is, Bf = 1) led to a per capita population growth rate of zero for all abundances.
Extended multispecies model M2
However, we wanted to extend our model M1 (equations (11a) and (11b)) to include a dependence of aggregation on abundance as new aspect. To introduce this new model, we first defined \({k}_{{ff}}^{* }\) and \({k}_{{fh}}^{* }\) as the observed value of aggregation and heterospecific association, respectively, and kff as aggregation that depended on abundance. We then assumed that heterospecific association \({k}_{{fh}}^{* }\) is independent of abundance (as suggested by Extended Data Fig. 4) and that the quantity (kff – kff) follows a power law with respect to abundance (Extended Data Fig. 9 and the section ‘The corrected aggregation–abundance relationship’). To formulate our extended multispecies model (M2), we therefore rewrote the fitness factor of equation (11b) by adding and subtracting the term \({k}_{{fh}}^{* }{{N}}_{f,t}\):
We obtained our extended fitness factor
by replacing the quantity (\({k}_{{ff}}^{* }-{k}_{{fh}}^{* }\)) in equation (11e) with a new function L(Nf,t) depending on abundance:
where Nf* is the observed species abundance. Equation (12) simplifies for the observed abundance (that is, Nf,t = Nf*) to \(L({N}_{f}^{* })=({k}_{{ff}}^{* }-{k}_{{fh}}^{* })\), but otherwise represents the desired power-law with respect to abundance.
Decoupled multispecies models M3
The overall objective of our extended model was to study the effect that a possible dependence of aggregation on abundance (Fig. 2) has on the ability of a newly invading (or an almost extinct) species to increase its abundance. Therefore, we decoupled our extended multispecies model M2 into multiple single-species models (M3) by assuming approximate zero-sum dynamics2 (see the section ‘Zero-sum assumption’ above), in which the number of heterospecifics is given by \(\sum _{i\ne f}{{N}}_{i}={J}^{* }-{N}_{f,t}\). We obtained from equation (11f) the fitness function
that results together with equation (11a) in a closed-form expression for the per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{f})\) of the focal species f at low abundance, which nevertheless includes the key information on crowding competition with heterospecifics through the parameters Bf and \({k}_{{fh}}^{* }\), and total community size J*.
Model parameterization
Our extended multispecies model M2 (equations (11a) and (11f)) approximated an underlying individual-based model in the tradition of earlier spatially explicit work7,35,44,46,73 (Extended Data Fig. 5), but used the empirically observed spatial patterns instead of explicitly modelling their dynamics8. In the most general case, the models M2 and M3 have therefore seven parameters per species: three demographic parameters (rf, sf and βff); three parameters that quantify the spatial patterns (that is, \({k}_{{ff}}^{* },{k}_{{fh}}^{* },{B}_{f}\)); and the exponent bf of the aggregation–abundance relationship. The parameterization of the models depended on the objective of the model application and the available data. In the example application of our theory, we wanted to feature the effects of spatial patterns (Fig. 2) on coexistence, and we derived a specific parameterization adapted to our data and objective.
We estimated the species-specific measures \({k}_{{ff}}^{* }\) and \({k}_{{fh}}^{* }\) of the observed spatial patterns directly from the ForestGEO plot data based on equations (7) and (10). The exponent bf of the corrected (power law) aggregation–abundance relationship (equation (12)) was estimated by linear regression, where the x value was ln(Nf) and the y value the corresponding ln(kff − kfh) (Extended Data Fig. 9). The parameter bf captured for a given forest plot the average species response of aggregation to abundance (Extended Data Fig. 9), and we used this value as a parameter for all focal species. This approach is based on a species-for-time substitution, which assumes that the power-law exponent bf derived for multiple species of one census would be the same as the bf derived for the same species but at multiple points in time. The results of our individual-based simulations (Extended Data Fig. 5a–f) supported this assumption. Note that effects of habitat association or details of dispersal will influence the values of kff* (and kfh*) and contribute to the observed departures from the power law aggregation–abundance relationship, particularly for tropical forests (compare Extended Data Figs. 3 and 5).
We assumed that all mortality was driven by neighbourhood crowding and therefore set the background survival rate to sf = 1. For estimation of the parameter Bf, we used the matrix of βfi/βff (see the section ‘Proxy for pairwise competition strength between species’ above) and derived the crowding indices Hof and Iof for each individual o (equations (7b) and (7c)). The value of Bf was then given by \({B}_{f}=\bar{{I}_{f}}/\bar{{H}_{f}}\), where \(\bar{{I}_{f}}\) and \(\bar{{H}_{f}}\) are the population-level averages of the individual crowding indices Hof and Iof.
To determine the unknown value of βff, the neighbourhood-scale conspecific competition coefficients of species f that determines the strength of crowding competition, we assumed that the focal species is close to equilibrium (that is, \({\widetilde{\lambda }}_{f}({N}_{f}^{* })\) = 0) and obtained by rewriting equation (11a):
At equilibrium, we found with equation (11b) that \({W}_{f}({N}_{f}^{* })\,=\)\(c{k}_{ff}{N}_{f}+{\rm{constant}}\), thus all else equal, βff is negatively related to the observed abundance: species with lower observed abundance experienced stronger negative impacts on conspecifics than more common species, a pattern frequently observed in plant communities22,67,73,74,75,76,77. The effect of departures from the equilibrium assumption on the invasion criterion can be assessed by using equation (S17) in the Supplementary Text.
Inserting equation (14) into equation (11a) led to our final equation for the per capita population growth rate, for which the fitness function is given by equation (13):
For small per capita recruitment rates rf and large background survival (sf = 1), we obtained
which suggests the use of the scaled per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{f,t})/{r}_{f}\) to remove the deterministic effects of the recruitment rate rf. Note that the effect of individual variation in model parameters on the invasion criterion can be directly investigated by using equations (13) and (15).
Adding a small immigration rate
We reformulated our mathematical model (equation (15a)) in terms of the change in the number of individuals during one time step and added a small constant immigration rate rf vf (ref. 41) (rf is the reproduction rate), thus:
which is equivalent to adding the term vf rf/Nf,t to the per capita population growth rate (equation (15a)). This approach differs from the way immigration is usually modelled in neutral theory2,26,78. Note that the term vf rf/Nf,t decreases rapidly with increasing abundance Nf,t and does therefore influence the dynamics only for small abundance values, particularly if the values of vf is small as assumed here (Extended Data Fig. 7a–d).
The invasion criterion
Stable coexistence requires that the abundance of a newly invading (or an almost extinct) species increases1,5,8,9 (that is, a rare species advantage), and we wanted to investigate the effect of spatial patterns on the invasion criterion. On the basis of our closed-form expression for the per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{f})\) (equations (13) and (15a)), invasion analysis reduces to the task of identifying the conditions under which \({\widetilde{\lambda }}_{f}({N}_{s})\) maintains a sufficiently high value for low abundance Ns. We therefore demanded that \({\widetilde{\lambda }}_{f}({N}_{s})\) should be slightly larger than zero (\({\widetilde{\lambda }}_{\min }\)) so that species can escape the effects of demographic stochasticity; that is, our invasion criterion is given by \({\widetilde{\lambda }}_{f}({N}_{s}) > {\widetilde{\lambda }}_{\min }\).
From equation (15a) we found that the invasion condition \({\widetilde{\lambda }}_{f}({N}_{s}) > {\widetilde{\lambda }}_{\min }\) translated into the condition
with \(\delta =1-{\rm{ln}}(({\widetilde{\lambda }}_{\min }-{r}_{f}+1)/{s}_{f})/{\rm{ln}}((1-{r}_{s})/{s}_{f})\). For our case of small reproduction rates rf and high background survival (that is, sf = 1), we obtained \(\delta \approx {\widetilde{\lambda }}_{\min }/{r}_{f}\). We therefore investigated the ratio \({W}_{f}({N}_{s})/{W}_{f}({N}_{f}^{* })\) in more detail, which can be expressed as (equation (S14) in the Supplementary Text):
with
The condition in equation (17a) led together with equation (17b) to the invasion criterion
For the case without niche differences (that is, κf = 0), equation (18) suggests that two main mechanisms allow species to increase their rare species advantage: either a smaller negative value of the exponent bf of the aggregation–abundance relationship or a smaller value of ρf (Fig. 4c). We therefore name ρf ‘risk factor’, because larger values of ρf lead to smaller values of the per capita growth rate. The maximal value of the risk factor that satisfies the invasion criterion is given by
Thus, for each combination of \({N}_{s}/{N}_{f}^{* }\), bf, κf and δ, we obtained one critical value of ρf,max in which the species can invade if ρf is smaller than this critical value (Fig. 4c).
It is instructive to investigate the biological mechanism that determines the values of the three factors ρf, \({({N}_{s}/{N}_{f}^{* })}^{{b}_{f}+1}\) and κf that determine the invasion criterion (equations (18) and (19)). First, the risk factor ρf (equation (17d)) becomes smaller if the observed relative species abundance Nf*/J* of the focal species in the community becomes larger (this favours forests at higher latitudes; Extended Data Fig. 8a), if aggregation increases (that is, \({k}_{{fh}}^{* }/({k}_{{ff}}^{* }-{k}_{{fh}}^{* })\) decreases; Extended Data Fig. 8d), and if niche differences become larger (that is, Bf decreases). Because of the overwhelming effect of the relative abundance, the risk factor ρf decreases strongly with latitude for our dataset (Extended Data Fig. 8c) and for species with lower abundance, aggregation \({k}_{{ff}}^{* }\) would increase and reduce ρf. Second, a smaller negative value of the exponent bf of the aggregation–abundance relationship leads to a smaller value of \({({N}_{s}/{N}_{f}^{* })}^{{b}_{f}+1}\) (this favours forests at lower latitudes; Extended Data Fig. 8b).
Finally, as expected, it is more likely that the invasion criterion is fulfilled if niche differences become larger (that is, Bf becomes smaller). In this case, ρf becomes smaller (equation (17d)) and κf becomes larger (equation (17c)), and because \({N}_{s}/{N}_{f}^{* }\) << (1 – δ), the inequality of equation (19) is easier to satisfy. The invasion criterion (equation (19)) can be satisfied for niche differences even if bf < −1. Indeed, some species of the CBS plot, which showed an exponent of bf = −1.077, fulfil the invasion criterion, mostly owing to weak aggregation that led to large values of kfh/(kff – kfh) and larger κf (Extended Data Fig. 7e,f). Thus, as expected, the stabilizing mechanism of niche differences (Bf < 1) has a key role if spatial patterns alone provide only weak stabilization.
Scenarios investigated
We considered four scenarios to investigate the effects of the spatial mechanism of neighbourhood crowding and immigration on the ability of the 720 study species to increase when having low abundance. Scenario 1 assumed that conspecifics and heterospecifics compete equally (that is, no niche differences; βfi = βff, Bf = 1), and scenario 2 considers niche differences between species approximated by phylogenetic dissimilarity (see the section ‘Proxy for pairwise competition strength between species’ above). Scenarios 3 and 4 are the same as scenarios 1 and 2, but also assume a small constant immigration with parameter vf = 0.1. For the mean reproduction rate of rf = 0.1 per time step across all plots, this results in an immigration rate of rf vf = 0.01, or 1 immigrant every 100 time steps.
Spatially explicit simulation model
Model description
This model description was adapted from a previous publication8. We used the individual-based simulations to verify that the observed patterns (that is, the power law of the abundance–aggregation relationship) can emerge in principle from the minimal mechanisms included in the spatial multispecies model (equations (8) and (11a)).
The individual-based model considered only reproductive (adult) trees, but no size differences. During a given 5-year time step, the model simulated first stochastic recruitment of reproductive trees and placement of recruits, and second, stochastic survival of adults as given by equation (8), depending on their neighbourhood crowding indices (equations (7a)–(7c)) estimated from the community of adult trees. In the next time step, the recruits counted as reproductive adults and were subject to mortality. No immigration from a metacommunity was considered (that is, vf = 0; equations (1a)). To avoid edge effects, torus geometry was assumed.
Each individual produced rf recruits on average, and their locations were determined by a type of Thomas process25 to obtain a clustered distribution of recruits. In our model, the spatial position of the recruits was determined using two independent mechanisms. First, a proportion 1 – pd of recruits was placed stochastically around randomly selected conspecific adults (parents) by using a two-dimensional kernel function (here a Gaussian with variance σ2). This is the most common way in most spatially explicit models to generate species clustering. Technically, we first selected for each of these recruits randomly one parent among the conspecific adults and then determined the position of the recruit by sampling from the kernel. Second, the remaining proportion pd of recruits was distributed in the same way, but around randomly placed cluster centres that are located independently of conspecific adults.
Parameterization of the simulation model
The simulation model used here is described in detail in a previous study8. However, we used here distance-weighting for the estimation of the crowding indices. Thus, in the source code (the supplementary information in ref. 8) we used DistanceWeighting = 1 instead of DistanceWeighting = 0.
The simulations of the individual-based forest model were conducted in 5-year time steps over 25,000 years (equivalent to 5,000 census periods) in an area of A = 200 ha, and comprised approximately 86,000 trees with initially 80 species. The model parameters were the same for all species, and all species followed exactly the same model rules. We selected βfi = βff to obtain no differences in conspecific and heterospecific interactions and sf = 1 (no background mortality), a standard deviation of σ = 10 m of the kernel function, and we adjusted the parameters βff = 0.02 and rf = 0.1 to produce tree densities (430 per ha) and an overall 5-year mortality rate (10%) similar to that of trees with d.b.h. ≥ 10 cm of the BCI plot. The radius of the neighbourhood used to estimate the crowding indices was r = 20 m, and the number of randomly assigned cluster centres was 16.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The minimum datasets that enable the repetition, interpretation, verification and extension of the research is provided in Supplementary Table 1. This table provides for all species the indices of spatial patterns and all other quantities used in the model analysis and to generate Table 1, Figs. 2–4 and Extended Data Figs. 2–4 and 6–9. Additional raw data for Fig. 3 are shown in Extended Data Table 1. The output data of the individual-based forest simulation model used to generate Extended Data Fig. 5 is provided in Supplementary Table 2. To estimate the spatial pattern indices, we used the raw census data of the ForestGEO network. The raw census data of several sites are publicly available (https://forestgeo.si.edu/explore-data). For the other sites, they are available upon reasonable request and with permission of the principal investigators of the corresponding ForestGEO sites (https://forestgeo.si.edu/sites-all). Mycorrhizal association types were based on available global datasets53,54,55 and website sources (https://www.mycorrhizas.info). To determine whether a species is mainly dispersed by animals (zoochory), we used the Seed Information Database (https://ser-sid.org) of the Society for Ecological Restoration and the Royal Botanic Gardens Kew. Source data are provided with this paper.
References
Chesson, P. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366 (2000).
Hubbell, S. P. The Unified Neutral Theory of Biodiversity and Biogeography (Princeton Univ. Press, 2001).
Wright, J. S. Plant diversity in tropical forests: a review of mechanisms of species coexistence. Oecologia 130, 1–14 (2002).
Kraft, N. J. B., Godoy, O. & Levine, J. M. Plant functional traits and the multidimensional nature of species coexistence. Proc. Natl Acad. Sci. USA 112, 797–802 (2015).
Barabás, G., D’Andrea, R. & Stump, S. M. Chesson’s coexistence theory. Ecol. Monogr. 88, 277–230 (2018).
Davies, S. J. et al. ForestGEO: understanding forest diversity and dynamics through a global observatory network. Biol. Conserv. 253, 108907 (2021).
Detto, M. & Muller-Landau, H. C. Stabilization of species coexistence in spatial models through the aggregation–segregation effect generated by local dispersal and nonspecific local interactions. Theor. Pop. Biol. 112, 97–108 (2016).
Wiegand, T. et al. Consequences of spatial patterns for coexistence in species-rich plant communities. Nat. Ecol. Evol. 5, 965–973 (2021).
Ellner, S. P., Snyder, R. E., Adler, P. B. & Hooker, G. Toward a “modern coexistence theory” for the discrete and spatial. Ecol. Monogr. 92, e1548 (2022).
Yamawo, A. & Ohno, M. Joint evolution of mutualistic interactions, pollination, seed dispersal mutualism, and mycorrhizal symbiosis in trees. New Phytol. 243, 1586–1599 (2024).
Rogers, H. S., Donoso, I., Traveset, A. & Fricke, E. C. Cascading impacts of seed disperser loss on plant communities and ecosystems. Annu. Rev. Ecol. Evol. Syst. 52, 641–666 (2021).
Tedersoo, L., Bahram, M. & Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 367, eaba1223 (2020).
Sasaki, T. et al. Role of mycorrhizal associations in tree spatial distribution patterns based on size class in an old-growth forest. Oecologia 189, 971–980 (2019).
Delavaux, C. S. et al. Mycorrhizal feedbacks influence global forest structure and diversity. Commun. Biol. 6, 1066 (2023).
Stone, L. The feasibility and stability of large complex biological networks: a random matrix approach. Sci. Rep. 8, 8246 (2018).
Gibbs, T., Levin, S. A. & Levine, J. M. Coexistence in diverse communities with higher-order interactions. Proc. Natl Acad. Sci. USA 119, e2205063119 (2022).
Janzen, D. H. Herbivores and the number of tree species in tropical forests. Am. Nat. 104, 501–528 (1970).
Connell, J. H. Diversity in tropical rain forests and coral reefs. Science 199, 1302–1310 (1978).
Hubbell, S. P. Tree dispersion, abundance, and diversity in a tropical dry forest. Science 203, 1299–1309 (1979).
Condit, R. et al. Spatial patterns in the distribution of tropical tree species. Science 288, 1414–1418 (2000).
Morlon, H. et al. A general framework for the distance–decay of similarity in ecological communities. Ecol. Lett. 11, 904–917 (2009).
Detto, M., Visser, M. D., Wright, S. J. & Pacala, S. W. Bias in the detection of negative density dependence in plant communities. Ecol. Lett. 22, 1923–1939 (2019).
Seidler, T. G. & Plotkin, J. B. Seed dispersal and spatial pattern in tropical trees. PLoS Biol. 4, e344 (2006).
McFadden, I. R. et al. Disentangling the functional trait correlates of spatial aggregation in tropical forest trees. Ecology 100, e02591 (2019).
Wiegand, T. & Moloney, K. A. A Handbook of Spatial Point Pattern Analysis in Ecology (Chapman and Hall/CRC, 2014).
May, F., Wiegand, T., Lehmann, S. & Huth, A. Do abundance distributions and species aggregation correctly predict macroecological biodiversity patterns in tropical forests? Glob. Ecol. Biogeogr. 25, 575–585 (2016).
Davis, M. A., Curran, C., Tietmeyer, A. & Miller, A. Dynamic tree aggregation patterns in a species-poor temperate woodland disturbed by fire. J. Veg. Sci. 16, 167–174 (2005).
Gilbert, S. et al. Beyond the tropics: forest structure in a temperate forest mapped plot. J. Veg. Sci. 21, 388–405 (2010).
He, F., Legendre, P. & LaFrankie, J. V. Distribution patterns of tree species in a Malaysian tropical rain forest. J. Veg. Sci. 8, 105–114 (1997).
Muller-Landau, H. C. et al. Interspecific variation in primary seed dispersal in a tropical forest. J. Ecol. 96, 653–667 (2008).
Wright, J. S. et al. Interspecific associations in seed arrival and seedling recruitment in a neotropical forest. Ecology 97, 2780–2790 (2016).
Chanthorn, W., Getzin, S., Wiegand, T., Brockelman, W. Y. & Nathalang, A. Spatial patterns of local species richness reveal importance of frugivores for tropical forest diversity. J. Ecol. 106, 925–935 (2018).
Getzin, S., Wiegand, T. & Hubbell, S. P. Stochastically driven adult–recruit associations of tree species on Barro Colorado Island. Proc. R. Soc. B 281, 20140922 (2014).
Pontarp, M. et al. The latitudinal diversity gradient: novel understanding through mechanistic eco-evolutionary models. Trends Ecol. Evol. 34, 211–223 (2019).
Detto, M. & Muller-Landau, H. C. Rates of formation and dissipation of clumping reveal lagged responses in tropical tree populations. Ecology 97, 1170–118 (2016).
Uriarte, M. et al. Trait similarity, shared ancestry and the structure of neighbourhood interactions in a subtropical wet forest: implications for community assembly. Ecol. Lett. 13, 1503–1514 (2010).
Canham, C. D. et al. Neighborhood analyses of canopy tree competition along environmental gradients in New England forests. Ecol. Appl. 16, 540–554 (2006).
Hülsmann, L. et al. Latitudinal patterns in stabilizing density dependence of forest communities. Nature 627, 564–571 (2024).
Lewis, S. L. & Tanner, E. V. J. Effects of above- and belowground competition on growth and survival of rain forest tree seedlings. Ecology 81, 2525–2538 (2000).
Bennett, J. A. et al. Plant–soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184 (2017).
Azaele, S., Pigolotti, S., Banavar, J. R. & Maritan, A. Dynamical evolution of ecosystems. Nature 444, 926–928 (2006).
Chesson, P. Scale transition theory: its aims, motivations and predictions. Ecol. Complex. 10, 52–68 (2012).
O’Dwyer, J. & Chisholm, R. A mean field model for competition: from neutral ecology to the red queen. Ecol. Let. 17, 961–969 (2014).
Murrell, D. When does local spatial structure hinder competitive coexistence and reverse competitive hierarchies? Ecology 91, 1605–1616 (2010).
Chesson, P. & Neuhauser, C. Intraspecific aggregation and species coexistence. Trends Ecol. Evol. 17, 210–211 (2002).
Bolker, B. & Pacala, S. W. Spatial moment equations for plant competition: understanding spatial strategies and the advantage of short dispersal. Am. Nat. 153, 575–602 (1999).
Broekman, M. J. E. et al. Signs of stabilisation and stable coexistence. Ecol. Lett. 22, 1957–1975 (2019).
Beckman, N. G. & Sullivan, L. L. The causes and consequences of seed dispersal. Annu. Rev. Ecol. Evol. Syst. 54, 403–427 (2023).
Serván, C. A., Capitán, J. A., Grilli, J., Morrison, K. E. & Allesina, S. Coexistence of many species in random ecosystems. Nat. Ecol. Evol. 2, 1237–1242 (2018).
Loreau, M. From Populations to Ecosystems (Princeton Univ. Press, 2010).
Flügge, A. J., Olhede, S. C. & Murrell, D. J. The memory of spatial patterns: changes in local abundance and aggregation in a tropical forest. Ecology 93, 1540–1549 (2012).
Wang, X. et al. Phylogenetic and functional area relationships in two temperate forests. Ecography 36, 883–893 (2013).
Soudzilovskaia, N. A. et al. FungalRoot v.2.0—an empirical database of plant mycorrhizal traits. N. Phytol. 235, 1689–1691 (2022).
Akhmetzhanova, A. A. et al. A rediscovered treasure: mycorrhizal intensity database for 3000 vascular plant species across the former Soviet Union. Ecology 93, 689–690 (2012).
Wang, B. & Qiu, Y.-L. Phylogenetic distribution and evolution of mycorrhizas in land plants. Mycorrhiza 16, 299–363 (2006).
Jin, J. et al. Stronger latitudinal phylogenetic patterns in woody angiosperm assemblages with higher dispersal abilities in China. J. Biogeogr. 51, 269–279 (2024).
Fortunel, C. et al. Topography and neighborhood crowding can interact to shape species growth and distribution in a diverse Amazonian forest. Ecology 99, 2272–2283 (2018).
Fortunel, C., Valencia, R., Wright, S. J., Garwood, N. C. & Kraft, N. J. B. Functional trait differences influence neighbourhood interactions in a hyperdiverse Amazonian forest. Ecol. Letters 19, 1062–1070 (2016).
Erickson, D. L. et al. Comparative evolutionary diversity and phylogenetic structure across multiple forest dynamics plots: a mega-phylogeny approach. Front. Genet. 5, 358 (2014).
Webb, C. & Donoghue, M. Phylomatic: tree assembly for applied phylogenetics. Mol. Ecol. Notes 5, 181–183 (2005).
Jin, Y. & Qian, H. V. PhyloMaker2: an updated and enlarged R package that can generate very large phylogenies for vascular plants. Plant Divers. 44, 335–339 (2022).
Smith, S. A. & Brown, J. W. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 105, 302–314 (2018).
Kress, W. J. et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl Acad. Sci. USA 106, 18621–18626 (2009).
Kembel, S. W. et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464 (2010).
Cadotte, M. W., Davies, T. J. & Peres-Neto, P. R. Why phylogenies do not always predict ecological differences. Ecol. Monogr. 87, 535–551 (2017).
The R Development Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2022).
Mangan, S. A. et al. Negative plant–soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755 (2010).
Wiegand, T. et al. Spatially explicit metrics of species diversity, functional diversity, and phylogenetic diversity: insights into plant community assembly processes. Annu. Rev. Ecol. Syst. 48, 329–351 (2017).
Lasky, J. R., Uriarte, M., Boukili, V. K. & Chazdon, R. L. Trait-mediated assembly processes predict successional changes in community diversity of tropical forests. Proc. Natl Acad. Sci. USA 111, 5616–5621 (2014).
Uriarte, M., Condit, R., Canham, C. D. & Hubbell, S. P. A spatially explicit model of sapling growth in a tropical forest: does the identity of neighbours matter? J. Ecol. 92, 348–360 (2004).
Fung, T., O’Dwyer, J. & Chisholm, R. Effects of temporal environmental stochasticity on species richness: a mechanistic unification spanning weak to strong temporal correlations. Oikos https://doi.org/10.1111/oik.08667 (2022).
Shigesada, N., Kawasaki, K. & Teramoto, E. The effects of interference competition on stability, structure and invasion of a multi-species system. J. Math. Biol. 21, 97–113 (1984).
Bolker, B., Pacala, S. W. & Neuhauser, C. Spatial dynamics in model plant communities: what do we really know? Am. Nat. 162, 135–148 (2003).
Comita, L., Muller-Landau, H. C., Aguilar, S. & Hubbell, S. P. Asymmetric density dependence shapes species abundances in a tropical tree community. Science 329, 330–332 (2010).
Chisholm, R. A. & Muller-Landau, H. C. A theoretical model linking interspecific variation in density dependence to species abundances. Theor. Ecol. 4, 241–253 (2011).
Yenni, G., Adler, P. B. & Ernest, S. K. M. Strong self-limitation promotes the persistence of rare species. Ecology 93, 456–461 (2012).
Mack, K. M. L. & Bever, J. D. Coexistence and relative abundance in plant communities are determined by feedbacks when the scale of feedback and dispersal is local. J. Ecol. 102, 1195–1201 (2014).
Chisholm, R. A. & Lichstein, J. W. Linking dispersal, immigration and scale in the neutral theory of biodiversity. Ecol. Lett. 12, 1385–1393 (2009).
Acknowledgements
We thank numerous local field and laboratory staff, technicians, interns, volunteers and researchers for their contributions to the collection and management of the forest inventory data used in the analyses; and D. Alonso, J. Capitan, members of the Kraft laboratory and R. Condit for feedback on earlier drafts. A detailed list of funding sources, acknowledgements and references for each forest site is available in the Supplementary Notes. T.W. and A.H. were supported by an ERC Advanced Grant (SpatioCoexistence, project 101141989). Xugao Wang was supported by the National Key Research and Development Programme of China (2022YFF1300501), the CAS Project for Young Scientists in Basic Research (YSBR-108) and the Key Research Programme of Frontier Sciences, Chinese Academy of Sciences (ZDBS-LY-DQC019). S.M.F. and A.H. received funding from the Deutsche Forschungsgemeinschaft (DFG; grant number 431504473).
Author information
Authors and Affiliations
Contributions
T.W., Xugao Wang, S.M.F. and A.H. conceived and designed the project. T.W. implemented the models, conducted the simulations, analysed the results and prepared figures and tables. T.W., A.H., S.M.F., Xugao Wang and N.J.B.K. led the writing of the manuscript. Xugao Wang assembled and analysed the plot data and conducted the spatial analyses. The other authors contributed forest census data. All authors reviewed, approved and had the opportunity to comment on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Ryan Chrisholm, Fangliang He and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Sensitivity of the aggregation-abundance relationships to the measure of aggregation.
a, Condits’s Ω1-10 aggregation measure for the species analysed in Wiegand et al. (ref. 8) plotted over the corresponding values of the distance-weighted aggregation measure kff(r = 15 m) used in the present study. b, exponents of the aggregation-abundance power-law derived with Condits’s Ω1-10 plotted over the corresponding values of the distance-weighted aggregation measure kff(r = 15 m). c, comparison of the distance-weighted aggregation measure kff(r) for the neighborhood radii 10 m vs. 15 m. d, same as c), but for the neighborhood radii 10 m vs. 20 m. e, same as c), but for the neighborhood radii 10 m vs. 25 m. To outline the overall tendency in the data we fitted linear regressions to the data. Note the double-logarithmic scale of panels a), c), d), and e).
Extended Data Fig. 2 Latitudinal gradients in the exponent of the abundance-aggregation relationship.
a, the exponent e of the power law kff(Nf) = a Nf e of aggregation kff with abundance Nf over latitude. b, the R2 of the linear regression between ln(kfh) and ln(Nf) for each species f. c, the Pearson correlation coefficient between the power law exponent e and the plot properties species richness, latitude and mean annual temperature for the scaling of kff with Nf. d, same as a) but for the exponent of the abundance scaling of the difference between aggregation kff and heterospecific association kfh, e, same as b) but for the linear regression between ln(kff − kfh) and ln(Nf). f, same as c, but for the scaling of kff − kfh with Nf. Our measures of aggregation kff and heterospecific association kfh are based on neighbourhood crowding indices that count the number of neighbours within distance r of the focal individual, but each neighbour is weighted by the inverse of its distance to the focal individual (Box 1). The neighbourhood distance was in all panels r = 15 m. For details on the forest plots see Extended Data Table 1 and for data Supplementary Data Table 1. To show the overall tendency in the data we fitted in a), b), d) and e) linear regressions. The plot acronyms: BCI: Barro Colorado Island (tropical forest); BDGS: Badagongshan (subtropical forest); BHI: Baihua (temperate forest); CBL: Chebaling (subtropical forest); CBS: Changbaishan (temperate forest); DHS: Dinghushan (subtropical forest); DLS: Donglingshan (temperate forest); FL: Fenglin (temperate forest); FS: Fushan (subtropical forest); GTS: Gutianshan (subtropical forest); HF: Harvard Forest (temperate forest); HSD: Heishiding (tropical–subtropical forest); MST: Mo Singto (tropical forest); NBH: Nabanhe (tropical forest); QL: Qinling (subtropical–temperate forest); SCBI: Smithsonian Conservation Biology Institute (temperate forest); SHJ or SIN: Sinharaja (tropical forest); TRC: Tyson Research Center (temperate forest); TTS: Tiantong (subtropical forest); WAB: Wabikon (temperate forest); XSBN: Xishuangbanna (tropical forest).
Extended Data Fig. 3 Aggregation − abundance relationships for the 21 ForestGEO plots.
The panels (a – u) show the relationship between aggregation kff (y-axis) and abundance per ha nf (x-axis) for each plot. The value of e is the exponent of the power law kff(nf) = a nfe (red lines; see also Fig. 2c), estimated by linear regression analysis of log(kff) over log(nf). The neighbourhood distance was in all panels r = 15 m. We analysed focal species f with more than 50 large individuals (i.e., dbh ≥ 10 cm). (a – f): tropical, (f – m): subtropical, and (m – u) temperate forests.
Extended Data Fig. 4 Abundance-dependency of the association kfh to heterospecifics.
a, exponent of the power law of kfh with respect to species abundance Nf for the 21 the ForestGEO data sets. b, the R2 of the linear regression between ln(kfh) and ln(Nf). The neighbourhood distance was in all panels r = 15 m. For plot acronyms see Extended Data Fig. 2.
Extended Data Fig. 5 The aggregation-abundance relationship and the rare species advantage in individual-based simulations.
We used an individual-based implementation of the community model of eqs. 1 and 4 with initially 80 species on an area of 200 ha without immigration, simulated for 5000 time steps (25,000 years, ∆t = 5 years). A proportion pd of recruits was placed with a Gaussian kernel around randomly distributed cluster centres, and the rest with the same kernel around their parents (see methods). The different simulations differ only in the parameter pd = 0.1, 0.2, 0.35, 0.6, 0.8 (from top to bottom). a–e: temporal aggregation-abundance relationship for 2 or 3 selected species, taken every 50 years (black disks), and for all 80 species at year 25,000 (red discs). Fit of the temporal relationship with a power law with exponent e (blue line), f–j: time series showing the abundances of the first 25 (out of 80) species, where one species (red) invades at year 1000 (and 2 timesteps after extinction) starting with 50 individuals. The average abundance is shown as blue line. k–o: Total crowding index \(\bar{{C}_{f}}+\bar{{H}_{f}}\) (i.e., distance-weighted number of neighbours) of the invading species in dependence on abundance [in k) we used a species that went extinct]. The red line shows the fit with a linear regression. p–t: same as k)-o), but only for conspecific crowding \(\bar{{C}_{f}}\). The model parameters were the same for all species: rf = 0.1/∆t, sf = 0, Bf = 1, c = 0.000063, r = 20 m, βff = 0.02, σ = 10 m (see methods), and Nf* = 1070 and J* = 86,000 emerged. For comparison with Table 1, the normalized per capita population growth rates \({\widetilde{\lambda }}_{f}({N}_{s}=10)/{r}_{f}\) at the last timestep were 0.019, 0.012, 0.022, 0.031, and 0.038 for parameter values of pd = 0.1, 0.2, 0.35, 0.6, and 0.8, respectively. The raw data with the output of the individual-based model can be found in the Supplementary Information as Supplementary Data Table 2.
Extended Data Fig. 6 The scaled per-capita population growth rate \({\tilde{{\boldsymbol{\lambda }}}}_{{\boldsymbol{f}}}({{\boldsymbol{N}}}_{{\boldsymbol{s}}})/{{\boldsymbol{r}}}_{{\boldsymbol{f}}}\), as influenced by spatial structure, niche differences and immigration for 21 ForestGEO plots.
a, the scaled per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{s})/{r}_{f}\) for a small abundance of Ns = 10 individuals, per forest plot for scenario 1 (no immigration: vf = 0, no niche differences: βfi = βff) that represents the “pure” effect of spatial structure. b, same as a), but for scenario 2 that adds niche differences (vf = 0, βfi < βff). c, same as a), but for scenario 3 adds immigration to scenario 1 (vf = 0.1, βfi = βff). d, scenario 4 adds niche differences and immigration (vf = 0.1, βfi < βff). e: \({\widetilde{\lambda }}_{f}({N}_{s})/{r}_{f}\) for scenario 1 in dependence on abundance Nf, averaged separately over all species of tropical, subtropical and temperate forests. The dashed vertical line indicates the small abundance of Ns = 10. f, same as e), but for scenario 2. g, same as e), but for scenario 3, and h, same as e), but for scenario 4. In scenarios 2 and 4 we assumed that more closely related species compete more strongly. The models were parameterized for 720 species of the 21 ForestGEO plots. We excluded the temperate forest at CBS with exponent bf < −1. To outline the overall tendency in the data we fitted in panels a) to d) a linear regression to the data.
Extended Data Fig. 7 Dependency of the scaled per capita population growth rate on the exponent of the aggregation-abundance relationship.
a, Examples for the influence of the exponent bf on the scaled per capita population growth rate of the species Castanopsis acuminatissima from the Mo Singto (MST) plot in Thailand with parameters bf = –0.3, Nf* = 291, J = 15,665, rf = 0.136/∆t, kff = 6.27, kfh = 1.02, Bf = 1 and c = 0.000314. The dashed vertical line indicates the small abundance of Ns = 10. b, same as a), but for niche differences (Bf = 0.692). c, and d, same as a) and b), but with immigration parameter vf = 0.1. e, and f, examples for the scaled per capita population growth rate of species at the CBS plot, which showed a power law exponent of bf = −1.077, for scenario 2 with niche differences (i.e., Bf < 1). We assumed that individuals compete at the individual scale more strongly if they are phylogenetically more similarity. The species acronyms: ACEMON (Acer mono), ACEPSE (A. pseudosieboldianum), ACETEG (A. tegmentosum), FRAMAN (Fraxinus mandshurica), PINKOR (Pinus koraiensis), QUEMON (Quercus mongolica), TILAMU (Tilia amurensis), ULMJAP (Ulmus japonica).
Extended Data Fig. 8 Properties of species leading to a high per capita population growth rate at low abundances.
The panels a) – d) show different quantities that determine the per capita population growth rate \({\widetilde{\lambda }}_{f}({N}_{f})\) (eq. 11a, 13). a, the mean relative abundance Nf*/J*, which drives the risk factor ρf, averaged over all species of a given plot in dependence on latitude, b, same as a), but for the quantity \({({N}_{s}/{N}_{f}^{* })}^{{b}_{f}+1}\) as it appears in eq. 2. c, same as a), but for the mean of the risk factor \({\rho }_{f}\) for niche differences (red) and no niche differences (black), d, same as a), but for the mean of the quantity kfh/(kff –kfh,). To outline the overall tendency in the data we fitted in panels a) a polynomial regression of order 2, and in panels b) and c) an exponential regression. For plot acronyms see Extended Data Fig. 2.
Extended Data Fig. 9 Aggregation − abundance relationships for the 21 ForestGEO plots.
The panels (a – u) show the relationships between corrected aggregation L(Nf) = (kff – kfh) (y-axis) and abundance Nf (x-axis) (eq. 8). The value of bf is the slope of the power law L(Nf) = af Nfbf (red lines), estimated by linear regression analysis of log(kff – kfh) over log(Nf). We used for the analysis focal species f with more than 50 individuals. (a – f): tropical, (f – m): subtropical, and (m – u) temperate forests.
Supplementary information
Supplementary Information
Supplementary Methods, Supplementary Table 1 and Supplementary Notes. Supplementary Table 1 explains the mathematical symbols used in the text and methods, and the Supplementary Text includes sections on studies investigating the aggregation–abundance relationship, details of the spatial analysis, model equations for alternative models and references. Supplementary Notes contain a detailed list of funding sources, acknowledgements and references for each forest site.
Supplementary Table 1
This table shows for each focal species the input quantities of the model (observed abundance, indices of spatial patterns, and so on), key quantities of the model analysis and estimation of the per capita population growth rate, and it provides per plot (and per forest type) summaries to generate most of the figures.
Supplementary Table 2
This table shows the output of the individual-based forest simulation model and the raw data for Extended Data Fig. 5. Each of the five sheets corresponds to one model simulation, the name of the sheet is the parameter pd, which was the only parameter that changes among the simulation runs.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wiegand, T., Wang, X., Fischer, S.M. et al. Latitudinal scaling of aggregation with abundance and coexistence in forests. Nature 640, 967–973 (2025). https://doi.org/10.1038/s41586-025-08604-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-025-08604-z