Abstract
Given the large number of genes significantly associated with risk for neuropsychiatric disorders, a critical unanswered question is the extent to which diverse mutations—sometimes affecting the same gene—will require tailored therapeutic strategies. Here we consider this in the context of rare neuropsychiatric disorder-associated copy number variants (2p16.3) resulting in heterozygous deletions in NRXN1, which encodes a presynaptic cell-adhesion protein that serves as a critical synaptic organizer in the brain. Complex patterns of NRXN1 alternative splicing are fundamental to establishing diverse neurocircuitry, vary between the cell types of the brain and are differentially affected by unique (non-recurrent) deletions1. We contrast the cell-type-specific effect of patient-specific mutations in NRXN1 using human-induced pluripotent stem cells, finding that perturbations in NRXN1 splicing result in divergent cell-type-specific synaptic outcomes. Through distinct loss-of-function (LOF) and gain-of-function (GOF) mechanisms, NRXN1+/− deletions cause decreased synaptic activity in glutamatergic neurons, yet increased synaptic activity in GABAergic neurons. Reciprocal isogenic manipulations causally demonstrate that aberrant splicing drives these changes in synaptic activity. For NRXN1 deletions, and perhaps more broadly, precision medicine will require stratifying patients based on whether their gene mutations act through LOF or GOF mechanisms, to achieve individualized restoration of NRXN1 isoform repertoires by increasing wild-type and/or ablating mutant isoforms. Given the increasing number of mutations predicted to engender both LOF and GOF mechanisms in brain disorders, our findings add nuance to future considerations of precision medicine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
199,00 € per year
only 3,90 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All source donor hiPS cells have been deposited at the National Institute of Mental Health/Sampled Repository (https://studyreg.nimhgenetics.org/ListOfStudies.jsp; study 160). All bulk and single-cell transcriptome sequencing data can be accessed via the Gene Expression Omnibus under the accession codes GSE288880, GSE288881 and GSE288964. Source data will be provided on request from the corresponding authors.
Code availability
To facilitate improved reproducibility of our data, analytical scripts have been deposited to GitHub (https://github.com/mbfernando/NRXN1).
References
Sudhof, T. C. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell 171, 745–769 (2017).
De Wit, J. & Ghosh, A. Specification of synaptic connectivity by cell surface interactions. Nat. Rev. Neurosci. 17, 22–35 (2016).
Gomez, A. M., Traunmüller, L. & Scheiffele, P. Neurexins: molecular codes for shaping neuronal synapses. Nat. Rev. Neurosci. 22, 137–151 (2021).
Schreiner, D. et al. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 84, 386–398 (2014).
Fuccillo, M. V. et al. Single-cell mRNA profiling reveals cell-type-specific expression of neurexin isoforms. Neuron 87, 326–340 (2015).
Marshall, C. R. et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 49, 27–35 (2017).
Matsunami, N. et al. Identification of rare recurrent copy number variants in high-risk autism families and their prevalence in a large ASD population. PLoS ONE 8, e52239 (2013).
Moller, R. S. et al. Exon-disrupting deletions of NRXN1 in idiopathic generalized epilepsy. Epilepsia 54, 256–264 (2013).
Ching, M. S. L. et al. Deletions of NRXN1 (neurexin‐1) predispose to a wide spectrum of developmental disorders. Am. J. Med. Genet. B Neuropsychiatr. Genet. 153B, 937–947 (2010).
Huang, A. Y. et al. Rare copy number variants in NRXN1 and CNTN6 increase risk for Tourette syndrome. Neuron 94, 1101–1111.e7 (2017).
Grayton, H. M., Missler, M., Collier, D. A. & Fernandes, C. Altered social behaviours in neurexin 1α knockout mice resemble core symptoms in neurodevelopmental disorders. PLoS ONE 8, e67114 (2013).
Pak, C. et al. Human neuropsychiatric disease modeling using conditional deletion reveals synaptic transmission defects caused by heterozygous mutations in NRXN1. Cell Stem Cell 17, 316–328 (2015).
Pak, C. et al. Cross-platform validation of neurotransmitter release impairments in schizophrenia patient-derived NRXN1-mutant neurons. Proc. Natl Acad. Sci. USA 118, e2025598118 (2021).
Sebastian, R. et al. Schizophrenia-associated NRXN1 deletions induce developmental-timing- and cell-type-specific vulnerabilities in human brain organoids. Nat. Commun. 14, 3770 (2023).
Flaherty, E. et al. Neuronal impact of patient-specific aberrant NRXN1α splicing. Nat. Genet. 51, 1679–1690 (2019).
Boxer, E. E. & Aoto, J. Neurexins and their ligands at inhibitory synapses. Front. Synaptic Neurosci. 14, 1087238 (2022).
Sohal, V. S. & Rubenstein, J. L. R. Excitation–inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol. Psychiatry 24, 1248–1257 (2019).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Yang, N. et al. Generation of pure GABAergic neurons by transcription factor programming. Nat. Methods 14, 621–628 (2017).
Li, Y. I. et al. Annotation-free quantification of RNA splicing using LeafCutter. Nat. Genet. 50, 151–158 (2018).
Iijima, T. et al. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell 147, 1601–1614 (2011).
Iijima, T., Iijima, Y., Witte, H. & Scheiffele, P. Neuronal cell type-specific alternative splicing is regulated by the KH ___domain protein SLM1. J. Cell Biol. 204, 331–342 (2014).
Traunmuller, L., Gomez, A. M., Nguyen, T. M. & Scheiffele, P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science 352, 982–986 (2016).
Traunmüller, L. et al. A cell-type-specific alternative splicing regulator shapes synapse properties in a trans-synaptic manner. Cell Rep. 42, 112173 (2023).
Virtanen, M. A., Uvarov, P., Mavrovic, M., Poncer, J. C. & Kaila, K. The multifaceted roles of KCC2 in cortical development. Trends Neurosci. 44, 378–392 (2021).
Willsey, H. R. et al. Parallel in vivo analysis of large-effect autism genes implicates cortical neurogenesis and estrogen in risk and resilience. Neuron 109, 1409 (2021).
Gegenhuber, B., Wu, M. V., Bronstein, R. & Tollkuhn, J. Gene regulation by gonadal hormone receptors underlies brain sex differences. Nature 606, 153–159 (2022).
Roberts, T. C., Langer, R. & Wood, M. J. A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 19, 673–694 (2020).
Saito, Y. et al. Differential NOVA2-mediated splicing in excitatory and inhibitory neurons regulates cortical development and cerebellar function. Neuron 101, 707–720.e5 (2019).
Paz, I., Kosti, I., Ares, M., Cline, M. & Mandel-Gutfreund, Y. RBPmap: a web server for mapping binding sites of RNA-binding proteins. Nucleic Acids Res. 42, W361–W367 (2014).
Ghanbarian, H., Aghamiri, S., Eftekhary, M., Wagner, N. & Wagner, K.-D. Small activating RNAs: towards the development of new therapeutic agents and clinical treatments. Cells 10, 591 (2021).
Subramanian, A. et al. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452.e17 (2017).
Yilmaz, C. et al. Neurosteroids as regulators of neuroinflammation. Front. Neuroendocrinol. 55, 100788 (2019).
Brunet De Courssou, J.-B., Durr, A., Adams, D., Corvol, J.-C. & Mariani, L.-L. Antisense therapies in neurological diseases. Brain 145, 816–831 (2022).
Ingusci, S., Verlengia, G., Soukupova, M., Zucchini, S. & Simonato, M. Gene therapy tools for brain diseases. Front. Pharmacol. 10, 724 (2019).
Lin, H.-C. et al. NGN2 induces diverse neuron types from human pluripotency. Stem Cell Rep. 16, 2118–2127 (2021).
Trotter, J. H. et al. Compartment-specific neurexin nanodomains orchestrate tripartite synapse assembly. Preprint at bioRxiv https://doi.org/10.1101/2020.08.21.262097 (2020).
Zafra, F. & Piniella, D. Proximity labeling methods for proteomic analysis of membrane proteins. J. Proteomics 264, 104620 (2022).
Bell, J. Stratified medicines: towards better treatment for disease. Lancet 383, S3–S5 (2014).
Tsimberidou, A. M. et al. Molecular tumour boards — current and future considerations for precision oncology. Nat. Rev. Clin. Oncol. 20, 843–863 (2023).
Zhang, H., Colclough, K., Gloyn, A. L. & Pollin, T. I. Monogenic diabetes: a gateway to precision medicine in diabetes. J. Clin. Invest. 131, e142244 (2021).
Sullivan, P. F. & Geschwind, D. H. Defining the genetic, genomic, cellular, and diagnostic architectures of psychiatric disorders. Cell 177, 162–183 (2019).
Gerasimavicius, L., Livesey, B. J. & Marsh, J. A. Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure. Nat. Commun. 13, 3895 (2022).
Wang, L. et al. Analyses of the autism-associated neuroligin-3 R451C mutation in human neurons reveal a gain-of-function synaptic mechanism. Mol. Psychiatry 29, 1620–1635 (2024).
Pinggera, A. et al. New gain-of-function mutation shows CACNA1D as recurrently mutated gene in autism spectrum disorders and epilepsy. Hum. Mol. Genet. 26, 2923–2932 (2017).
Clark, M. B. et al. Long-read sequencing reveals the complex splicing profile of the psychiatric risk gene CACNA1C in human brain. Mol. Psychiatry 25, 37–47 (2020).
Brunklaus, A. et al. Gene variant effects across sodium channelopathies predict function and guide precision therapy. Brain 145, 4275–4286 (2022).
Sanders, S. J. et al. Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci. 41, 442–456 (2018).
Kim, G., Gautier, O., Tassoni-Tsuchida, E., Ma, X. R. & Gitler, A. D. ALS genetics: gains, losses, and implications for future therapies. Neuron 108, 822–842 (2020).
Balendra, R. & Isaacs, A. M. C9orf72-mediated ALS and FTD: multiple pathways to disease. Nat. Rev. Neurol. 14, 544–558 (2018).
Prakasam, R. et al. LSD1/PRMT6-targeting gene therapy to attenuate androgen receptor toxic gain-of-function ameliorates spinobulbar muscular atrophy phenotypes in flies and mice. Nat. Commun. 14, 603 (2023).
Chintalaphani, S. R., Pineda, S. S., Deveson, I. W. & Kumar, K. R. An update on the neurological short tandem repeat expansion disorders and the emergence of long-read sequencing diagnostics. Acta Neuropathol. Commun. 9, 98 (2021).
Ho, S.-M. et al. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods 101, 113–124 (2016).
Rhee, H. J. et al. An autaptic culture system for standardized analyses of iPSC-derived human neurons. Cell Rep. 27, 2212–2228.e7 (2019).
Barretto, N. et al. ASCL1- and DLX2-induced GABAergic neurons from hiPSC-derived NPCs. J. Neurosci. Methods 334, 108548 (2020).
Sloan, S. A., Andersen, J., Pașca, A. M., Birey, F. & Pașca, S. P. Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Hoffman, G. E. et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 8, 2225 (2017).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773–782 (2019).
Hoffman, G. E. & Schadt, E. E. variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483 (2016).
Ritchie, M. E. et al. limma Powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Seah, C. et al. Modeling gene × environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression. Nat. Neurosci. 25, 1434–1445 (2022).
De Marinis, I., Lo Surdo, P., Cesareni, G. & Perfetto, L. SIGNORApp: a Cytoscape 3 application to access SIGNOR data. Bioinformatics 38, 1764–1766 (2022).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019).
Urresti, J. et al. Cortical organoids model early brain development disrupted by 16p11.2 copy number variants in autism. Mol. Psychiatry 26, 7560–7580 (2021).
Yang, X. et al. Identification and validation of genes affecting aortic lesions in mice. J. Clin. Invest. 120, 2414–2422 (2010).
Birey, F. et al. Assembly of functionally integrated human forebrain spheroids. Nature 545, 54–59 (2017).
Kurtenbach, S. & Harbour, J. W. SparK: a publication-quality NGS visualization tool. Preprint at bioRxiv https://doi.org/10.1101/845529 (2019).
Acknowledgements
M.B.F. was supported by a Gilliam Fellowship from the Howard Hughes Medical Institute. This work was supported by the National Institute of Mental Health grants RO1 MH121074 (to K.J.B., G.F. and P.A.S.), RO1 MH125579 (to G.F. and K.J.B.), R01MH123155 (to K.J.B.) and RM1MH132648 (to K.J.B.). S.G. is supported by grants from the Trond Mohn Research Foundation (grant numbers TMS2021TMT07 and TMS2023TMT06). D.A.K. is supported by the National Science Foundation under grant number DBI2146398. Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. We thank the Stem Cell Engineering Core at the Icahn School of Medicine at Mount Sinai; the laboratories of N. Yang (R. Hu and X. Zhou) and S. Marro (M. Durens) for assistance in primary glial preparations; K. G. Townsley and M. G. Baxter for advice on statistical testing; L. Yang for critical advice on single-cell RNA-seq analyses; D. Weinberger and the Lieber Institute for Brain Development at Johns Hopkins School of Medicine for sharing post-mortem materials; and all members of the Brennand, Slesinger and Fang laboratories for critical feedback and discussions throughout the course of this work.
Author information
Authors and Affiliations
Contributions
M.B.F., S.K., A.N.M., R.O., C.P. and A.P. performed and/or analysed the experiments supervised by P.A.S. and K.J.B.; Y.Z. and Y.F. performed the bioinformatic analyses supervised by G.F. A.T. performed the bioinformatic analysis of alternative splicing estimates, supervised by D.A.K. S.E.W. produced virus for the generation of iGABA neurons. S.G. and L.C. made substantial contributions to experimental design, data analysis and interpretation of results. P.J.M.D. performed high-content imaging experiments. E.K.F. processed post-mortem tissue and generated long-read data. I.A.P. assisted in statistical analyses. M.B.F., G.F., P.A.S. and K.J.B. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Lee Rubin and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Transcriptomic analysis of differentially expressed genes in iGLUT and iGABA neurons.
(a) Splicegraph displaying significant gene wide splicing clusters at NRXN1 SS3 (p = 0.0196), compared via Dirichlet-multinomial generalized linear model with Bonferroni corrections. (b, d) Volcano plots of differential gene expression (DE) analysis across both genotypes in iGLUT and (i, k) iGABA neurons, significant genes defined by FDR correction. Vertical dashed lines represent DE genes ±1.5 Log2FC. Horizontal dashed lines represent FDR = 0.1 cutoff (lower) and Bonferroni corrected cutoff (upper). (c, e) Sunburst plots of all FDR corrected DEGs with SynGO annotated synapse function for iGLUT and (j, l) iGABA neurons. (f, m) Overlap of DEGs and (g,n) gene set enrichment analysis (GSEA) between genotypes. (h,o) Distinct gene expression patterns by hierarchal clustering of all patient specific DEGs. Sample information correspond to Fig. 1.
Extended Data Fig. 2 Transcriptomic profile of NRXN1 RNA-binding proteins.
(a) Gene expression fold-change of select NRXN1 predicted RNA-binding proteins (RBP) across patients and control iGLUT neurons with statistical comparisons for STAR-Family RBPs, (n/d: Control = 6/2; 5′-Del = 6/2; 3′-Del = 6/2 | 1 batch), compared via two-tailed 1-way ANOVA, with Dunnett’s test (Sam68 F2, 15 = 15.36, 5′-Del p = 0.0001, 3′-Del p = 0.0181; Slm1 F2, 15 = 15.29, 5′-Del p = 0.0002, 3′-Del p = 0.0014; Slm2 F2, 15 = 20.22, 5′-Del p = 5.34E-05, 3′-Del p = 0.0003. (b) Gene expression fold-change of select NRXN1 predicted RNA-binding proteins (RBP) across patients and control iGABA neurons with statistical comparisons for STAR-Family RBPs, (n/d: Control = 5/2; 5′-Del = 6/2; 3′-Del = 6/2 | 1 batch), compared via two-tailed 1-way ANOVA, with Dunnett’s test (Sam68 F2, 14 = 1.449, 5′-Del p = 0.8826, 3′-Del p = 0.2172; Slm1 F2, 14 = 6.421, 5′-Del p = 0.0109, 3′-Del p = 0.0158; Slm2 F2, 14 = 21.06, 5′-Del p = 5.53E-05, 3′-Del p = 0.0002. n reported as samples/donors | independent batches.
Extended Data Fig. 3 Extended transcriptomics analysis on disease risk associated genes.
(a) Summary table of overlapping DEGs with risk enrichments across publicly curated datasets for autism (ASD), bipolar disorder (BD) and schizophrenia. (b, e) Enrichment of genes across neuropsychiatric disorders for iGLUT and iGABA neurons. (c) Interaction maps of risk genes for 5′-Del iGLUT, (d) 3′-Del iGLUT, (f) 5′-Del iGABA and (g) 5′-iGABA. Sample information correspond to Fig. 1.
Extended Data Fig. 4 Extended data on human organoid generation and characterization.
(a) Timeline of dorsal organogenesis for hCOs and (b,c) representative images of hiPSC aggregation and immature spheroids post dislodging and normalized organoid perimeters over time, relative to averaged control (n/d: Control = 136/2; 5′-Del = 128/2; 3′-Del = 150/2 | 2 batches) (d) hCO RT-qPCR results from 4-month organoids of genes for pluripotency, neuronal, and cell-type specific markers (n/d: Control = 4/2; 5′-Del = 4/2; 3′-Del = 4/2 | 2 batches), compared via two-tailed 1-way ANOVA, with Dunnett’s test (SOX2 F2, 9 = 3.921, 5′-Del p = 0.065, 3′-Del p = 0.9981; MAP2AB F2, 9 = 0.7201, 5′-Del p = 0.5479, 3′-Del p = 0.4688; vGLUT1 F2, 9 = 1.594, 5′-Del p = 0.6353, 3′-Del p = 0.6331; vGLUT2 F2, 9 = 1.710, 5′-Del p = 0.3992, 3′-Del p = 0.7901). (e,f) hCO RT-qPCR results of NRXN1 WT and MT expression (n/d: Control = 4/2; 5′-Del = 4/2; 3′-Del = 4/2 | Representative batch) compared via two-tailed 1-way ANOVA, with Dunnett’s test (NRXN1-WT F2, 9 = 0.5873, 5′-Del p = 0.9256, 3′-Del p = 0.6961). (g) Timeline of ventral organogenesis for hSOs and (h,i) representative images of hiPSC aggregation and immature spheroids post dislodging and normalized organoid perimeters over time, relative to averaged control (n/d: Control = 118/2; 5′-Del = 92/2; 3′-Del = 120/2 | 2 batches). (j) hSO RT-qPCR results from 4-month organoids of genes for pluripotency, neuronal, and cell-type specific markers (n/d: Control = 4/2; 5′-Del = 4/2; 3′-Del = 4/2 | 2 batches), compared via 1-way ANOVA, with Dunnett’s test (SOX2 F2, 9 = 5.683, 5′-Del p = 0.1563, 3′-Del p = 0.0152; MAP2AB F2, 9 = 4.800, 5′-Del p = 0.0875, 3′-Del p = 0.0285; GAD65 F2, 9 = 1.641, 5′-Del p 0.3573, 3′-Del p = 0.8806; DLX5 F2, 9 = 0.01755, 5′-Del p = 0.981, 3′-Del p = 0.9809). (k,l) hSO RT-qPCR results of NRXN1 WT and MT expression (n/d: Control = 4/2; 5′-Del = 4/2; 3′-Del = 4/2 | Representative batch) compared via 1-way ANOVA, with Dunnett’s test (NRXN1-WT F2, 9 = 4.818, 5′-Del p = 0.7447, 3′-Del p = 0.0824). Data represented as mean ± sem. n reported as samples/donors | independent batches.
Extended Data Fig. 5 Single cell characterization of NRXN1+/− organoids.
(a, f), UMAPs of hCO and hSO organoid samples sequenced at 6 months, annotated by cell clusters. (b, g) Extended gene expression panel across sub-clusters of hCO and hSO samples across neuronal, cortical, subpallial and astroglia markers. (c, h) Validation of regionalization across forebrain (FOXG1), dorsal (EMX1) and ventral (DLX2) regions, with NRXN1 expression across all cells. (d, i) Relative proportions of cell clusters across pooled and individual genotypes, (hCO = 47,460 cells) and (hSO = 35,563 cells). (e, j) Gene ontological analysis results using DEGs from scRNASeq. *P < 0.05, **P < 0.01, ***P <0.001, two-tailed Wilcoxon’s rank sum test, FDR = 0.05.
Extended Data Fig. 6 Extended data on electrophysiological properties of 5′-Del and 3′-Del neurons.
(a) Voltage-gated potassium and channel kinetics across genotypes for iGLUT neurons (n/d = Control = 16/2; 5′-Del = 13/2; 3′-Del = 16/2 | 2 batches). (b) Comparative GLUT mEPSC kinetics of IEI (n/d: Control = 22/2; 5′-Del = 18/2; 3′-Del = 7/2 | 4 batches) compared via two-tailed 1-way ANOVA, with Dunnett’s test (F2,44 = 2.880; 5′-Del p = 0.0411, 3′-Del p = 0.8229, and (c) amplitude size (n/d: Control = 24/2; 5′-Del = 22/2; 3′-Del = 10/2 | 4 batches) compared via two-tailed 1-way ANOVA, with Dunnett’s test (F2,53 = 0.1996; 5′-Del p = 0.7806, 3′-Del p = 0.9972). (d) Voltage-gated potassium and channel kinetics across genotypes for iGABA neurons (n/d = Control = 11/2; 5′-Del = 12/2; 3′-Del = 11/2 | 2 batches). (e) Comparative iGABA mIPSC kinetics of IEI (n/d: Control = 10/2; 5′-Del = 9/2; 3′-Del = 8/2 | 3 batches) compared via two-tailed 1-way ANOVA, with Dunnett’s test (F2,24 = 0.9632; 5′-Del p = 0.6452, 3′-Del p = 0.7565, and (f) amplitude size (n/d: Control = 13/2; 5′-Del = 10/2; 3′-Del = 9/2 | 3 batches) compared via two-tailed 1-way ANOVA, with Dunnett’s test (F2,29 = 0.5097; 5′-Del p = 0.5141, 3′-Del p = 0.8618). Data represented as mean ± sem. n reported as samples/donors | independent batches.
Extended Data Fig. 7 Extended KCC2 related data from immature GABA neurons.
(a) Transcriptomic comparison of SLC12A5 expression across DIV14 and DIV35 RNASeq timepoints, compared via a two-tailed mixed effects model-time (F1,28 = 499.1 p <2.2E-16), sample information correspond Fig. 1. (b,c) MEA tests from pre- and post- treatment of 10uM GABAzine (n/d = 28/2 | 1 representative batch) compared via two-tailed paired student’s t-test for time-linked comparison (t = 9.739, df = 27; p = 2.497E-10) and two-tailed unpaired student’s t-test for pre/post activity foldchange, (t = 2.380, df = 54; p = 0.0209). Data represented as mean ± sem. n reported as samples/donors | independent batches.
Extended Data Fig. 8 shRNA knockdown validation.
(a) Extent of shRNA knockdown on WT and in iGLUT neurons (n: shNT = 5/2; shWT = 6/2 | Representative) compared via two-tailed t-test (t = 4.186, df = 9, p = 0.0024), and (b) MT NRXN1 (n/d: shNT = 8/2; shMT = 7/2 | Representative) compared via two-tailed t-test (t = 9.342, df = 13, p = 4.0E-07). (c, d) iGLUT RNASeq DEG plots of WT (n/d: shNT = 5/2; shWT = 5/2 | Representative) and MT (n/d: shNT = 4/2; shMT = 5/2 | Representative) knockdowns performed in isogenic analyses. (e) iGABA RT-qPCR of shRNA knockdown on WT NRXN1 (n/d: shNT = 9/3; shWT = 8/3 | Representative) compared via two-tailed t-test (t = 5.294, df = 15, p = 9.00E-05), and (f) MT NRXN1 (n/d: shNT = 9/2; shMT = 10/2 | Representative) compared via two-tailed t-test (t = 2.011, df = 17, p = 0.0604). (g,h) iGABA RNASeq DEG plots of WT (n/d: shNT = 3/1; shWT = 3/1) and MT (n/d: shNT = 2/1; shMT = 2/1 | 2-4 wells pooled across 2 batches) knockdowns performed in isogenic analyses. Data represented as mean ± sem. n reported as samples/donors | independent batches.
Extended Data Fig. 9 ChIP-seq enrichment of ER1 binding at NRXN1 loci in rodent brain.
(a) Female and (b) male mus musculus ChIP tracts of NRXN1 locus, with red dashed areas highlighting binding enrichment across vehicle and estradiol treated mice. (c) Effect of β-estradiol on control donors (n = 16/4 | Representative, (n/d: Control-Vehicle = 8/4; Control-β-estradiol = 8/4 | Representative) compared via two-sided t-test (t = 1.270, df = 14, p = 0.2248).
Extended Data Fig. 10 In-vivo validation of MT isoform expression from an unrelated autism NRXN1+/− patient.
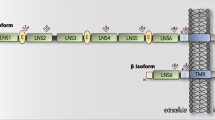
(a) Schematic of novel NRXN1 autism patient, and GOF therapeutic targeting pipeline, with (b) schematic of the NRXN1α isoform structures, with each row representing a unique NRXN1α isoform and each column representing a NRXN1 exon. The colored isoforms (navy, wildtype; peach, patient-specific) are spliced into the transcript while the blank exons are spliced out. The schematic in panel a was created using BioRender. Fernando, M. (2025) https://BioRender.com/f78d262. (c) The abundance of each NRXN1α isoform by sample.
Supplementary information
Supplementary Note
Generation and characterization of NRXN1+/− sub-regional organoids
Supplementary Table 1
Clinical information of all hiPSC and post-mortem donors and detailed summary of hiPSC lines used per experiment. Adapted from ref. 15, Springer Nature.
Supplementary Table 2
Leafcutter results of differential splicing at neurexin loci
Supplementary Table 3
Leafcutter results of genome-wide differential splicing
Supplementary Table 4
SynapseGO results of shRNA experiments
Supplementary Table 5
Standard Gene Ontology and SynapseGO of ASO experiment
Supplementary Table 6
Details on oligonucleotide sequences and antibodies used throughout the study
Supplementary Table 7
Concise summary of all statistical information presented throughout the study
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Fernando, M.B., Fan, Y., Zhang, Y. et al. Phenotypic complexities of rare heterozygous neurexin-1 deletions. Nature 642, 710–720 (2025). https://doi.org/10.1038/s41586-025-08864-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-025-08864-9