Abstract
Type 2 diabetes (T2D) genome-wide association studies (GWASs) often overlook rare variants as a result of previous imputation panels’ limitations and scarce whole-genome sequencing (WGS) data. We used TOPMed imputation and WGS to conduct the largest T2D GWAS meta-analysis involving 51,256 cases of T2D and 370,487 controls, targeting variants with a minor allele frequency as low as 5 × 10−5. We identified 12 new variants, including a rare African/African American-enriched enhancer variant near the LEP gene (rs147287548), associated with fourfold increased T2D risk. We also identified a rare missense variant in HNF4A (p.Arg114Trp), associated with eightfold increased T2D risk, previously reported in maturity-onset diabetes of the young with reduced penetrance, but observed here in a T2D GWAS. We further leveraged these data to analyze 1,634 ClinVar variants in 22 genes related to monogenic diabetes, identifying two additional rare variants in HNF1A and GCK associated with fivefold and eightfold increased T2D risk, respectively, the effects of which were modified by the individual’s polygenic risk score. For 21% of the variants with conflicting interpretations or uncertain significance in ClinVar, we provided support of being benign based on their lack of association with T2D. Our work provides a framework for using rare variant GWASs to identify large-effect variants and assess variant pathogenicity in monogenic diabetes genes.
Similar content being viewed by others
Main
Large genome-wide association meta-analyses have allowed the discovery of hundreds of genetic variants, mostly common (minor allele frequency (MAF) > 0.05), associated with altered risk for T2D1,2,3,4,5. Genotype imputation, which predicts indirectly measured genotypes based on a reference panel, has been widely used for the largest GWAS meta-analyses, but has typically limited imputation to variants with MAF > 0.005. As a result, large-scale meta-analyses to date focused on variants with MAF > 0.005. Until recently, the only approach to analyze variants with MAF below this threshold has been either via WGS or whole-exome sequencing (WES). WGS and WES datasets are still limited in sample size and WES focuses only on the ~1% of the genome that codes for protein exons. The National Heart, Lung, and Blood Institute Trans-Omics for Precision Medicine (TOPMed) imputation reference panel has demonstrated the ability to accurately impute variants with a frequency as low as 5 × 10−5 (refs. 1,6,7), providing a unique opportunity to explore the contribution of rare and low-frequency variants at scale.
Approximately 0.4% of all cases of diabetes are caused by rare variants with very large-effect sizes (odds ratios (ORs) often >20)8,9,10 and are classified as monogenic diabetes. Many monogenic diabetes subtypes are well suited for precision medicine interventions11, but the vast majority of patients with monogenic diabetes are undiagnosed12, with populations of non-European genetic ancestry more vulnerable to misdiagnosis13. A large percentage of people have probably inherited rare variants with intermediate penetrance, which increase diabetes risk by approximately five- to tenfold14; however, without clear evidence for this, clinicians are unable to incorporate genetic considerations in patient care or clinical decision-making. The ability to estimate the pathogenicity of such variants, which are reported as variants of uncertain significance (VUSs)15, partly relied on population-based allele frequencies from databases, such as gnomAD16, or bioinformatics tools that are used to predict pathogenicity. Segregation in families or enrichment of carriers among those with the disease is used, but this information is often not available or not generated in a systematic way.
Estimating the risk of certain variants found in patients of non-European ancestry is particularly limited as a result of the deficit of genomic data available, such as population-specific allele frequencies17. In addition, it is increasingly recognized that disease risk owing to variants of intermediate penetrance can be modified by an individual’s polygenic background, meaning that risk estimation can be improved by combining rare variants with polygenic risk scores (PRSs) constructed based on common variants8,14.
Beyond monogenic diabetes, recent studies using large-scale WGS data have shown that rare variants account for a substantial proportion of the heritability of common complex traits18. There is clear evidence for the contribution of rare variants to T2D and even the convergence of signals from rare and common variants19,20,21. The discovery of large-effect, rare and population-specific variants associated with T2D3,4,16,17 suggests a continuum of diabetes subtypes and degrees of severity, rather than a categorical distinction between common forms of diabetes and monogenic diabetes18.
We hypothesized that analyzing low-frequency and rare variants at scale would allow for (1) the identification of rare variants of large-effect size across diverse ancestries, (2) the assessment of the pathogenicity of variants in known monogenic diabetes genes and (3) the investigation of the interplay between rare variants and PRS.
In the present study, we combined TOPMed-imputed data7 from the UK Biobank (UKB)22, the Mass General Brigham Biobank (MGBB)23 and the Genetic Epidemiology Research on Adult Health and Aging (GERA)24 cohort with WGS data from the All of Us Research Program (AoU)25 to perform the largest and most diverse T2D GWAS meta-analysis incorporating variants with MAF as low as 5 × 10−5. We identified large-effect variants that are rare or enriched in non-European populations. Finally, we provided a framework for informing pathogenicity in monogenic diabetes genes based on effect size for diabetes susceptibility, with consideration of additional risk stratification using background common genetic variation.
Results
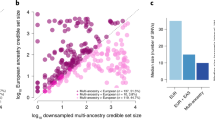
After quality control and imputation of each of the cohorts with the TOPMed reference panel (except AoU, as it is WGS data), we conducted a meta-analysis comprising a total of 51,256 cases and 370,487 controls with 12.2% cases of non-European ancestry (Fig. 1a, Extended Data Fig. 1 and Supplementary Table 1). Genotype imputation with TOPMed resulted in a much larger fraction of variants imputed with higher quality, especially at lower allele frequencies, with tenfold more imputed variants for MAF between 5 × 10−5 and 1 × 10−4 (Extended Data Fig. 2) compared with imputation with the Haplotype Reference Consortium (HRC)26 and 1000 Genomes Project (1000G)27. We also demonstrate that there is ~75% minor allele concordance of variants at a MAF between 5 × 10−5 and 1 × 10−4 when comparing with data from WESs from UKB (Extended Data Fig. 3). A comparison of the effect sizes of established variants shows a strong positive correlation (r2 ≈ 0.88) between our meta-analysis and published studies (Extended Data Fig. 4a). Of note, for low-frequency variants (MAF < 0.05), we observed larger effect sizes in our meta-analysis and P values of similar significance despite the effective sample size of our meta-analysis being smaller (172,321 in our study versus 765,591 in ref. 5), suggesting an improvement of power for low-frequency variants, because of either the more accurate imputation of the TOPMed reference panel or more stringent definition of cases and controls (Extended Data Fig. 4b–d).
a, Overview of the cohorts, sample size and pre-processing steps for each cohort included in the T2D GWAS meta-analysis. b, Manhattan plots for variants with an overall study MAF > 0.001 (bottom) and MAF < 0.001 (top). The y axis shows the −log10(P) from the meta-analysis of two-sided logistic regression models, weighting the cohorts by the inverse of the s.e. for each variant. The dashed horizontal line represents the genome-wide significance threshold (P < 5 × 10−8). The x axis represents the genomic position (GRCh38). c, ORs for all genome-wide, significant, conditionally independent variants plotted across MAF. New and known variants are represented, with primary signals denoted by stars and secondary signals by points. d, Overview of the downstream analyses that use the rare variant meta-analysis GWAS results to inform the classification of variants in monogenic diabetes genes within ClinVar groups. We selected all the variants reported in ClinVar in genes involved in monogenic diabetes. For those that are present in our meta-analysis, we categorized them as ‘VIP’, ‘supporting benign’ or ‘inconclusive’ according to the OR and CIs of their association with T2D. We then validated the GWAS-based classification in the AoU external dataset, assessing the aggregate effect of the variants on T2D risk. Finally, we stratified the carriers and the noncarriers of the variants within the VIP category based on their PRS and assessed their risk of T2D. QC, quality control.
TOPMed imputation combined with WGS increases power to detect rare variant associations
Through approximate conditional and joint association analysis (COJO)28, we identified 284 distinct signals in 214 loci. Of those, 239 were common (MAF > 0.05), 37 were low frequency (MAF 0.05-0.001) and 8 were rare (MAF < 0.001). Of the 284 total signals, 34 were new, of which 8% (19 out of 239 variants), 19% (7 out of 37 variants) and 100% (8 variants) were common, low frequency and rare, respectively. Of the 34 new variants, 24 were the lead signals and 10 were secondary because they remained significant (P < 5 × 10−8) after approximate conditional analysis (Fig. 1b,c, Extended Data Fig. 5 and Supplementary Tables 2 and 3).
Of the 26 common and low-frequency new variants, 11 were replicated in two previous T2D GWAS meta-analyses5,29 including independent samples (P < 0.0015 from 0.05 of 34 tested new variants) (Supplementary Table 4). As rare variants are not considered in previous meta-analyses, we looked for replication of the eight new rare variants in independent TOPMed-imputed, WES and WGS data for a total of 73,088 cases with T2D and 79,827 controls from the GEISINGER MyCode cohort and participants from GERA (GERA_REP) and AoU (AoU_REP) cohorts not overlapping and unrelated to discovery (Supplementary Table 1). In our replication dataset, all the rare variants showed a consistent direction of effect with the discovery (binomial P = 0.007). We had sufficient statistical power and imputation quality to test seven of them, of which we replicated two variants (Supplementary Table 5): one noncoding near LEP and a missense variant in HNF4A.
Variant 7:128323039-G-A is prevalent in African/African American (AFA) populations (rs147287548, MAFAFA = 0.002; discovery: OR = 10.4, 95% confidence interval (CI) = 4.5–24.2, P = 4.53 × 10−8; discovery + replication: OR = 4.7, 95% CI = 2.9–7.6, P = 8.8 × 10−10) and is located in an enhancer active in adipose tissue and adipose tissue-derived mesenchymal stem cells that interact with the LEP gene30 (Fig. 2a–c, Extended Data Fig. 6 and Supplementary Table 6). The AFA-specific allele of this variant disrupts a binding motif for NFATc transcription factor, which is implicated in adipogenesis31 (Fig. 2d). Using luciferase reporter assays, we demonstrated that the risk allele reduces the regulatory potential of the enhancer in adipocytes in vitro (Fig. 2e). Moreover, 7:128323039-G-A is associated with lower apolipoprotein A levels (beta (β) = −0.084 g l−1, P = 0.003) and lower high-density lipoprotein (HDL) cholesterol levels (β = −0.117 mmol l−1, P = 0.002) in participants without diabetes from the UKB (Supplementary Table 7 and Extended Data Fig. 7a,b).
a, LocusZoom plots for the rs147287548 region. Each point represents a variant, with its P value (on a −log10 scale, y axis) derived from the meta-analysis of two-sided logistic regression models, weighting the cohorts by the inverse of the s.e. for each variant. The x axis represents the genomic position (GRCh38). b, Representation of chromatin interactions (enhancer-capture HiC), accessibility (assay for transposable-accessible chromatin with sequencing), H3K27ac and H3K4me1 chromatin immunoprecipitation sequencing signal coverage in T2D-relevant tissues. The box with the dashed line highlights the chromatin fragment that contains rs147287548, which shows significant long-range chromatin interactions with the promoter of the LEP gene in mesenchymal stem cells (MSCs) and throughout in vitro adipogenesis30. The wider chromatin landscape of this locus and chromatin interactions detected by enhancer-capture HiC are shown in Extended Data Fig. 6. Details of the datasets shown are provided in Supplementary Table 6. c, Forest plot showing the carrier counts and ORs of rs147287548 in the discovery, replication and overall datasets. The ORs from each cohort from the discovery and replication datasets are denoted by boxes and the 95% CIs by horizontal lines. Arrows were added for 95% CI LB < 0.3 and 95% CI UB > 40. The center of the diamonds represents the OR of the meta-analysis, with the horizontal extremities indicating the 95% CI. Statistical significance is from the meta-analysis of two-sided logistic regression models, weighting the cohorts by the inverse of the s.e. for each variant. d, Transcription factor motif disruption results. The minor allele of rs147287548 is predicted to disrupt an NFATc-binding site. e, Luciferase reporter assay in mouse 3T3-L1-derived adipocytes showing allele-dependent activity of the enhancer harboring the rs147287548 variant. The data are represented as the fold change in relative luciferase signal over the average activity of the negative controls (empty pGL4.23) ± s.e.m. (n = 3 independent experiments with four independent transfections). Statistical significance was determined using a two-tailed Student’s t-test. Alt., alternative; Ref., reference.
The second is a nonsynonymous variant (20:44413714-C-T, p.Arg114Trp) in HNF4A, a known gene causing maturity-onset diabetes of the young (MODY). During the preparation of this work, it was classified as having ‘conflicting interpretations of pathogenicity’ (CIP) in ClinVar (accessed July 2023)32 and was associated with ~8-fold increased risk of T2D (rs137853336; MAF = 0.0001; discovery: OR = 8.3, 95% CI = 4.7–14.14, P = 1.08 × 10−13; discovery + replication: OR = 7.9, 95% CI = 4.9–12.7, P = 3.1 × 10−18). This variant has previously been reported as a mutation causing a distinct clinical subtype of monogenic diabetes with reduced penetrance, reduced sensitivity to sulfonylurea treatment and no effect on birth weight33. In participants without diabetes from UKB, p.Arg114Trp is associated with lower apolipoprotein A (β = −0.150 g l−1, P = 4.9 × 10−11), aspartate aminotransferase (β = −1.508 U l−1, P = 0.036), HDL-cholesterol (β = −0.167 mmol l−1, P = 6.9 × 10−9) and sex hormone-binding globulin (SHBG; β = −20.38 nmol l−1, P = 7.8 × 10−18), as well as with higher levels of glucose (β = 0.265 mmol l−1, P = 0.024), triglycerides (β = 0.266 mmol l−1, P = 0.0005), total cholesterol (β = 0.314 mmol l−1, P = 0.005), low-density lipoprotein (LDL)-cholesterol (β = 0.381 mmol l−1, P = 2.9 × 10−6), apolipoprotein B (β = 0.141 g l−1, P = 2.2 × 10−11), lipoprotein A (β = 1.78 nmol l−1, P = 0.008) and urea (β = 3.03 mmol l−1, P = 0.031) (Supplementary Table 7 and Extended Data Fig. 7d–m).
Association results inform pathogenicity of rare variants in monogenic diabetes genes
Motivated by the identification of the HNF4A p.Arg114Trp variant, we analyzed the effect of rare variants in genes known to be involved in monogenic forms of diabetes, including MODY, neonatal diabetes and rare forms of syndromic diabetes. In a meta-analysis that included UKB, MGBB and GERA, we evaluated the effect of 1,634 well-imputed variants (MAF < 0.001) in 22 monogenic diabetes genes reported in ClinVar (Supplementary Table 8). Of those, 1,007 of the variants (61.6%) had MAF < 0.0001. Based on the recommendations by the American College of Medical Genetics’ guidelines15, we defined those with OR > 5 and lower bound (LB) of the 95% CI > 2 as ‘variants of intermediate penetrance’ (VIPs), variants with a 95% CI upper bound (UB) <2 as ‘benign’ and variants with 95% CI UB > 2 and LB < 2 as ‘inconclusive’ because they are still probably too rare to have sufficient power to identify an association if present (Figs. 1d and 3a).
a, Overview of the variant classification strategy according to the meta-analyses results in UKB/GERA/MGBB (excluding AoU). We extracted variants in monogenic diabetes genes from ClinVar labeled as ‘uncertain significance’, ‘conflicting interpretations of pathogenicity’, ‘likely benign’, ‘likely pathogenic’ and ‘pathogenic’. We then classified these variants based on the UKB/GERA/MGBB meta-analysis OR and 95% CI LB and UB. Variants with a meta-analytic OR > 5 and an OR 95% CI LB > 2 are classified as VIP. Variants with an OR 95% UB < 2 are classified as supports benign. Variants with an OR 95% CI UB > 2 and LB < 2 are classified as inconclusive. b, Results of this analysis for the variants of CIP and uncertain significance according to ClinVar. Only variants with MAF < 0.001 were considered for this analysis. The x axis represents the MAF. Along the y axis, the OR for each variant is denoted by the points and the 95% CI by the vertical lines. The P values are from the meta-analysis of two-sided logistic regression models, weighting the cohorts by the inverse of the s.e. for each variant. c, Variants aggregated in a single burden variable according to the ClinVar- and GWAS-based classifications to test for their cumulative effects on T2D in the full AoU cohort (n T2D cases = 26,271, n controls = 43,174). The forest plots represent each combination of ClinVar groups and GWAS-based classifications. The OR for each burden test is denoted by the points and the 95% CI by the horizontal lines. The P values are from two-sided logistic regression models.
Among the 381 variants within the ClinVar category of CIP (64.3% with MAF < 0.0001), we identified 5 VIPs with OR > 5 and 95% CI LB > 2 (Fig. 3b and Extended Data Fig. 8a–d). Including the HNF4A p.Arg114Trp described above, we replicated three of the five CIP VIPs in the full AoU and GEISINGER cohorts (Fig. 4a–c and Supplementary Table 9). They are in the known MODY genes HNF1A (12:120997588:C:T, p.Pro475Leu, enriched in Ashkenazi Jewish individuals, maxMAFAJ = 0.00086; discovery: OR = 6.9, 95% CI = 3.2–14.8, P = 8.8 × 10−7; discovery + replication: OR = 5.4, 95% CI = 2.9–10.2, P = 1.8 × 10−7) and GCK (7:44145170:A:T, p.Val455Glu, maxMAFEUR = 0.00002; discovery: OR = 8.7, 95% CI = 3.2–23.6, P = 2 × 10−5; discovery + replication: OR = 7.9, 95% CI = 3.5–18.3, P = 9.4 × 10−7). The p.Val455Glu is also associated with glucose (β = 0.781 mmol l−1, P = 8.1 × 10−5) and glycated hemoglobin (Hb1Ac; β = 5.11 mmol l−1, P = 7.4 × 10−7) in the UKB cohort (Extended Data Fig. 7n,o). Although not reaching statistical significance after correcting for multiple comparisons (P < 3.1 × 10−5 from 0.05 of 1,634 tested ClinVar variants), we also identified two VIPs in POLD1 (19:50413456:G:A, p.Glu475Lys, maxMAFFIN = 0.00019, OR = 13.3, 95% CI = 2.2–81.7, P = 5 × 10−3; 19:50402602:A:G, c.841-10A>G, maxMAFEUR = 0.00008, OR = 7.1, 95% CI = 2.4–21.1, P = 4 × 10−4). Mutations in POLD1 have been observed in a multisystem disorder with lipodystrophy causing diabetes34. Of the 381 CIP variants, 109 (28.6%) showed ORs with a 95% CI UB < 2, suggesting that these variants could be considered as benign despite being currently in the category CIP in ClinVar. Of these 109 variants, 88.1% had >80% power to detect T2D association with OR > 5 (Supplementary Table 8).
a–c, Forest plots showing the carrier counts and ORs of p.Arg114Trp (a), p.Pro475Leu (b) and p.Val455Glu (c) in the discovery, replication and overall datasets. The ORs of each cohort from the discovery and replication datasets are denoted by boxes and the 95% CIs by horizontal lines. Arrows were added for 95% CI LB < 0.5 and 95% CI UB > 4. The center of the diamonds represents the OR of the meta-analysis, with the horizontal extremities indicating the 95% CI. The P values are from the meta-analysis of two-sided logistic regression models, weighting the cohorts by the inverse of the s.e. for each variant. After correcting for multiple comparisons, the three variants showed significance (P < 3.1 × 10−5 from 0.05 of 1,634 tested ClinVar variants).
Within the 533 variants in the ClinVar category of uncertain significance (75.6% with MAF < 0.0001), we identified 3 additional VIPs (Fig. 3b and Extended Data Fig. 8e–g) in WFS1 (4:6302287:G:A, p.Gly831Asp, maxMAFEUR = 0.00002, OR = 15.3, 95% CI = 2.7–85.8, P = 2 × 10−3), KCNJ11 (11:17388128:G:A, 3′-UTR, maxMAFAFA = 0.00002, OR = 17.7, 95% CI = 2.7–113.5, P = 2 × 10−3), observed in only one carrier in gnomAD in the AFA subgroup, and POLD1 (19:50402228:G:A, Gly205Ser, maxMAFEUR = 0.00001, OR = 20.3, 95% CI = 4.0–102.4, P = 2 × 10−4). None of these VIPs reached statistical significance after multiple comparisons and we were unable to replicate them because they were absent or showed a lack of statistical power to detect an association (power < 67% for an OR = 5) in the replication cohorts (Supplementary Table 8). In addition, 79 (14.8%) of the 533 VUSs had association results supporting benign, based on the lack of association with T2D. Of these 79 variants, 97.4% had >80% power to detect an association with OR > 5 (Supplementary Table 8).
We identified one additional VIP (POLD1 19:50409504:C:T, c.2007-15C>T, maxMAFEUR = 0.00002, OR = 9.9, 95% CI = 2.1–47.8, P = 4 × 10−3) in the 546 variants classified as likely benign according to ClinVar groups (Fig. 1d, Extended Data Fig. 9a and Supplementary Table 8). Based on the GWAS meta-analysis results, 216 out of the total 546 likely benign ClinVar variants (39.6%) supported a benign category and 96.8% had >80% power to detect an association with OR > 5 (Supplementary Table 8). As expected, within the ClinVar benign category, 73.9% of the variants are supported as benign and none supported pathogenicity according to our association data (Extended Data Fig. 9b). There were no variants with association results supporting pathogenicity in the likely pathogenic and pathogenic categories (Extended Data Fig. 9c,d), probably because such variants have much lower allele frequency and there is still not enough power to detect association in our dataset. However, our results supported evidence of being benign for two variants categorized as pathogenic in ClinVar, showing no association with T2D and having >80% power to detect an association with OR > 5 (Supplementary Table 8).
To further validate the effects of the VIPs, we tested the association of carriers of VIPs in aggregate with T2D in the AoU cohort using a burden test approach. We compared the association results using the whole set of ClinVar variants and stratified by the ClinVar categories (Figs. 1d and 3c). Except for the VUS POLD1 p.Gly205Ser, for which no carriers were present in the AoU cohort, carriers of the remaining eight VIPs exhibited a 3.4-fold increased risk for T2D (OR = 3.4, 95% CI = 1.82–6.40, P = 9.5 × 10−5), in contrast to variants identified as supporting benign (OR = 1.0, 95% CI = 0.95–1.06, P = 0.911) and inconclusive variants (OR = 1.06, 95% CI = 1.01–1.11, P = 0.02), which, in aggregate, showed no significant effect on T2D risk (Fig. 3c).
When aggregating the variants according to the ClinVar criteria, only the VUS (OR = 1.1, 95% CI = 1.04–1.19, P = 0.002) and the likely pathogenic and pathogenic groups (OR = 2.15, 95% CI = 1.23–3.80, P = 0.007) showed an association with T2D. However, VIPs showed consistent direction when stratifying by ClinVar categories, with VIP in CIP ClinVar category showing significantly increased risk (OR = 3.59, 95% CI = 1.78–7.23, P = 2.7 × 10−4) and likely benign VIP (OR = 2.01, 95% CI = 0.44–9.23, P = 0.369) and VUS-VIP (OR = 5.39, 95% CI = 0.43–68.10, P = 0.193) groups not reaching statistical significance (Fig. 3c).
Common variant PRSs modulate the effect of rare variants in the monogenic diabetes genes
To understand potential causes for the conflicting interpretation of pathogenicity of the identified and replicated VIPs in the three established MODY genes (that is, HNF4A, HNF1A and GCK), we tested how a PRS for T2D influences diabetes risk pathogenicity and compared the effects with those of established confirmed MODY variants identified using WES data from the UKB35. For the HNF4A p.Arg114Trp variant, when compared with noncarriers in the middle tertile of the PRS, carriers in the highest tertile had a higher OR (=18.3, 95% CI = 7.2–46.9, P = 1.2 × 10−9), with an effect size that was comparable to the observed in carriers of confirmed pathogenic MODY variants (OR = 17.7, 95% CI = 6.5–48.1, P = 7.8 × 10−6). However, HNF4A p.Arg114Trp carriers within the lowest tertile of the PRS showed a much smaller OR (=2.62, 95% CI = 0.97–7.09, P = 0.06) (Fig. 5a).
a–c, Forest plots showing the effect of p.Arg114Trp, p.Pro475Leu and p.Val455Glu, stratified by PRS tertiles. The ORs are denoted by boxes and the 95% CIs by horizontal lines. The P values are from the meta-analysis of two-sided logistic regression models, weighting the cohorts by the inverse of the s.e. for each variant. The ORs are relative to the noncarriers in the middle tertile of the PRS. On the top of each Forest plot, the effects of being a carrier for a confirmed pathogenic variant for HNF4A (a), HNF1A (b) and GCK (c) MODY genes are also represented, using data identified via exome sequencing in UKB34. For each effect estimate, the diabetes case definition included individuals with T1D or T2D. d–f, Boxplots of HbA1c (%), random glucose (mg dl−1) and BMI (kg m−2) in cases with diabetes and noncases among noncarriers (NCs, left), carriers of variants with intermediate penetrance (middle) and carriers of confirmed pathogenic MODY variants (right) in HNF4A (d), HNF1A (e) and GCK (f). The covariate-adjusted P value is included for comparisons with significant differences (*P < 0.05, **P < 0.001, ***P < 0.0001) between groups using two-sided Wilcoxon’s rank-sum tests. Boxplots indicate the group median (central line), first and third quartiles (bounds of box) and 1.5× interquartile range (whiskers).
Similar to what we observed for the HNF4A variant, carriers of either p.Pro475Leu in HNF1A or p.Val455Glu in GCK who also were in the top tertile of the PRS had markedly increased risks of diabetes (OR > 10). However, carriers in the top tertile had ORs smaller than those observed with confirmed HNF1A or GCK MODY variants (Fig. 5b,c).
VIP carriers show phenotypic characteristics intermediate between T2D and MODY
We then tested whether carriers of VIPs in established MODY genes showed a different phenotypic pattern compared with T2D and MODY. For the three variants, the effect on HbA1c and random glucose was in between those of noncarriers and carriers of confirmed MODY variants, supporting that these may be variants with intermediate penetrance. Carriers of HNF4A p.Arg114Trp and HNF1A p.Pro475Leu with diabetes had significantly lower body mass index (BMI) than the noncarriers (Fig. 5d,e). Carriers of the HNF4A p.Arg114Trp variant had a significantly lower age at onset of diabetes compared with noncarriers (Extended Data Fig. 10a). These results support these variants being in a continuum spectrum of pathogenicity in monogenic diabetes.
Discussion
Genetic studies for T2D have been usually categorized as rare variant association studies (RVASs) or common variant association studies (CVASs)36, with RVASs typically performed via WES or WGS and CVASs typically done via GWAS meta-analyses because the genotyping array data and imputation panels used cover only relatively common frequencies. In the present study, we performed the largest T2D GWAS meta-analysis, including variants with allele frequencies <0.001, by combining TOPMed imputation from three large and diverse cohorts and WGS data from the AoU cohort. We demonstrated that TOPMed imputation provides accurate genotypes for variants with MAF as low as 5 × 10−5 and is a reliable strategy to analyze low-frequency and rare genetic variation in cohorts for which WGS data are unavailable. Despite new WGS data at scale becoming available, there are still substantial extant data that can benefit from imputation with large reference panels such as the TOPMed panel, especially from non-European ancestries.
The present study has enabled several discoveries. First, we identified and replicated 12 new T2D-associated variants, of which 4 were low frequency (MAF < 0.05) and 1 was rare and population specific (rs147287548) near LEP, which codes for leptin, a hormone/adipokine primarily secreted by white adipose tissue and the deficiency of which causes insulin resistance37. The rs147287548 overlaps with an enhancer active in adipocyte-derived mesenchymal stem cells and adipose tissue that interacts with a region proximal to the LEP transcription start site by enhancer-capture HiC30 and is predicted to disrupt an NFATc motif. Accordingly, we demonstrate that the allele predicted to disrupt NFATc binding associates with reduced enhancer activity in adipocytes. NFATc transcription factors have been implicated in adipogenesis, are modulated in obesity and regulate glucose and insulin homeostasis31. Homozygous mutations in LEP are additionally known to cause extreme obesity in humans and mice38.
The present study also represented a proof of concept of how use of a large-scale biobank or population-based data can provide additional evidence for pathogenicity of variants within genes known to cause monogenic diabetes. Monogenic diabetes exemplifies precision medicine39, because identifying the gene causing diabetes guides the management of the disease. For example, individuals with MODY caused by HNF1A mutations can often be transitioned from insulin therapy to oral hypoglycemic agents such as sulfonylureas40 and those with MODY caused by GCK mutations develop stable mild hyperglycemia and do not need pharmacological therapy because they are often refractory to it41, so their risk of microvascular and macrovascular complications is considered low42. The current assessment of the pathogenicity of genetic variants relies on bioinformatic prediction, population allele frequency and expert panel curation. The presence of carriers among patients with diabetes or segregation analyses is also considered, but these data are usually unavailable. For this reason, many of the missense variants identified in these genes remain as VUSs or conflicting interpretations. By testing rare variant associations at scale using TOPMed imputation, we identified variants in known monogenic diabetes genes with intermediate pathogenicity, which we defined as having an OR > 5 and a 95% CI LB > 2. Additional evidence may be needed to confirm the pathogenicity and clinical utility of such variants, such as testing whether carriers of HNF1A or HNF4A variants of intermediate penetrance have heightened responses to sulfonylureas. Nevertheless, we propose that our approach could serve as a tool to prioritize variants for future functional validation or deep phenotypic characterization.
Some of the variants that we identified as having intermediate penetrance have had evidence published of functional consequences. For example, GCK p.Val455Glu is in the same position as a GCK-activating mutation (p.Val455Met) causing familial hyperinsulinism43. It has been reported previously in individuals with impaired glucose tolerance or fasting hyperglycemia44 and two independent studies have demonstrated that the variant is kinetically inactivating, consistent with loss of function45,46. Identifying carriers of GCK-inactivating mutations is important, not only because these carriers may not need treatment but also because of possible complications in pregnancy if the risk allele is not inherited by the fetus of the carrier mother, resulting in hyperglycemia and excessive growth in the fetus47.
We also showed that, in aggregate, variants of intermediate penetrance confer a 3.4-fold increased risk of T2D in an independent WGS-based cohort, demonstrating that our approach provides robust identification of rare variants with a large effect and that rare variants contribute to the overall burden of T2D.
Although we were able to identify only a handful of variants that support intermediate penetrance, we also found a much larger fraction of variants that are sufficiently powered and which, based on their lack of association with diabetes, support being benign. These results can de-prioritize candidate variants and genes in patients suspected of having monogenic diabetes. An illustrative example is a variant in HNF1A, currently classified as VUS (12-120994274-A-C, p.Glu275Ala, MAF = 0.00002, OR = 0.65, 95% CI = 0.23–1.86, P = 0.42). This variant is rare and predicted to be deleterious by several bioinformatic tools and therefore remains as a VUS in ClinVar. However, knowing that carriers of this variant are not at increased risk for T2D can add additional evidence for this variant being benign.
Our study design also allowed us to assess the interplay between common and rare variants in monogenic diabetes genes. We observed that stratifying carriers of such variants by their PRS identifies carriers that can have the same magnitude of increased risk of developing diabetes as the carriers of well-established MODY variants. For example, our results showed that a third of the carriers of the HNF4A p.Arg114Trp variant, particularly those in the highest tertile of the PRS, had the same risk as those carrying confirmed MODY variants. This is in line with previous studies that have shown how a polygenic background can influence the penetrance of pathogenic mutations related to obesity, cancer susceptibility, lipid disorders and coronary artery disease8,48,49,50,51. Future work should leverage these data to develop PRSs with the inclusion of rare variants. However, new methods that allow modeling linkage disequilibrium (LD) between rare variants and common variants are needed to take full advantage of both common and rare genetic variation.
Previous studies have proposed using endophenotype biomarker association data, such as HbA1c, to support pathogenicity52. However, association with common forms of diabetes (that is, T2D) has not been previously used to assess pathogenicity owing to the lack of power of WGS- or WES-based studies.
Our study has several limitations. First, we acknowledge that the standard genome-wide significant threshold (P < 5 × 10−8), developed initially as a genome-wide significant threshold for common variants, may not be sufficiently stringent because many more variants, including those that are rare and population specific, are being tested. However, we provide extensive replication analyses and only claim as new those that replicate. In addition, although we have used a broad definition of T2D as a surrogate phenotype to assess the potential pathogenicity of monogenic diabetes genes, we cannot discard the possibility that some of the variants have a phenotype that is closer to prediabetes or T1D rather than T2D. Although power is limited to testing the association of such rare variants with T1D, our definition of controls excluded those participants who met the criteria for diagnosing T1D or prediabetes. This could only bias the association results toward the null and not increase our false-positive rate.
The implementation of these data as an additional metric to assess variant pathogenicity will require additional investigation involving expertise from clinicians, genetic counselors and the ClinGen Monogenic Diabetes Expert Panel. Nevertheless, we make the full summary statistics available to the scientific community through the Type 2 Diabetes Knowledge Portal as a resource that can be used for further investigations53.
In summary, our work underscores the value of combining WGS data with low-frequency and rare genotype imputation in cohorts for which WGS or WES data still do not exist, not only to discover new associations but also to guide the interpretation of variant pathogenicity. Expanding this work to larger and more diverse populations will contribute to reducing health disparities in the application of precision medicine in diabetes. The framework provided in the present study will serve as an example for the study of genetic variation associated with common and monogenic forms of disease.
Methods
Description of discovery cohorts and case–control definitions
The discovery association analyses were conducted using genetic data from the UKB, the MGBB, the GERA and the AoU. Overall, the discovery sample included 51,256 cases of T2D and 370,487 controls (Supplementary Table 1).
The UKB is a multi-ancestry prospective cohort study with genetic and phenotypic data collected on approximately 500,000 individuals across the United Kingdom22. Participants agreed to provide detailed information about their lifestyle, environment and medical history and biological samples (for genotyping and biochemical assays), undergo measures and have their health followed (http://www.ukbiobank.ac.uk). The UKB has obtained ethical approval covering the present study from the National Research Ethics Committee (REC ref. no. 11/NW/0382) and the data were accessed through application no. 27892. For this analysis, we included a total of 27,323 cases of T2D and 259,916 controls, and defined cases of T2D and controls using an algorithm designed specifically for the UKB54.
The MGBB (formerly Partners HealthCare Biobank)23 is a large repository of biospecimens and data linked to extensive electronic health records and survey data. Its objective is to support and enable translational research on genomic, environmental, biomarker and family history associations with disease phenotypes. MGBB has enrolled more than 135,000 participants and has generated genomic data on >65,000 participants of multiple ancestries. MGBB consists of consented patients seen at various US hospitals. Patients are recruited in the context of clinical care appointments at >40 sites. MGBB participants provide consent for the use of their samples and data in broad-based research. The approval for the analysis of MGBB data was obtained from the MGB Institutional Review Board (IRB; study no. 2016P001018). We included a total of 6,623 cases of T2D and 41,411 controls. T2D status was defined based on ‘curated phenotypes’ developed by the MGBB Portal team using structured and unstructured electronic medical record data and clinical, computational and statistical methods55.
The GERA cohort was created by an RC2 Grand Opportunity grant that was awarded to the Kaiser Permanente Research Program on Genes, Environment, and Health (RPGEH) and the Univerity of California, San Francisco (UCSF) Institute for Human Genetics (AG036607; Schaefer/Risch, PIs). The RC2 project enabled GWASs to be conducted on a cohort of >100,000 adults who were members of the Kaiser Permanente Medical Care Plan, Northern California Region and participating in its RPGEH. The GERA cohort data was obtained through dbGaP under accession no. phs000674.v1.p1. Further information about the specific phenotypes (International Classification of Diseases, 9th revision, Clinical Modification codes; https://www.cdc.gov/nchs/icd/) included in GERA is available on its website at dbGaP (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/GetPdf.cgi?id=phd004308). We included a total of 7,498 cases of T2D and 53,212 controls of European ancestry.
The AoU is a large, US-funded biobank developed to leverage the diversity of the United States of America for facilitating and improving high-powered genetic and epidemiological studies25. We used the AoU short-read WGS data from 22 June 2022 for 98,590 participants (release v.6). Cases with T2D and controls were defined using a modified version of the Northwestern T2D algorithm56,57. In total, we identified 9,812 cases with T2D and 15,948 controls with complete genetic data and covariate data, which we used in our discovery analysis.
The Supplementary Note provides a detailed description of the cases with T2D and controls used for each cohort.
Imputation and accuracy assessment
We accessed genotyping data from the UKB, MGBB and GERA cohorts. In each, we applied a quality control, phased and imputed to the TOPMed reference panel freeze 8, separately. A full description of the procedures by cohort is in the Supplementary Note.
To assess the accuracy of the TOPMed imputation, we compared the UKB data imputed with the TOPMed reference panel with that of a previous imputation of UKB, which was based on the HRC, 1000G and UK10K Project. Specifically, we compared the average INFO score provided by REGENIE and the total number of variants in the TOPMed imputation versus the HRC–1000G–UK10K imputation across the allele frequency spectrum, before and after filtering by an INFO score of 0.7 (Extended Data Fig. 2). In addition, in a subset of 40,000 UKB samples with WES data, we evaluated the average percentage of carriers identified in the WES who are also identified with the TOPMed imputation across the allele frequency spectrum (Extended Data Fig. 3).
Association analysis
We performed whole-genome logistic regression analyses using REGENIE v.2 for UKB, GERA and MGBB and v.3 for AoU. We used T2D as a binary outcome and included age, sex, BMI, ten principal components (PCs) and the imputation batch for each cohort. We used a block size of 1,000 for step 1 and 400 for step 2. All variants with minor allele count <3 for UKB, GERA and MGBB and <5 for AoU among cases with T2D and controls were excluded from the analysis. To provide better-calibrated test statistics, REGENIE supports the option (--firth --approx --firth-se) to use the Firth correction for variants where the P value from the standard logistic regression test is below a threshold, as defined by pThresh. For UKB, GERA and MGBB, we set pThresh to 0.999, causing the Firth correction to be applied to all variants. For AoU, we used the default pThresh of 0.05. After generating summary statistics using REGENIE for each cohort, we used METAL v.2011-03-25 (ref. 58) to meta-analyze the results, weighting cohorts by the inverse of the s.e. for each variant. Our threshold for genome-wide significance was P < 5 × 10−8.
Post-association analysis
Identification of the independent set of T2D-associated markers
We used the conditional regression-GCTA (cojo-GCTA v.1.94.1)28 method on the GWAS meta-analysis summary data. This approach allowed us to assess each locus with a joint combination of several independent markers, corrected for LD between the markers. We constructed an external reference sample with individual genotypes from the UKB and MGBB cohorts to obtain an approximate LD structure. Our reference sample comprised 45,918 samples, ensuring that the proportions of genetic ancestry diversity matched those included in the meta-analysis. Before the analysis, we filtered the genotypes for imputation quality, retaining those with r2 ≥ 0.8, a minimum dosage certainty ≥0.8 and an MAF ≥ 0.0001%. After filtering, we kept a subset of 78,867,233 markers for performing the conditional analyses. We assumed that markers on different chromosomes or those located >500 kb distant from each other were uncorrelated.
Criteria for definition of new variants
To define a new variant, we followed a two-step procedure. We first looked for each independent signal in five different T2D GWAS meta-analyses: the MVP5, the DIAMANTE EUR59 and multi-ancestry3, the FinnGen r9 (ref. 29) and the T2DGGI2. Second, for those index variants that were absent or that did not have genome-wide significance (P < 5 × 10−8) in any of the above five T2D GWAS meta-analyses, we used our reference sample to extract all tag variants in LD r2 > 0.8 within 2 Mb of the index variant. We then performed lookups of all the tag variants in the T2D GWAS meta-analyses listed above. We defined a new variant when (1) neither the index nor their tag variants were genome-wide significant (P < 5 × 10−8) in any of the T2D GWAS meta-analyses listed above or (2) the index variant was not genome-wide significant in any of the T2D GWAS meta-analyses listed above, and no tag variant was identified.
Biomarker analysis in the UKB
To provide additional supporting evidence for the new variants identified in the T2D GWAS meta-analysis, we used REGENIE v.2 to test their association with 30 biomarkers in all individuals without a diagnosis of T2D in the UKB. Therefore, we included the 175,039 individuals who were identified as T2D controls, along with the 259,916 individuals who were not identified as either a case with T2D or a control. The 30 biomarkers included albumin, alkaline phosphatase, alanine aminotransferase, apolipoprotein A, apolipoprotein B, aspartate aminotransferase, direct bilirubin, urea, calcium, cholesterol, creatinine, C-reactive protein, cystatin C, γ-glutamyl transferase, glucose, HbA1c, HDL-cholesterol, insulin growth factor-1, LDL direct, lipoprotein A, estradiol, phosphate, rheumatoid factor, SHBG, total bilirubin, testosterone, total protein, triglycerides, urate and vitamin D. To ensure normality, we applied logarithmic normal transformation to each of the biomarkers and additional ranked inverse normal transformation to direct bilirubin, lipoprotein A and estradiol traits. Models were adjusted for age, sex, BMI, ten PCs and the imputation batch.
Description of replication cohorts
We first tested the 34 new variants for replication in two previous T2D GWAS meta-analyses, including independent samples: the MVP5 and the FinnGen r9 (ref. 29) (Supplementary Table 4). As rare variants are not considered in previous meta-analyses, we tested the eight new rare or population-specific variants for replication in three independent datasets not included in our discovery meta-analysis: the Geisinger MyCode Community Health Initiative (GEISINGER), as well as the discovery nonoverlapping individuals from the GERA and the AoU cohorts. Overall, the replication sample for the rare variants included 73,088 cases with T2D and 79,827 controls (Supplementary Table 1).
The GEISINGER is a biorepository of blood, serum and DNA samples that are linked to electronic health records for the purpose of broad research use. Participants were recruited from Geisinger, an integrated healthcare system located in central and north-eastern Pennsylvania. The study sample consisted of 172,366 individuals with exome sequence and microarray genotype data. Available clinical data include clinical diagnoses, procedures, medications and laboratory results, which are updated daily35. Participation in MyCode is done through written consent under Geisinger IRB study no. 2016-0269. The results reported here were determined by the Geisinger IRB to meet the criteria for ‘nonhuman subject research’ as defined in 45CFR46.102(e). All research was performed in accordance with relevant guidelines and regulations. We included 52,658 cases with T2D and 41,639 controls (Supplementary Note).
The GERA replication cohort (GERA_REP) included individuals from African American, admixed American and east Asian ancestry24 who were not previously included in the discovery GWAS. In total, GERA_REP included 2,737 cases with T2D and 9,270 controls.
The AoU replication cohort (AoU_REP) included nonoverlapping individuals and nonfamily members (pairwise kinship score > 0.1) to those included in the discovery meta-analysis because they were released in the posterior v.7 data freeze. In total, we included 17,693 independent cases with T2D and 28,918 controls.
The genotypes for the three cohorts were quality controlled, imputed and analyzed to test the association with T2D. After generating the T2D summary statistics for each replication cohort, we used METAL to meta-analyze the results, weighting the cohorts by the inverse of the s.e. for each variant. We considered replication when the variant showed evidence of association with T2D at P < 0.0015 (0.05 of 34 total new variants) and consistent direction of effect with the T2D discovery GWAS meta-analysis.
Analysis of variants in monogenic diabetes genes
Although >40 genes are known to cause monogenic diabetes, distinguishing pathogenic variants from those that are benign in these genes remains a challenge. Databases such as ClinVar32 provide designations of variant pathogenicity as determined by submitting laboratories and researchers. However, misclassification of variant pathogenicity is not uncommon, for example, in entries that predated the current gold-standard curation approach, as well as for variants that are enriched in understudied populations. The ClinVar designations of variant pathogenicity include benign, likely benign, pathogenic, likely pathogenic, uncertain significance and CIP. We evaluated the T2D association results in our meta-analysis for variants reported in ClinVar (accessed July 2023) that are in 22 known monogenic diabetes genes and have an MAF < 0.001, with a particular interest in VUSs and CIP. We focused on monogenic diabetes-related genes with a nonautoimmune autosomal dominant mechanism for causing diabetes (Supplementary Table 8).
We classified these variants based on the effect observed in the meta-analysis of UKB, GERA and MGBB, with AoU held out to serve as a validation cohort. We classified the variants based on the meta-analytic OR and 95% CI LB and UB (Fig. 3). Variants with an OR > 5 and 95% CI LB > 2 were classified as VIP, variants with a 95% CI UB < 2 were classified as supports benign and variants with a 95% CI UB > 2 and 95% CI LB < 2 were classified as inconclusive.
Testing a rare variant burden in an independent sample in AoU
We aggregated the variants from each of the three classes described above (VIP, supports benign and inconclusive) as either a whole or stratified by ClinVar groups into single burden variables, and then regressed the T2D phenotype in the burden variable to test for the cumulative effects of all possible combinations of ClinVar and GWAS-based classified variants, using the AoU cohort as a validation dataset. We ran burden tests using T2D as a binary outcome and included age, sex, BMI and ten PCs as covariates with REGENIE v.2. We used a block size of 1,000 for step 1 and 400 for step 2.
PRSs to stratify carriers of monogenic diabetes variants
We generated a T2D PRS using the PRS-CS software60 with the auto option to set the global shrinkage parameter. As input to the software, we meta-analyzed summary statistics from the European ancestry subset GWAS meta-analysis of the T2D published by Vujkovic et al.5 and the T2D GWAS made publicly available by the FinnGen Consortium29 (release 6), which did not overlap with any of the samples included in our meta-analysis. The summary statistics had a final sample size of 277,802 cases with T2D and 1,434,249 controls. The meta-analysis was performed using METAL, with inverse-variance weighting. As an LD reference panel for PRS under continuous shrinkage (PRS-CS), we used the LD panel of European ancestry from the United Kingdom, available from the PRS-CS GitHub webpage (https://github.com/getian107/PRScs). After applying the PRS, we tested the PRS-stratified effect of three rare coding variants in well-established MODY genes—20:44413714:C:T (HNF4A), 12:120997588:C:T (HNF1A) and 7:44145170:A:T (GCK), (Fig. 4)—which are considered CIP from the ClinVar designation, but were classified as VIP from the analysis of variants in monogenic diabetes genes described above. Individuals were stratified based on PRS tertiles, with individuals in the top tertile considered to have a high PRS and individuals in the bottom tertile to have a low PRS. For each effect estimate, the diabetes case definition included individuals with T1D or T2D and controls were defined as described above. Individual noncases who do not carry any of the three variants and have a PRS in the middle tertile were treated as the reference group. This analysis was done separately in the UKB, MGBB and GERA cohorts and meta-analyzed with inverse-variance weighting. We also compared the PRS-stratified effects of the three variants with the effect of being a carrier for any confirmed pathogenic variant for HNF4A, HNF1A and GCK MODY35 using data from UKB exome sequencing. We also compared the levels of HbA1c, random glucose, BMI and age at diabetes onset in cases with diabetes and noncases (those with no diagnosis of any type of diabetes) who carry either the VIPs or confirmed MODY variants in HNF4A, HNF1A and GCK versus noncarriers.
Functional analysis of noncoding variants
In silico interpretation of new noncoding variants
Lead variants from the GWASs were investigated in combination with all other variants in high LD (r2 > 0.8 in all populations). This expanded set of variants was intersected with ENCODE’s collection of candidate cis regulatory elements (cCREs) (GRCh38, SCREEN Registry v.3), which represented 1,063,878 cCREs across 1,518 cell types. To ascertain tissue and/or cell type-specific cCREs hosting T2D variants, we used SCREEN (https://screen.wenglab.org) and the Roadmap Epigenomics 127-reference chromatin states (12-mark, 25-state imputation-based, lift-over to GRCh38), which were visualized using the WashU Epigenome Browser61. We also interrogated human pancreatic islet epigenomic datasets from Miguel-Escalada et al.62, which were realigned to hg38 assembly for this analysis, and epigenomic datasets from human mesenchymal stem cells undergoing adipocyte differentiation from Madsen et al.30 and Rauch et al.63, which had coordinates lifted from hg19 to hg38 for the main figure (Fig. 2b) and are shown for hg19 in Extended Data Fig. 6. A list of the representative epigenomic datasets from diabetes-relevant tissues used to generate Fig. 2b and Extended Data Fig. 6 using the UCSC Genome Browser64 is provided in Supplementary Table 6.
The R package motifbreakR (v.2.13.7)65 was used to carry out motif disruption analysis of variants residing in CREs of diabetes-relevant tissues. The variant data were downloaded from the dbSNP155 database (v.GRCh38_0.99.23) in the human genome (v.hg38_1.4.5) using the snps.from.rsid() command. Transcription factor-binding motif data (ENCODE, HOMER, Hocomoco and FactorBook) were downloaded from the MotifDB database (v.1.42.0)66. The default, weighted-sum method was used for disruption analysis where the difference of the probabilities for the two bases of the variant was calculated. The results were queried for the disruptive effect of the variant with P value maximum 0.001 and plotted in R (v.4.3.1). Disrupted motifs with the flag for strong effect and corresponding to transcription factors expressed in the tissues of interest (gene expression queried using the TIGER Data Portal, https://tiger.bsc.es) were retained.
Cell culture and in vitro differentiation of adipocytes
The 3T3-L1 mouse fibroblasts were obtained from the European Collection of Authenticated Cell Cultures. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, cat. no. D6326) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, cat. no. 10500-064), 2 mM glutamine, 100 U ml−1 penicillin and 100 µg ml−1 streptomycin (Sigma-Aldrich, cat. nos. G7513 and P4333). The cells were differentiated to adipocytes by first growing to confluence for 48 h before commencing the differentiation protocol. On differentiation day 0, cells were treated with basal medium supplemented with 0.5 mM 3-isobutyl-1-methylxanthine (Sigma-Aldrich, cat. no. I5879), 1 µM dexamethasone (Sigma-Aldrich, cat. no. D4902) and 1 µg ml−1 of insulin (Sigma-Aldrich, cat. no. I9278). After 48 h, medium was replaced by medium supplemented with 1 µg ml−1 of insulin only for the next 4 d, refreshing the medium every 48 h. Mature adipocytes were assessed for accumulation of lipid droplets by visual inspection using a brightfield microscope on day 8 and experiments were performed on day 10.
Luciferase reporter assays
The LEP enhancer region containing the variant 7:128323039-G-A was amplified from human genomic DNA. The PCR amplicon was then cloned into the pGL4.23(luc2/minP) vector (Promega, cat. no. E8411) at KpnI and HindIII, upstream of a minimal promoter and the Firefly luciferase coding sequence, using Gibson Assembly Master Mix67 (New England Biolabs, cat. no. E2611L). The cloned enhancer sequence contained the nonrisk allele (7:128323039-G); therefore, the enhancer sequence containing the risk allele (7:128323039-A) was produced via site-directed mutagenesis, which was performed using a Q5 Site-Directed Mutagenesis Kit (New England Biolabs, cat. no. E0554). Correct cloning was confirmed by Sanger sequencing (Genewiz). Plasmids were amplified in DH5α cells and purified using PureYield Plasmid Midiprep System (Promega, cat. no. A2496). All primers are listed in Supplementary Table 10. To test constructs, 40,000 3T3-L1-derived adipocytes were reverse transfected in 24-well plates with 1 µg of tested construct and 0.1 µg of the internal control, Renilla-expressing pGL7.74 vector (Promega, cat. no. E6921), using 1 µl of Lipofectamine 2000 (Thermo Fisher Scientific, cat. no. 11668019) per 1 µg of construct. Luciferase activity was measured 24 h post-transfection with Dual-Luciferase Reporter Assay System (Promega, cat. no. E1980) on a GloMax-Multi Microplate Multimode Reader (Promega, cat. no. GM3510). The vectors pGL4.23 and pGL4.13 (Promega, cat. nos. E8411 and E6681) were used as internal negative and positive controls, respectively. Firefly luciferase measurements were normalized to Renilla luciferase. Three experiments with four independent transfections were performed per construct. A two-sided Student’s t-test was used to calculate significance.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Full summary statistics of the T2D case–control meta-analysis are available through the Common Metabolic Diseases Knowledge Portal (https://t2d.hugeamp.org/downloads.html) and through the GWAS catalog (https://www.ebi.ac.uk/gwas/studies/GCST90444202, accession no. GCST90444202).
Change history
22 October 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41588-024-01986-2
25 March 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41588-025-02175-5
References
Huerta-Chagoya, A. et al. The power of TOPMed imputation for the discovery of Latino-enriched rare variants associated with type 2 diabetes. Diabetologia 66, 1273–1288 (2023).
Suzuki, K. et al. Genetic drivers of heterogeneity in type 2 diabetes pathophysiology. Nature 627, 347–357 (2024).
Mahajan, A. et al. Multi-ancestry genetic study of type 2 diabetes highlights the power of diverse populations for discovery and translation. Nat. Genet. 54, 560–572 (2022).
Spracklen, C. N. et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature 582, 240–245 (2020).
Vujkovic, M. et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 5, 680–691 (2020).
Sengupta, D. et al. Performance and accuracy evaluation of reference panels for genotype imputation in sub-Saharan African populations. Cell Genom. 3, 100332 (2023).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature 590, 290–299 (2021).
Goodrich, J. K. et al. Determinants of penetrance and variable expressivity in monogenic metabolic conditions across 77,184 exomes. Nat. Commun. 12, 3505 (2021).
Zhang, H., Colclough, K., Gloyn, A. L. & Pollin, T. I. Monogenic diabetes: a gateway to precision medicine in diabetes. J. Clin. Invest. 131, e142244 (2021).
Shields, B. M. et al. Population-based assessment of a biomarker-based screening pathway to aid diagnosis of monogenic diabetes in young-onset patients. Diabetes Care 40, 1017–1025 (2017).
Murphy, R., Ellard, S. & Hattersley, A. T. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat. Clin. Pr. Endocrinol. Metab. 4, 200–213 (2008).
Riddle, M. C. et al. Monogenic diabetes: from genetic insights to population-based precision in care. Reflections from a Diabetes Care editors’ expert forum. Diabetes Care 43, 3117–3128 (2020).
Mifsud, F. et al. Monogenic diabetes in adults: a multi-ancestry study reveals strong disparities in diagnosis rates and clinical presentation. Diabetes Res. Clin. Pr. 188, 109908 (2022).
Lango Allen, H. et al. Polygenic risk variants for type 2 diabetes susceptibility modify age at diagnosis in monogenic HNF1A diabetes. Diabetes 59, 266–271 (2010).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Karczewski, K. J. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020).
Lumaka, A. et al. Increasing African genomic data generation and sharing to resolve rare and undiagnosed diseases in Africa: a call-to-action by the H3Africa rare diseases working group. Orphanet J. Rare Dis. 17, 230 (2022).
Wainschtein, P. et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat. Genet. 54, 263–273 (2022).
Sigma Type 2 Diabetes Consortium et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 311, 2305–2314 (2014).
Flannick, J. et al. Exome sequencing of 20,791 cases of type 2 diabetes and 24,440 controls. Nature 570, 71–76 (2019).
Flannick, J., Johansson, S. & Njolstad, P. R. Common and rare forms of diabetes mellitus: towards a continuum of diabetes subtypes. Nat. Rev. Endocrinol. 12, 394–406 (2016).
Sudlow, C. et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015).
Karlson, E. W., Boutin, N. T., Hoffnagle, A. G. & Allen, N. L. Building the Partners HealthCare Biobank at Partners personalized medicine: informed consent, return of research results, recruitment lessons and operational considerations. J. Pers. Med. 6, 2 (2016).
Banda, Y. et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. Genetics 200, 1285–1295 (2015).
All of Us Research Program Investigators et al. The ‘All of Us’ research program. N. Engl. J. Med. 381, 668–676 (2019).
McCarthy, S. et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283 (2016).
1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Madsen, J. G. S. et al. Highly interconnected enhancer communities control lineage-determining genes in human mesenchymal stem cells. Nat. Genet. 52, 1227–1238 (2020).
Yang, T. T. et al. Role of transcription factor NFAT in glucose and insulin homeostasis. Mol. Cell. Biol. 26, 7372–7387 (2006).
Landrum, M. J. et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 46, D1062–D1067 (2018).
Laver, T. W. et al. The common p.R114W HNF4A mutation causes a distinct clinical subtype of monogenic diabetes. Diabetes 65, 3212–3217 (2016).
Weedon, M. N. et al. An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat. Genet. 45, 947–950 (2013).
Mirshahi, U. L. et al. Reduced penetrance of MODY-associated HNF1A/HNF4A variants but not GCK variants in clinically unselected cohorts. Am. J. Hum. Genet. 109, 2018–2028 (2022).
Zuk, O. et al. Searching for missing heritability: designing rare variant association studies. Proc. Natl Acad. Sci. USA 111, E455–E464 (2014).
German, J. P. et al. Leptin deficiency causes insulin resistance induced by uncontrolled diabetes. Diabetes 59, 1626–1634 (2010).
Zhang, Y. et al. Positional cloning of the mouse obese gene and its human homologue. Nature 372, 425–432 (1994).
Florez, J. C. & Pearson, E. R. A roadmap to achieve pharmacological precision medicine in diabetes. Diabetologia 65, 1830–1838 (2022).
Pearson, E. R. et al. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet 362, 1275–1281 (2003).
Stride, A. et al. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia 57, 54–56 (2014).
Steele, A. M. et al. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA 311, 279–286 (2014).
Glaser, B. et al. Familial hyperinsulinism caused by an activating glucokinase mutation. N. Engl. J. Med. 338, 226–230 (1998).
Gloyn, A. L. et al. Prevalence of GCK mutations in individuals screened for fasting hyperglycaemia. Diabetologia 52, 172–174 (2009).
Gloyn, A. L., Tribble, N. D., van de Bunt, M., Barrett, A. & Johnson, P. R. Glucokinase (GCK) and other susceptibility genes for beta-cell dysfunction: the candidate approach. Biochem. Soc. Trans. 36, 306–311 (2008).
Langer, S., Waterstradt, R., Hillebrand, G., Santer, R. & Baltrusch, S. The novel GCK variant p.Val455Leu associated with hyperinsulinism is susceptible to allosteric activation and is conducive to weight gain and the development of diabetes. Diabetologia 64, 2687–2700 (2021).
Dickens, L. T. et al. Management and pregnancy outcomes of women with GCK-MODY enrolled in the US Monogenic Diabetes Registry. Acta Diabetol. 56, 405–411 (2019).
Chami, N., Preuss, M., Walker, R. W., Moscati, A. & Loos, R. J. F. The role of polygenic susceptibility to obesity among carriers of pathogenic mutations in MC4R in the UK Biobank population. PLoS Med. 17, e1003196 (2020).
Fahed, A. C. et al. Polygenic background modifies penetrance of monogenic variants for tier 1 genomic conditions. Nat. Commun. 11, 3635 (2020).
Oetjens, M. T., Kelly, M. A., Sturm, A. C., Martin, C. L. & Ledbetter, D. H. Quantifying the polygenic contribution to variable expressivity in eleven rare genetic disorders. Nat. Commun. 10, 4897 (2019).
Mars, N. et al. The role of polygenic risk and susceptibility genes in breast cancer over the course of life. Nat. Commun. 11, 6383 (2020).
Halford, J. L. et al. Endophenotype effect sizes support variant pathogenicity in monogenic disease susceptibility genes. Nat. Commun. 13, 5106 (2022).
Costanzo, M. C. et al. The Type 2 Diabetes Knowledge Portal: an open access genetic resource dedicated to type 2 diabetes and related traits. Cell Metab. 35, 695–710 e6 (2023).
Eastwood, S. V. et al. Algorithms for the capture and adjudication of prevalent and incident diabetes in UK Biobank. PLoS ONE 11, e0162388 (2016).
Yu, S. et al. Toward high-throughput phenotyping: unbiased automated feature extraction and selection from knowledge sources. J. Am. Med Inf. Assoc. 22, 993–1000 (2015).
Hripcsak, G. et al. Facilitating phenotype transfer using a common data model. J. Biomed. Inf. 96, 103253 (2019).
Pacheco, J. & Thompson, W. Type 2 Diabetes Mellitus. PheKB https://phekb.org/phenotype/18 (2012).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Mahajan, A. et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat. Genet. 50, 1505–1513 (2018).
Ge, T., Chen, C. Y., Ni, Y., Feng, Y. A. & Smoller, J. W. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat. Commun. 10, 1776 (2019).
Li, D., Harrison, J. K., Purushotham, D. & Wang, T. Exploring genomic data coupled with 3D chromatin structures using the WashU Epigenome Browser. Nat. Methods 19, 909–910 (2022).
Miguel-Escalada, I. et al. Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat. Genet. 51, 1137–1148 (2019).
Rauch, A. et al. Osteogenesis depends on commissioning of a network of stem cell transcription factors that act as repressors of adipogenesis. Nat. Genet. 51, 716–727 (2019).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Coetzee, S. G., Coetzee, G. A. & Hazelett, D. J. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics 31, 3847–3849 (2015).
Shannon, P. & Richards, M. MotifDb: an annotated collection of protein-DNA binding sequence motifs. R package version 1.42.0 (2023).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009).
Acknowledgements
This research has been conducted using the UKB resource under application no. 27892. We thank the MGBB for providing genomic data and the contributions of MGBB participants, without whom this work would not be possible. The AoU is supported by the National Institutes of Health (NIH), Office of the Director, regional medical centers: grant nos. 1 OT2 OD026549, 1 OT2 OD026554, 1 OT2 OD026557, 1 OT2 OD026556, 1 OT2 OD026550, 1 OT2 OD 026552, 1 OT2 OD026553, 1 OT2 OD026548, 1 OT2 OD026551 and 1 OT2 OD026555; IAA #: AOD 16037; federally qualified health centers: HHSN 263201600085U; data and research center: 5 U2C OD023196; Biobank: 1 U24 OD023121; participant center: U24 OD023176; participant technology systems center: 1 U24 OD023163; communications and engagement: 3 OT2 OD023205 and 3 OT2 OD023206; and community partners: 1 OT2 OD025277, 3 OT2 OD025315, 1 OT2 OD025337 and 1 OT2 OD025276. In addition, the AoU would not be possible without the partnership of its participants. We thank A. Pollard and S. Yu from D. Carling and S. Vernia’s groups, respectively (Imperial, Laboratory of Medical Sciences), for technical assistance during experimental validations. The present study was funded by the American Diabetes Association (grant no. 11-23-PDF-35 to A.H.-C.), the American Diabetes Association Innovative and Clinical Translational Award (no. 1-19-ICTS-068), the American Diabetes Association (grant no. 11-22-ICTSPM-16) and National Human Genome Research Institute (grant no. U01HG011723) and Medical University of Bialystok (MUB) grant from the Ministry of Science and Higher Education (Poland) to J.M.M., the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK; grant no. UM1-DK078616 to A.K.M. and J.M.M.), a NIDDK grant (no. K23 DK131345) and MGH ECOR Fund for Medical Discovery Clinical Research Award to J.H.L., a British Heart Foundation 4-year PhD studentship (no. FS/4yPhD/F/20/34128) to D.N., a Sir Henry Dale Fellowship and the Wellcome Trust and Royal Society (grant no. 224662/Z/21/Z) to I.C., an NC3Rs PhD studentship (no. NC/Y500628/1) to L.M., a Wellcome Trust Senior Fellow (grant no. 200837/Z/16/Z) and NIDDK grants (nos. UM-1DK126185, R01DK140555) to A.L.G. and the Ministry of Education and Science of Poland within the project ‘Excellence Initiative—Research University’, the Ministry of Health of Poland within the project ‘Center of Artificial Intelligence in Medicine at the MUB’, the Medical Research Agency within the project ‘Regional Center for Digital Medicine at the MUB’ (grant no. 2023.ABM.02.00008), the American Diabetes Association (grant no. 11-22-PDFPM-03) to L.S., the Foundation of National Institutes of Health and the Accelerated Medicines Partnership Common Metabolic Diseases initiative to A.K.M., the Novo Nordisk Foundation (no. NNF21SA0072102 and NNF21OC0068929) to J.G.S.M. and the Doris Duke Foundation (grant no. 2020096) and American Diabetes Association (grant no. 7-22-ICTSPM-23) to A.L.
Author information
Authors and Affiliations
Contributions
J.M.M. conceived the project. P.S. and R.M. imputed and performed the association analysis. A.H.-C., P.S. and R.M. carried out the data analysis. M.V., U.L.M. and J.L. ran replication analyses of new variants. L.M. performed experimental validations under the supervision of I.C. J.M.M., A.H.-C., P.S. and R.M. wrote the manuscript. A.A. identified MODY carriers in the UKB. J.B.C., B.P., J.H.L., T.I.P., J.C.F., A.L.G., L.S., K.W. and D.J.C. contributed clinical and technical expertise. J.G.S.M., D.N., I.C., S.B.-G. and F.M. performed in silico analysis of noncoding associations. A.K.M. supervised the association analysis in the All of Us cohort. U.L.M., A.M., A.L., M.U. and J.M.M. supervised the study. J.M.M. is the guarantor of this work and, as such, has full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Corresponding author
Ethics declarations
Competing interests
A.L.G.’s spouse is employed by Genentech and holds stock options in Roche. A.K.M. is an unpaid research collaborator with AstraZeneca. J.M.M. has research funded in collaboration with Novo Nordisk. The other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Timothy Frayling and Constantin Polychronakos for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 QQ plots from the discovery T2D GWAS meta-analysis, including UKB, GERA, MGBB, and AoU v5 cohorts.
a, QQ plot including variants with minor allele frequency ≥ 0.05. b, QQ plot including variants with minor allele frequency between 0.01 and 0.05. c, QQ plot including variants with minor allele frequency between 0.01 and 0.005. d, QQ plot including variants with minor allele frequency between 0.005 and 0.001. e, QQ plot including variants with minor allele frequency between 0.001 and 0.0005. f, QQ plot including variants with minor allele frequency between 0.0005 and 0.00005.
Extended Data Fig. 2 Comparison of UKB data imputed with TOPMed versus HRC-1000G-UK10K (original imputation release).
a,b, The line graphs show the average INFO score, and the bar plots show the total number of variants in the TOPMed imputation versus the HRC-1000G-UK10K imputation across the minor allele frequency (MAF) spectrum before (a) and after (b) filtering for variants with an INFO score greater than 0.7. MAC, minor allele count.
Extended Data Fig. 3 Benchmark of TOPMed imputation accuracy across the allele frequency spectrum.
The average percentage of carriers of variants identified in Monogenic Diabetes whole-exome sequencing in Goodrich et al.8 identified with imputation in a subset of 40 K UKB samples. The y-axis represents the average proportion of carriers identified among variants with imputation INFO > 0.8 in the imputed data from TOPMed vs HRC-1000G-UK10K (original UKB imputation release). The x-axis represents the different allele frequency bins. MAF, minor allele frequency; MAC, minor allele count.
Extended Data Fig. 4 Comparison of UKB/GERA/MGBB/AoU results for lead variants from largest T2D GWAS meta-analysis.
Comparison of effect estimates and -log10(P) values from Vujkovic et al.1 (x-axis, n T2D cases = 228,499, n controls = 1,178,783) and UKB/GERA/MGBB/AoU meta-analysis (y-axis, n T2D cases = 51,256, n controls = 370,487) for lead variants from Vujkovic et al.1. a,b, Comparison of the beta and standard error values for variants with minor allele frequency (MAF) > 0.05 (a) and MAF < 0.05 (b), respectively. Each point represents the beta value for each variant. The standard errors from the UKB/GERA/MGBB/AoU results are represented by the blue vertical bars, while the standard errors from Vujkovic et al.1 are represented by the black horizontal bars. c,d, Comparison of the -log10(P) values for variants with MAF > 0.05 (c) and MAF < 0.05 (d), respectively. Each point represents the -log10(P) values for each variant.
Extended Data Fig. 5 Locuszoom plots of novel-identified variants at genome-wide significance (p < 5 ×10−8) and corresponding forest plots from the discovery T2D GWAS meta-analysis.
a-h, Rare (MAF < 0.001) variants identified in autosomes: 2:27425274:C:T (a), 2:213085963:G:A (b), 4:23750157:G:A (c), 6:99432794:G:A (d), 7:128323039:G:A (e), 8:110165438:T:C (f), 14:73781721:C:T (g), 20:44385421:G:A (h). i-k, Variants identified in chrX, sex-combined analysis: X:9605153:C:T (i), X:19361522:G:C (j), X:45923705:A:C (k). l, Variant identified in chrX, female-only analysis. The meta-analysis included 51,256 T2D cases and 370,487 controls. The forest plots show the carrier counts and odds ratios for each cohort in which the variant was present. The odds ratio (OR) from each cohort from the discovery dataset is denoted by boxes proportional to the size of the cohort, and the 95% confidence intervals (CI) are denoted by the horizontal lines. Sample sizes for each cohort are detailed in Supplementary Table 1. MAF, minor allele frequency.
Extended Data Fig. 6 Epigenomic landscape of the LEP locus.
a, Colored tracks show Roadmap Epigenomics 12-mark, 25-state imputation-based chromatin state models (GRCh38 lift-over version) for 127 human tissues and cell types. The zoomed inset at the bottom highlights the only tissues (out of 127) in which the region where rs147287548 resides is annotated as an enhancer. b, Chromatin landscape of the LEP locus throughout in vitro adipogenesis29. The left panel shows all enhancer-capture HiC29 chromatin interactions stemming from the fragment containing the rs147287548 variant, which resides in an active enhancer in mesenchymal stem cells and throughout adipogenesis (see also Fig. 2b, and panel a of this figure). The right panel shows a zoomed-in region, revealing more clearly chromatin interactions between the rs147287548-enhancer and the promoter of the LEP gene.
Extended Data Fig. 7 Plots of relevant metabolic traits in individuals free of diabetes who are carriers and non-carriers of the LEP rare variant or the variants with intermediate penetrance (VIPs) in Monogenic Diabetes genes.
a-b, Effect of LEP, rs147287548, chr7:128323039 on the levels of apolipoprotein A (a) and HDL cholesterol (b). c-m, Effect of HNF4A, chr20:44413714, p.Arg114Trp on the levels of apolipoprotein a (c), apolipoprotein b (d), aspartate aminotransferase (e), glucose (f), HDL cholesterol (g), lipoprotein A (h), triglycerides (i), total cholesterol (j), LDL cholesterol (k), sex hormone binding globulin (l) and urea (m). n-o, Effect of GCK, chr7:44145170, p.Val455Glu on the levels of glucose (n) and hba1c (o). Data from heterozygous carriers and from homozygous non-carriers of the variants. Individuals from UKB were considered for this analysis. Each violin plot represents the distribution of the metabolic trait values by genotype, with the width of the violin indicating the density of the data. The inner box plots indicate the group median (central line), first and third quartiles (bounds of box), and 1.5x interquartile range (whiskers).
Extended Data Fig. 8 Forest plots showing the carrier counts and odds ratios of the variants with intermediate penetrance (VIPs) (odds ratio > 5, and 95% confidence interval lower-bound 95% > 2) identified in the analysis of variants from ClinVar in Monogenic Diabetes genes.
This analysis included the UKB (n = 27,323 cases and 259,916 controls), MGBB (n = 6,623 cases and 41,411 controls), and GERA (n = 7,498 cases and 53,212 controls) cohorts. The odds ratio (OR) from each cohort from the discovery dataset is denoted by boxes proportional to the size of the cohort, and the 95% confidence intervals (CI) are denoted by the horizontal lines. a-d, Variants with conflicting interpretations of pathogenicity in ClinVar: 7:44145170:A:T (a), 12:120997588:C:T (b), 19:50402602:A:G (c), 19:50413456:G:A (d). e-g, Variants of uncertain significance in ClinVar: 4:6302287:G:A (e), 11:17388128:G:A (f), 19:50402228:G:A (g). h, Variant classified as likely benign in ClinVar: 19:50409504:C:T.
Extended Data Fig. 9 Classification of variants in 22 Monogenic Diabetes genes.
a, Variants classified as “likely benign” in Clinvar. b, Variants classified as “benign” in ClinVar. c, Variants classified as “likely pathogenic” in ClinVar. d, Variants classified as “pathogenic” in ClinVar. Variants classified as “conflicting interpretations of pathogenicity” or “uncertain significance” in ClinVar are shown in Fig. 3b. The x-axis represents the MAF. Along the y-axis, the odds ratio (OR) for each variant is denoted by the points, and the 95% confidence interval (CI) is denoted by the vertical lines. Only variants with MAF < 0.001 were considered for this analysis. Variants with a meta-analytic OR > 5 and an OR 95% LB > 2 are classified as “intermediate penetrance”. Variants with an OR 95% UB < 2 are classified as “supports benign”. Variants with an OR 95% UB > 2 and LB < 2 are classified as “inconclusive”. This analysis included the UKB (n = 27,323 cases and 259,916 controls), MGBB (n = 6,623 cases and 41,411 controls), and GERA (n = 7,498 cases and 53,212 controls) cohorts.
Extended Data Fig. 10 Boxplots of the age of diabetes diagnosis among non-carriers, carriers of variants with intermediate penetrance (VIPs), and carriers of confirmed pathogenic MODY variants.
a, Data for VIP in HNF4A. b, Data for VIP in HNF1A. c, Data for VIP in GCK. The age of diabetes diagnosis is expressed in years. Box plots indicate the group median (central line), first and third quartiles (bounds of box), and 1.5x interquartile range (whiskers). The covariate-adjusted P is included for comparisons with significant differences (P < 0.05) between groups.
Supplementary information
Supplementary Information
Supplementary Note.
Supplementary Tables
Supplementary Tables 1–10.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Huerta-Chagoya, A., Schroeder, P., Mandla, R. et al. Rare variant analyses in 51,256 type 2 diabetes cases and 370,487 controls reveal the pathogenicity spectrum of monogenic diabetes genes. Nat Genet 56, 2370–2379 (2024). https://doi.org/10.1038/s41588-024-01947-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41588-024-01947-9
This article is cited by
-
Using large-scale population-based data to improve disease risk assessment of clinical variants
Nature Genetics (2025)
-
Investigating the sources of variable impact of pathogenic variants in monogenic metabolic conditions
Nature Communications (2025)
-
Diabetes mellitus polygenic risk scores: heterogeneity and clinical translation
Nature Reviews Endocrinology (2025)
-
Bariatric Surgery for Diabetic Obesity: Insights and Challenges
Obesity Surgery (2024)