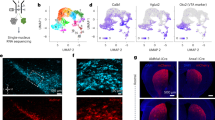
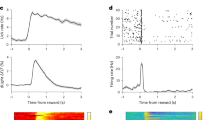
Abstract
The hippocampus1,2,3,4,5,6, as well as dopamine circuits7,8,9, coordinates decision-making in anxiety-eliciting situations. Yet, little is known about how dopamine modulates hippocampal representations of emotionally salient stimuli to inform appropriate resolution of approach versus avoidance conflicts. Here we studied dopaminoceptive neurons in the male mouse ventral hippocampus (vHipp), molecularly distinguished by their expression of dopamine D1 or D2 receptors. We show that these neurons are transcriptionally distinct and topographically organized across vHipp subfields and cell types. In the ventral subiculum where they are enriched, both D1 and D2 neurons are recruited during anxiogenic exploration, yet with distinct profiles related to investigation and behavioural selection. In turn, they mediate opposite approach–avoidance responses, and are differentially modulated by dopaminergic transmission in that region. Together, these results suggest that vHipp dopamine dynamics gate exploratory behaviours under contextual uncertainty, implicating dopaminoception in the complex computation engaged in the vHipp to govern emotional states.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
199,00 € per year
only 3,90 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All snRNA-seq data reported in this study have been deposited in the Gene Expression Omnibus under the accession number GSE227313. Source data for all statistical analyses are provided with this article. All other data, including raw photometry data, are available on request from the corresponding authors. Source data are provided with this paper.
Code availability
Sample code and data for the fibre photometry signal analysis are publicly available on GitHub (https://github.com/arthurgodino/Photometry-analysis). All other custom scripts and code utilized in this study, including for statistical analysis, are available on request from the corresponding authors.
References
Gray, J. A. Précis of the neuropsychology of anxiety: an enquiry into the functions of the septo-hippocampal system. Behav. Brain Sci. 5, 469–484 (1982).
Moser, M. B. & Moser, E. I. Functional differentiation in the hippocampus. Hippocampus 8, 608–619 (1998).
Fanselow, M. S. & Dong, H.-W. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron 65, 7–19 (2010).
Strange, B. A., Witter, M. P., Lein, E. S. & Moser, E. I. Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 15, 655–669 (2014).
Bryant, K. G. & Barker, J. M. Arbitration of approach-avoidance conflict by ventral hippocampus. Front. Neurosci. 14, 615337 (2020).
Turner, V. S., O’Sullivan, R. O. & Kheirbek, M. A. Linking external stimuli with internal drives: a role for the ventral hippocampus. Curr. Opin. Neurobiol. 76, 102590 (2022).
Zweifel, L. S. et al. Activation of dopamine neurons is critical for aversive conditioning and prevention of generalized anxiety. Nat. Neurosci. 14, 620–626 (2011).
Lammel, S., Lim, B. K. & Malenka, R. C. Reward and aversion in a heterogeneous midbrain dopamine system. Neuropharmacology 76, 351–359 (2014).
Morel, C. et al. Midbrain projection to the basolateral amygdala encodes anxiety-like but not depression-like behaviors. Nat. Commun. 13, 1532 (2022).
Robinson, O. J., Pike, A. C., Cornwell, B. & Grillon, C. The translational neural circuitry of anxiety. J. Neurol. Neurosurg. Psychiatry 90, 1353–1360 (2019).
Gergues, M. M. et al. Circuit and molecular architecture of a ventral hippocampal network. Nat. Neurosci. 23, 1444–1452 (2020).
Wee, R. W. S. & MacAskill, A. F. Biased connectivity of brain-wide inputs to ventral subiculum output neurons. Cell Rep. 30, 3644–3654 (2020).
Royer, S., Sirota, A., Patel, J. & Buzsáki, G. Distinct representations and theta dynamics in dorsal and ventral hippocampus. J. Neurosci. 30, 1777–1787 (2010).
Ciocchi, S., Passecker, J., Malagon-Vina, H., Mikus, N. & Klausberger, T. Brain computation. Selective information routing by ventral hippocampal CA1 projection neurons. Science 348, 560–563 (2015).
Jimenez, J. C. et al. Anxiety cells in a hippocampal-hypothalamic Circuit. Neuron 97, 670–683 (2018).
Verney, C. et al. Morphological evidence for a dopaminergic terminal field in the hippocampal formation of young and adult rat. Neuroscience 14, 1039–1052 (1985).
Gasbarri, A., Verney, C., Innocenzi, R., Campana, E. & Pacitti, C. Mesolimbic dopaminergic neurons innervating the hippocampal formation in the rat: a combined retrograde tracing and immunohistochemical study. Brain Res. 668, 71–79 (1994).
Park, A. J. et al. Reset of hippocampal-prefrontal circuitry facilitates learning. Nature 591, 615–619 (2021).
Witter, M. P. & Amaral, D. G. in The Rat Nervous System 3rd edn (ed. Paxinos, G.) 635–704 (Academic Press, 2004).
Bentivoglio, M. & Morelli, M. in Handbook of Chemical Neuroanatomy Vol. 21 (eds Dunnett, S. B. et al.) 1–107 (Elsevier, 2005).
Dubovyk, V. & Manahan-Vaughan, D. Less means more: the magnitude of synaptic plasticity along the hippocampal dorso-ventral axis is inversely related to the expression levels of plasticity-related neurotransmitter receptors. Hippocampus 28, 136–150 (2018).
Dubovyk, V. & Manahan-Vaughan, D. Gradient of expression of dopamine D2 receptors along the dorso-ventral axis of the hippocampus. Front. Synaptic Neurosci. 11, 28 (2019).
Wei, X. et al. Dopamine D1 or D2 receptor-expressing neurons in the central nervous system. Addict. Biol. 23, 569–584 (2018).
Salery, M., Trifilieff, P., Caboche, J. & Vanhoutte, P. From signaling molecules to circuits and behaviors: cell-type-specific adaptations to psychostimulant exposure in the striatum. Biol. Psychiatry 87, 944–953 (2020).
Gerfen, C. R. Segregation of D1 and D2 dopamine receptors in the striatal direct and indirect pathways: an historical perspective. Front. Synaptic Neurosci. 14, 1002960 (2022).
Berke, J. D. What does dopamine mean? Nat. Neurosci. 21, 787–793 (2018).
Bromberg-Martin, E. S., Matsumoto, M. & Hikosaka, O. Dopamine in motivational control: rewarding, aversive, and alerting. Neuron 68, 815–834 (2010).
Berry, A. S. et al. Dopaminergic mechanisms underlying normal variation in trait anxiety. J. Neurosci. 39, 2735–2744 (2019).
Kronman, H. et al. Biology and bias in cell type-specific RNAseq of nucleus accumbens medium spiny neurons. Sci. Rep. 9, 8350 (2019).
Gangarossa, G. et al. Characterization of dopamine D1 and D2 receptor-expressing neurons in the mouse hippocampus. Hippocampus 22, 2199–2207 (2012).
Puighermanal, E. et al. drd2-cre:ribotag Mouse line unravels the possible diversity of dopamine d2 receptor-expressing cells of the dorsal mouse hippocampus. Hippocampus 25, 858–875 (2015).
Puighermanal, E. et al. Anatomical and molecular characterization of dopamine D1 receptor-expressing neurons of the mouse CA1 dorsal hippocampus. Brain Struct. Funct. 222, 1897–1911 (2017).
Cembrowski, M. S., Wang, L., Sugino, K., Shields, B. C. & Spruston, N. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife 5, e14997 (2016).
Cembrowski, M. S. et al. Spatial gene-expression gradients underlie prominent heterogeneity of CA1 pyramidal neurons. Neuron 89, 351–368 (2016).
Cembrowski, M. S. et al. The subiculum is a patchwork of discrete subregions. eLife 7, e37701 (2018).
Harris, K. D. et al. Classes and continua of hippocampal CA1 inhibitory neurons revealed by single-cell transcriptomics. PLoS Biol. 16, e2006387 (2018).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021).
Smith, S. J. & von Zastrow, M. A molecular landscape of mouse hippocampal neuromodulation. Front. Neural Circuits 16, 836930 (2022).
Fenno, L. E. et al. Comprehensive dual- and triple-feature intersectional single-vector delivery of diverse functional payloads to cells of behaving mammals. Neuron 107, 836–853.e11 (2020).
Biane, J. S. et al. Neural dynamics underlying associative learning in the dorsal and ventral hippocampus. Nat. Neurosci. 26, 798–809 (2023).
Labouesse, M. A., Cola, R. B. & Patriarchi, T. GPCR-based dopamine sensors — a detailed guide to inform sensor choice for in vivo imaging. Int. J. Mol. Sci. 21, e8048 (2020).
Patriarchi, T. et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, eaat4422 (2018).
Patriarchi, T. et al. An expanded palette of dopamine sensors for multiplex imaging in vivo. Nat. Methods 17, 1147–1155 (2020).
Sun, F. et al. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 174, 481–496 (2018).
Tritsch, N. X. & Sabatini, B. L. Dopaminergic modulation of synaptic transmission in cortex and striatum. Neuron 76, 33–50 (2012).
Bravo-Rivera, C., Roman-Ortiz, C., Brignoni-Perez, E., Sotres-Bayon, F. & Quirk, G. J. Neural structures mediating expression and extinction of platform-mediated avoidance. J. Neurosci. 34, 9736–9742 (2014).
Diehl, M. M., Bravo-Rivera, C. & Quirk, G. J. The study of active avoidance: a platform for discussion. Neurosci. Biobehav. Rev. 107, 229–237 (2019).
Lisman, J. E. & Grace, A. A. The hippocampal-VTA loop: controlling the entry of information into long-term memory. Neuron 46, 703–713 (2005).
Duszkiewicz, A. J., McNamara, C. G., Takeuchi, T. & Genzel, L. Novelty and dopaminergic modulation of memory persistence: a tale of two systems. Trends Neurosci. 42, 102–114 (2019).
Forro, T. et al. Anxiety-related activity of ventral hippocampal interneurons. Prog. Neurobiol. 219, 102368 (2022).
Volitaki, E., Forro, T., Li, K., Nevian, T. & Ciocchi, S. Activity of ventral hippocampal parvalbumin interneurons during anxiety. Cell Rep. 43, 114295 (2024).
Li, K., Koukoutselos, K., Sakaguchi, M. & Ciocchi, S. Distinct ventral hippocampal inhibitory microcircuits regulating anxiety and fear behaviors. Nat. Commun. 15, 8228 (2024).
Padilla-Coreano, N. et al. Direct ventral hippocampal-prefrontal input is required for anxiety-related neural activity and behavior. Neuron 89, 857–866 (2016).
Glangetas, C. et al. NMDA-receptor-dependent plasticity in the bed nucleus of the stria terminalis triggers long-term anxiolysis. Nat. Commun. 8, 14456 (2017).
Sánchez-Bellot, C., AlSubaie, R., Mishchanchuk, K., Wee, R. W. S. & MacAskill, A. F. Two opposing hippocampus to prefrontal cortex pathways for the control of approach and avoidance behaviour. Nat. Commun. 13, 339 (2022).
Edelmann, E. & Lessmann, V. Dopaminergic innervation and modulation of hippocampal networks. Cell Tissue Res. 373, 711–727 (2018).
Goto, Y. & Grace, A. A. Limbic and cortical information processing in the nucleus accumbens. Trends Neurosci. 31, 552–558 (2008).
Morales, M. & Margolis, E. B. Ventral tegmental area: cellular heterogeneity, connectivity and behaviour. Nat. Rev. Neurosci. 18, 73–85 (2017).
Takeuchi, T. et al. Locus coeruleus and dopaminergic consolidation of everyday memory. Nature 537, 357–362 (2016).
Kempadoo, K. A., Mosharov, E. V., Choi, S. J., Sulzer, D. & Kandel, E. R. Dopamine release from the locus coeruleus to the dorsal hippocampus promotes spatial learning and memory. Proc. Natl Acad. Sci. USA 113, 14835–14840 (2016).
Godino, A. et al. Transcriptional control of nucleus accumbens neuronal excitability by retinoid X receptor α tunes sensitivity to drug rewards. Neuron 111, 1453–1467.e7 (2023).
Mi, H. et al. Protocol update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat. Protoc. 14, 703–721 (2019).
Calipari, E. S. et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc. Natl Acad. Sci. USA 113, 2726–2731 (2016).
Martianova, E., Aronson, S. & Proulx, C. D. Multi-fiber photometry to record neural activity in freely-moving animals. J. Vis. Exp. https://doi.org/10.3791/60278 (2019).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034 (2002).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995).
Acknowledgements
We thank S. Pirpinias, K. Beach, C. McManus, K. Schmidt, N. Pulido, K. Rosen, C. Blaschke and E. Mouzon for transgenics breeding and genotyping; and X. Qiao and E. Aritzia from the Dean’s Flow Cytometry CoRE at the Icahn School of Medicine at Mount Sinai for assistance in nuclei sorting. This work was supported by grants from the Boehringer Ingelheim Fonds (PhD Fellowship to A.G.), the US National Institutes of Health (RF1MH128970 and U01MH116442 to P.R., and R01DA014133 and R01MH051399 to E.J.N.) and the Hope for Depression Research Foundation (to E.J.N.).
Author information
Authors and Affiliations
Contributions
A.G. and E.J.N. conceived the project. A.G. developed all methodology, except for the PMA task, which was developed by A.M.M.-T. and F.J.M.-R. A.G. performed all behavioural experiments and photometry recordings, with help from A.M.M.-T. and V.K. for PMA testing and E.M.P. and V.K. for stereotaxic surgeries. M.S. performed nuclei sorting, FISH and confocal imaging. V.P. performed the electrophysiological recordings, with supervision by R.D.B. J.F.F. performed snRNA-seq library preparation. P.R. contributed resources for snRNA-seq and software for analysis. C.M. contributed software for photometry analysis. A.G. performed all formal analysis and visualization. A.G. wrote the manuscript, which was reviewed and edited by M.S., A.M.M.-T., J.F.F., F.J.M.-R., C.M., P.R., R.D.B. and E.J.N.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Anna Beyeler and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Visualization, validation and sorting of D1 and D2 vHipp cells.
a, Representative images after RNA fluorescence in situ hybridization (FISH) for eGFP, Drd1 and Drd2 transcripts in the DG (top) or vSub (bottom) of a D1-Cre (left) or D2-Cre (right) x fl/fleGFP::L10a male mouse. b, Drd1 and Drd2 combinatorial expression in eGFP-positive cells in the DG (top) or vSub (bottom) of D1-Cre (n = 4 male mice, 2 hemisections/mouse) and D2-Cre (n = 5 male mice, 2 hemisections/mouse) x fl/fleGFP::L10a male mice. Cells circled in yellow are Drd1+/Drd2+. LMM-ANOVA: D1/D2 F1,7 = 0 p = 1, region F1,49 = 0 p = 1, Drd1/Drd2 F3,49 = 45.86 p < 0.0001, D1/D2 x region F1,49 = 0 p = 1, D1/D2 x Drd1/Drd2 F3,49 = 238.30 p < 0.0001, region x Drd1/Drd2 F3,49 = 8.96 p < 0.0001, D1/D2 x region x Drd1/Drd2 F3,49 = 17.47 p < 0.0001; followed by FDR-adjusted post-hoc tests. c, Representative Fluorescence-Activated Nuclei Sorting (FANS) gating strategy from a D1-Cre x fl/fleGFP::L10a male sample. d, Representative sorting summary. e, Percent of GFP-positive nuclei for all D1-Cre (n = 4) and D2-Cre (n = 4) x fl/fleGFP::L10a sorted samples. Data represented as mean ± sem. Scale bars 100 µm.
Extended Data Fig. 2 Additional topography and distribution of vHipp D1 and D2 cells.
Same data as in Fig. 1: n = 23,809 GFP-positive cells from D1-Cre x fl/fleGFP::L10a mice (3 female, 4 male); n = 20,136 GFP-positive cells from D2-Cre x fl/fleGFP::L10a mice (4 female, 4 male), 3-5 hemisections/mouse. a, Density heatmaps of D1 (red) and D2 (green) cells along dorso-ventral (DV) and antero-posterior (AP) coordinates in the subiculum and CA1 pyramidal and stratum oriens layers (left) and in the DG hilus (right). b, Distribution of D1 and D2 GFP-positive cells, as a proportion of the total D1 or D2 GFP-positive cells detected. LMM-ANOVA: region F11,121 = 55.51 p < 0.0001, region x D1/D2 F11,121 = 13.95 p < 0.0001; followed by FDR-adjusted post-hoc tests. c, Segregation of D1 (left) and D2 (right) vHipp cells between GAD67+|CaMKIIα− inhibitory cells and GAD67−|CaMKIIα+ excitatory cells, as a proportion of the total D1 or D2 cells detected in each subregion. LMM-ANOVA: region F11,253 = 4.00 p < 0.0001, inh/exc F1,253 = 1320.13 p < 0.0001, D1/D2 x inh/exc F1,253 = 34.36 p < 0.0001, region x sex x inh/exc F11,253 = 2.34 p = 0.0094, region x D1/D2 x inh/exc F11,253 = 15.03 p < 0.0001; followed by FDR-adjusted post-hoc tests. Data represented as mean ± sem. FDR < 0.05*, <0.01**, <0.001***.
Extended Data Fig. 3 Sample integration and cell-type annotation of snRNAseq clusters.
a, Workflow for snRNAseq of vHipp from male D1-Cre (n = 4) and D2-Cre (n = 4) x fl/fleGFP::L10a. b, Sample integration. c, Proportion of all nuclei in individual clusters. d, D1-sorted or D2-sorted origin of individual nuclei, quantified in Fig. 2b. e, Expression of dopamine receptor Drd1 and Drd2 genes in individual nuclei. Nuclei are considered as co-expressing D1 and D2 receptors (yellow) either if both Drd1 and Drd2 are detected, if Drd1 is detected in a D2-sorted nucleus or if Drd2 is detected in a D1-sorted nucleus. Insert shows the repartition of D1-D2 co-expressing nuclei across clusters. f, Expression of published marker genes for different cell types across clusters. Full lists of cluster marker genes available in Supplementary Table 1.
Extended Data Fig. 4 Viral targeting of vSub.
a, Representative spread, fiber placements (red lines) and representative image of AAV-DIO-GCaMP6s expression in the vSub of D1-Cre (left) and D2-Cre (right) male mice. b, Representative spread, fiber placements (red lines) and representative image of AAV-dLight-1.1, AAV-GRABDA-1h expression in the vSub. Full hemisection illustrates spread of a control AAV-GFP virus, while zoomed insets depict faint membrane expression patterns of dopamine sensors. Note the difference in GFP signal intensity with neighboring entorhinal cortex (EC). FT fiber tip. c, Representative spread and representative image of AAV-DIO-mCherry, -hM3Dq and -hM4Di expression in the vSub of D1-Cre (left) and D2-Cre (right) male mice. Placement was verified in all mice either by confocal imaging or by dissection of fresh tissue under fluorescent illumination. d, Representative spread, fiber placements (red lines) and representative image of AAV-DIO-EYFP and -ChR2 expression in the vSub of D1-Cre (left) and D2-Cre (right) male mice. vSub was primarily targeted (dark shaded areas), but unavoidable viral spread encompassed various degrees of vCA3 and vDG (light shaded areas). Only animals with notable somatic expression in neighboring entorhinal cortex (<10%) were removed from analysis. Scale bars 500 µm.
Extended Data Fig. 5 Additional analyses of behavior and calcium imaging of vHipp D1 and D2 neuronal activity during EPM testing.
Data are obtained from the same mice as in Fig. 3 (D1: n = 11, D2: n = 10 male mice). a, Distance travelled (left; t-test: t14.63 = −0.73 p = 0.4783) and velocity (right; t-test: t14.70 = −0.71 p = 0.4885). b, Open arm (OA) exploration time (left; LMM-ANOVA: cell type F1,19 = 0.0013 p = 0.9713, compartment F2,38 = 498.02 p < 0.0001, cell type x compartment F2,38 = 8.12 p = 0.0012; followed by FDR-adjusted post-hoc tests), total number of OA entries (middle; LMM-ANOVA: cell type F1,19 = 1.87 p = 0.1876, compartment F2,38 = 64.02 p < 0.0001, cell type x compartment F2,38 = 2.51 p = 0.0950; followed by FDR-adjusted post-hoc tests) and average OA exploration bout length (right; LMM-ANOVA: cell type F1,19 = 0.011 p = 0.9167, compartment F2,38 = 36.85 p < 0.0001, cell type x compartment F2,38 = 0.14 p = 0.8673; followed by FDR-adjusted post-hoc tests). c, Number of head-dips (total: t-test: t18.945 = 0.83 p = 0.4170; per behavioral outcome: LMM-ANOVA: cell type F1,19 = 0.69 p = 0.4180, outcome F2,38 = 44.21 p < 0.0001, cell type x outcome F2,38 = 4.04 p = 0.0256; followed by FDR-adjusted post-hoc tests). d, Correlation of GCaMP6s signal intensity with individual OA (left, D1: n = 163; Pearson’s r = 0.0499, p = 0.5273; D2: n = 107; Pearson’s r = 0.3242, p = 0.0007) or closed arm (CA, right, D1: n = 197; Pearson’s r = −0.2383, p = 0.0007; D2: n = 198; Pearson’s r = −0.1984, p = 0.0051) exploration bout lengths. e, Average GCaMP6s signal in D1-Cre (n = 27 head-dips) and D2-Cre (n = 36 head-dips) mice around head-dips events with unclear outcome. Boxplot represents the median time ± inter-quartile range of the times of maximal acceleration (max. acc.) after each head-dip. f, Maximum (peak) GCaMP6s signal before (pre) or after (post) each head-dip event. LMM-ANOVA: cell type F1,22.22 = 0.27 p = 0.6069, outcome F2,375.77 = 7.06 p = 0.0010, pre-post F1,385 = 84.64 p < 0.0001, cell type x outcome F2,375.77 = 16.74 p < 0.0001, cell type x pre-post F1,385 = 0.17 p = 0.6775, outcome x pre-post F2,385 = 11.69 p < 0.0001, cell type x outcome x pre-post F2,385 = 34.37 p < 0.0001; followed by FDR-adjusted post-hoc tests. g, GCaMP6s signal change slope after head-dip events. LMM-ANOVA: cell type F1,23.29 = 1.29 p = 0.2679, outcome F2,380.68 = 6.90 p = 0.0011, cell type x outcome F2,380.68 = 28.50 p < 0.0001; followed by FDR-adjusted post-hoc tests. h, Time delay between the time of maximal GCaMP6s signal change (max. slope) and the time of movement initiation (max. acc.). LMM-summary: intercept ≠ 0 t67.07 = −3.68 p = 0.0005, and LMM-ANOVA: cell type F1,23.10 = 0.0053 p = 0.9427, outcome F2,383.84 = 0.19 p = 0.8241, cell type x outcome F2,383.84 = 0.46 p = 0.6310; followed by individual LMM-summary for intercept ≠ 0 for each cell type x outcome combination and FDR adjustment of p-values. i, SVM workflow schematic. j, Classification of unclear-outcome head-dips using a SVM trained on manually-annotated avoid/explore D1 and D2 time-series. Both average GCaMP6s signal (top) and heatmaps of individual time-series (bottom) are represented for both D1 and D2 signals, split by SVM outcome prediction. Data represented as mean ± sem, regression lines with their 95% confidence intervals.
Extended Data Fig. 6
Calcium imaging of vSub D1 and D2 excitatory neuron activity during EPM testing. a, Experimental schematic (n = 6 D1-Cre, n = 6 D2-Cre male mice). b, Representative images at the fiber tip (FT) in a D1-Cre (left) and D2-Cre (right) mouse vSub. GCaMP6f-positive cells in magenta if CaMKIIα+|GAD67−, in cyan if CaMKIIα−|GAD67+, in yellow if CaMKIIα+|GAD67+. EC entorhinal cortex. c, Quantification in D1-Cre (top, 1 hemisection/mouse) and D2-Cre (bottom, 1 hemisection/mouse) animals indicating GCaMP6f expression largely restricted to excitatory CaMKIIα+|GAD67− cells. d, Representative D1-Cre (top) and D2-Cre (bottom) GCaMP6f signal. e, Heatmaps of average GCaMP6f signal. f, Average GCaMP6f signal in during entries in the open arm (OA) or closed arm (CA). D1: n = 15 OA, n = 33 CA, D2: n = 18 OA, n = 32 CA. g, Signal intensity (AUC) by EPM compartment. LMM-ANOVA: compartment F2,20 = 58.21 p < 0.0001, cell type F1,10 = 1.12 p = 0.3142, compartment x cell type F2,20 = 1.04 p = 0.3724; followed by FDR-adjusted post-hoc tests. Data represented as mean ± sem. Scale bars 100 µm.
Extended Data Fig. 7 Calcium imaging of vSub D1 and D2 neuronal activity during anxiety-related testing.
a, Experimental schematic (n = 7 D1, n = 8 D2 male mice). b, Representative D1-Cre (top) and D2-Cre (bottom) GCaMP6s signal. c, Heatmaps of average GCaMP6s signal. d, Average GCaMP6s signal during entries in the arena center (D1: n = 50; D2: n = 149 entries). e, Average GCaMP6s signal around exits from immobility in arena corners (D1: n = 231; D2: n = 178 events). Dotted line indicates movement initiation. f, Signal intensity (AUC) by OFT zone. LMM-ANOVA: zone F3,39 = 6.40 p = 0.0012, cell type F1,13 = 0.33 p = 0.5742, zone x cell type F3,39 = 7.74 p = 0.0004; followed by FDR-adjusted post-hoc tests. g, Experimental schematic (n = 6 D1, n = 7 D2 male mice). h, Representative D1-Cre (top) and D2-Cre (bottom) GCaMP6s signal. The first (1) feeding bout is denoted. i, Heatmaps of average GCaMP6s signal. P food pellet. j, Average GCaMP6s signal around the initiation of feeding bouts (D1: n = 68; D2: n = 88 bouts). k, Average GCaMP6s signal around exits from immobility in arena corners (D1: n = 87; D2: n = 32 events). Dotted line indicates movement initiation. l, Signal intensity (AUC) by NSF arena zone and feeding activity in the center zone. LMM-ANOVA: zone F4,44 = 10.36 p < 0.0001, cell type F1,11 = 0.64 p = 0.4405, zone x cell type F4,44 = 1.65 p = 0.1785; followed by FDR-adjusted post-hoc tests. Data represented as mean ± sem.
Extended Data Fig. 8 Chemogenetic and optogenetic manipulation of vHipp D1 and D2 neurons during anxiety-related testing – controls and additional metrics.
Effects of CNO itself (n = 10 CNO, n = 10 vehicle male mice) on a, OA exploration time (t-test: t14.65 = −0.061 p = 0.9523); b, total number of open arm entries (left; t-test: t17.99 = −0.53 p = 0.6045) and average OA exploration bout length (right; t-test: t17.26 = −0.057 p = 0.9548). c, Experimental schematic. Data are obtained from the same mice as in Fig. 4a–c (n = 12 D1-mCherry, n = 8 D1-hM3Dq, n = 11 D2-mCherry, n = 12 D2-hM3Dq male mice). d, Total number of OA entries (left; t-test: t8.16 = 0.73 p = 0.4857) and average OA exploration bout length (right; t-test: t16.42 = 1.05 p = 0.3095) for D1-Cre mice. e, Total number of OA entries (left; t-test: t20.68 = −2.16 p = 0.0423) and average OA exploration bout length (right; t-test: t14.63 = −2.18 p = 0.0463) for D2-Cre mice. f, Experimental schematic. Data are obtained from the same mice as in Fig. 4d–f (n = 12 D1-mCherry, n = 11 D1-hM4Di, n = 12 D2-mCherry, n = 12 D2-hM4Di male mice). g, Total number of OA entries (left; t-test: t20.94 = 0.64 p = 0.5284) and average OA exploration bout length (right; t-test: t20.60 = −1.89 p = 0.0723) for D1-Cre mice. h, Total number of OA entries (left; t-test: t21.68 = 1.55 p = 0.1354) and average OA exploration bout length (right; t-test: t21.93 = 1.80 p = 0.0858) for D2-Cre mice. i, Experimental schematic. (n = 11 D1-mCherry, n = 11 D1-hM3Dq, n = 11 D2-mCherry, n = 12 D2-hM3Dq male mice for OFT, and n = 11 D1-mCherry, n = 11 D1-hM4Di, n = 12 D2-mCherry, n = 12 D2-hM4Di for NSF). j, Total locomotor activity in OFT for D1-Cre (left; t-test: t13.25 = −2.52 p = 0.0253) and D2-Cre (right; t-test: t20.99 = 0.08 p = 0.9374) mice. k, Time spent in the OFT center zone for D1-Cre (left; t-test: t18.84 = 2.6052 p = 0.0175) and D2-Cre (right; t-test: t19.62 = −0.63 p = 0.5379) mice. l, Latency to the first feeding bout during NSF testing for D1-Cre (left; t-test: t18.31 = 0.51 p = 0.6171) and D2-Cre (right; t-test: t21.82 = −2.32 p = 0.0303) mice. m, Experimental schematic. Data are obtained from the same mice as in Fig. 4g–i (n = 10 D1-EYFP, n = 9 D1-ChR2, n = 11 D2-EYFP, n = 10 D2-ChR2 male mice). n, Total number of OA entries (left; LMM-ANOVA: stimulation zone F1,17 = 0.67 p = 0.4232, virus F1,17 = 2.63 p = 0.1234, stimulation zone x virus F1,17 = 2.55 p = 0.1284; followed by FDR-adjusted post-hoc tests) and OA exploration bout length (right; LMM-ANOVA: stimulation zone F1,17 = 8.88 p = 0.0084, virus F1,17 = 0.58 p = 0.4580, stimulation zone x virus F1,17 = 5.8529 p = 0.0271; followed by FDR-adjusted post-hoc tests) for D1-Cre mice. o, Total number of open arm entries (left; LMM-ANOVA: stimulation zone F1,19 = 8.70 p = 0.0082, virus F1,19 = 7.94 p = 0.0110, stimulation zone x virus F1,19 = 0.036 p = 0.8526; followed by FDR-adjusted post-hoc tests) and OA exploration bout length (right; LMM-ANOVA: stimulation zone F1,19 = 2.62 p = 0.1221, virus F1,19 = 6.1645 p = 0.0225, stimulation zone x virus F1,19 = 9.55 p = 0.0060; followed by FDR-adjusted post-hoc tests) for D2-Cre mice. Data represented as mean ± sem.
Extended Data Fig. 9 In vivo dopamine sensing in vSub – other sensors.
a, Experimental schematic. (n = 13 RdLight−1, n = 15 GRABDA−1h). b, Representative RdLight-1 (top) and GRABDA−1h (bottom) signal. c, Heatmaps of average signal. d, Average RdLight−1 signal during entries to (left) and exits from (right) the open arm (OA) or closed arm (CA) (entry: n = 24 OA, n = 99 CA, exit: n = 42 OA, n = 97 CA). e, Average GRABDA-1h signal during entries to (left) and exits from (right) the OA or CA (entry: n = 28 OA, n = 92 CA, exit: n = 44 OA, n = 77 CA). f, Average control signal during entries to (left) and exits from (right) the OA or CA (entry: n = 33 OA, n = 112 CA, exit: n = 45 OA, n = 105 CA). g, Signal intensity by EPM compartment. Data analyzed together with Fig. 5e data, statistics in Fig. 5e legend. Data represented as mean ± sem.
Extended Data Fig. 10 Viral knock-down of Drd1 or Drd2 in vSub – validation and additional anxiety-related behaviors.
a, Experimental schematic. Data are obtained from the same mice as in Fig. 5i–l (n = 11 shDrd1, n = 11 shDrd2, n = 14 GFP male mice). b, Drd1 mRNA expression. LM-ANOVA: virus F2,32 = 3.60 p = 0.0389; followed by FDR-adjusted post-hoc tests. c, Drd2 mRNA expression. LM-ANOVA: virus F2,32 = 7.14 p = 0.0027; followed by FDR-adjusted post-hoc tests. d, Total number of OA entries in EPM. LM-ANOVA: virus F2,33 = 0.12 p = 0.8839. e, Total locomotor activity in OFT. LM-ANOVA: virus F2,33 = 0.67 p = 0.5190. f, Time spent in OFT center zone. LM-ANOVA: virus F2,33 = 1.66 p = 0.2049; followed by FDR-adjusted post-hoc tests. g, Latency to the first feeding bout during NSF testing. LM-ANOVA: virus F2,33 = 3.33 p = 0.0482; followed by FDR-adjusted post-hoc tests. Data represented as mean ± sem.
Extended Data Fig. 11 vSub dopamine, D1 and D2 correlates of approach and avoidance in PMA.
a, Experimental schematic (n = 4 RdLight−1, n = 13 D1-GCaMP6s, n = 8 D2-GCaMP6s male mice). b, Average GCaMP6s signal around electric foot-shock onset when on the grid (left, RdLight-1 n = 22, D1-GCaMP6s n = 69, D2-GCaMP6s n = 45 events), reward magazine entry (middle, RdLight−1 n = 25, D1-GCaMP6s n = 6, D2-GCaMP6s n = 11 events) or the first platform exit after tone/shock (right, RdLight-1 n = 18, D1-GCaMP6s n = 43, D2-GCaMP6s n = 40 events). c, Quantification of peri-event signal changes. LMM-ANOVAs (Foot-shock, DA: F7,147 = 18.64 p < 0.0001. Foot-shock, D1: F7,475.06 = 79.37 p < 0.0001. Foot-shock, D2: F7,308 = 48.69 p < 0.0001. Reward, DA: F7,161 = 6.88 p < 0.0001. Reward, D1: F7,35 = 0.27 p = 0.9605. Reward, D2: F7,70 = 3.15 p = 0.0059. First platform exit, DA: F7,119 = 7.64 p < 0.0001. First platform exit, D1: F7,294 = 1.19 p = 0.3109. First platform exit, D2: F7,273 = 9.72 p < 0.0001); followed by FDR-adjusted post-hoc tests (all pairwise comparisons were computed but only the ones significant vs the first 1.5 s time-bin are represented, p < 0.05*, <0.01**, <0.001***). d, Times of peak photometry RdLight−1 and D2-GCaMP6s signals after foot-shock (left, LMM-ANOVA: sensor F1,22.94 = 4.99 p = 0.0355) and first platform exit (right, LMM-ANOVA: sensor F1,19.468 = 1.49 p = 0.2376). e, Correlation of RdLight-1 and D2-GCaMP6s signal (n = 1 dual-injected male mouse) around foot-shocks (left; Pearson’s r = −0.18 p < 0.0001), reward consumption (Pearson’s r = −0.37 p < 0.0001) and first platform exit after tone end (right; Pearson’s r = −0.19, p < 0.0001). Data represented as mean ± sem, regression lines with their 95% confidence intervals.
Extended Data Fig. 12 Chemogenetic manipulation of vSub D1 and D2 neurons in the PMA task.
a, Experimental schematic (n = 10 D1-mCherry, n = 7 D1-hM4Di male mice). b, Approach behavior (lever presses) during extinction sessions, during tone presentation (left; LMM-ANOVA: session F4,60 = 6.50 p = 0.0002, DREADD F1,15 = 0.47 p = 0.5023, session x DREADD F4,60 = 0.31 p = 0.8692) and inter-trial intervals (ITI, right; LMM-ANOVA: session F4,60 = 1.10 p = 0.3644, DREADD F1,15 = 1.45 p = 0.2466, session x DREADD F4,60 = 1.35 p = 0.2623). c, Avoidance behavior (time on platform) during extinction sessions. LMM-ANOVA: session F4,60 = 21.56, p < 0.0001, DREADD F1,15 = 0.003 p = 0.9568, session x DREADD F4,60 = 0.36 p = 0.8348. d, Freezing upon tone presentation during extinction sessions. LMM-ANOVA: session F4,60 = 2.92 p = 0.0283, DREADD F1,15 = 2.73 p = 0.1191, session x DREADD F4,60 = 0.19 p = 0.9424. e, Approach behavior (lever presses) during conditioning sessions for D1-Cre mice during tone presentation (left; LMM-ANOVA: session F9,135 = 3.05 p = 0.0024, DREADD F1,15 = 0.04 p = 0.8533, session x DREADD F9,135 = 0.27 p = 0.9808) or during inter-tone intervals (ITI; right; LMM-ANOVA: session F9,135 = 5.61 p < 0.0001, DREADD F1,15 = 0.003 p = 0.9608, session x DREADD F9,135 = 1.08 p = 0.3846). f, Avoidance (time on platform) behavior during conditioning sessions for D1-Cre mice. LMM-ANOVA: session F9,133.1 = 15.04 p < 0.0001, DREADD F1,15.006 = 2.79 p = 0.1156, session x DREADD F9,133.1 = 1.67 p = 0.1027. g, Experimental schematic (n = 11 D2-mCherry, n = 9 D2-hM4Di male mice). h, Approach behavior (lever presses) during extinction sessions, during tone presentation (left; LMM-ANOVA: session F4,72 = 64.75 p < 0.0001, DREADD F1,18 = 5.27 p = 0.0339, session x DREADD F4,72 = 0.83 p = 0.5099) and inter-trial intervals (ITI, right; LMM-ANOVA: session F4,72 = 22.15 p < 0.0001, DREADD F1,18 = 0.006 p = 0.9379, session x DREADD F4,72 = 2.03 p = 0.0994). i, Avoidance behavior (time on platform) during extinction sessions. LMM-ANOVA: session F4,72 = 26.69, p < 0.0001, DREADD F1,18 = 6.13 p = 0.0234, session x DREADD F4,72 = 0.57 p = 0.6858. j, Freezing upon tone presentation during extinction sessions. LMM-ANOVA: session F4,72 = 4.25 p = 0.0038, DREADD F1,18 = 0.003 p = 0.9570, session x DREADD F4,72 = 1.45 p = 0.2278. k, Approach behavior (lever presses) during conditioning sessions for D2-Cre mice during tone presentation (left; LMM-ANOVA: session F9,162 = 10.30 p < 0.0001, DREADD F1,18 = 1.03 p = 0.3227, session x DREADD F9,162 = 1.54 p = 0.1373) or during inter-tone intervals (ITI; right; LMM-ANOVA: session F9,162 = 5.50 p < 0.0001, DREADD F1,18 = 4.12 p = 0.0575, session x DREADD F9,162 = 1.11 p = 0.3589). l, Avoidance behavior (time on platform) during conditioning sessions for D2-Cre mice (LMM-ANOVA: session F9,162 = 26.31 p < 0.0001, DREADD F1,18 = 0.44 p = 0.5170, session x DREADD F9,162 = 1.91 p = 0.05385). Data represented as mean ± sem.
Supplementary information
Supplementary Table 1
List of marker genes for snRNAseq cell type clusters
Supplementary Table 2
List of D1 vs D2 DEGs for each snRNAseq cell type cluster
Supplementary Table 3
List of D1 vs D2 DEGs grouped by gene expression pattern
Supplementary Table 4
List of gene ontology terms associated with DEG lists in each gene expression pattern
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Godino, A., Salery, M., Minier-Toribio, A.M. et al. Dopamine D1–D2 signalling in hippocampus arbitrates approach and avoidance. Nature (2025). https://doi.org/10.1038/s41586-025-08957-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41586-025-08957-5