Abstract
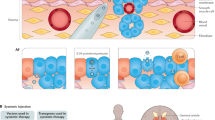
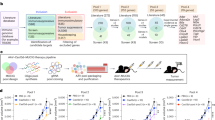
Immunotherapy has transformed cancer treatment. However, current immunotherapy modalities face various limitations. In the present study, we developed multiplexed activation of endogenous genes as an immunotherapy (MAEGI), a new form of immunotherapy that elicits antitumor immunity through multiplexed activation of endogenous genes in tumors. We leveraged CRISPR activation (CRISPRa) to directly augment the in situ expression of endogenous genes, and thereby the presentation of tumor antigens, leading to dramatic antitumor immune responses. Deploying this as a cell-based vaccination strategy showed efficacy in both prophylactic and therapeutic settings. Intratumoral adeno-associated virus delivery of CRISPRa libraries elicited strong antitumor immunity across multiple cancer types. Precision targeting of mutated gene sets eradicated a large fraction of established tumors at both local and distant sites. This treatment modality led to alterations in the tumor microenvironment, marked by enhanced T cell infiltration and antitumor immune signatures. Multiplexed endogenous gene activation is a versatile and highly scalable strategy to elicit potent immune responses against cancer, distinct from all existing cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
Source data and statistics for non-next-generation sequencing experiments such as tumor studies, flow cytometry, ELISpot and qPCR are provided in an Excel table (see Supplementary Table 1). Processed genomic sequencing data are also provided in an Excel table (see Supplementary Table 2). Original exome sequencing data are available via the National Center for Biotechnology Information Sequence Read Archive (PRJNA553203). Single-cell transcriptome sequencing data are available via the Gene Expression Omnibus (GSE133983). Other data, reagents, methods, computational codes and materials that support the findings of this research are available from the corresponding author upon reasonable request.
Code availability
Custom codes used to support the findings of this research are available from the corresponding author upon reasonable request.
References
Sharma, P., Hu-Lieskovan, S., Wargo, J. A. & Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553, 446–454 (2018).
Rosenberg, S. A. & Restifo, N. P. Adoptive cell transfer as personalized immunotherapy for human cancer. Science 348, 62–68 (2015).
Robert, C. et al. Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 372, 2521–2532 (2015).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015).
Ribas, A. & Wolchok, J. D. Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018).
June, C. H., O’Connor, R. S., Kawalekar, O. U., Ghassemi, S. & Milone, M. C. CAR T cell immunotherapy for human cancer. Science 359, 1361–1365 (2018).
Sahin, U. & Tureci, O. Personalized vaccines for cancer immunotherapy. Science. 359, 1355–1360 (2018).
Chen, D. S. & Mellman, I. Oncology meets immunology: the cancer-immunity cycle. Immunity 39, 1–10 (2013).
Chen, D. S. & Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 541, 321–330 (2017).
Mellman, I., Coukos, G. & Dranoff, G. Cancer immunotherapy comes of age. Nature 480, 480–489 (2011).
Hamid, O. et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 369, 134–144 (2013).
Hodi, F. S. et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363, 711–723 (2010).
Wolchok, J. D. et al. Nivolumab plus ipilimumab in advanced melanoma. N. Engl. J. Med. 369, 122–133 (2013).
Topalian, S. L. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366, 2443–2454 (2012).
Dunn, G. P., Bruce, A. T., Ikeda, H., Old, L. J. & Schreiber, R. D. Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 3, 991–998 (2002).
Schumacher, T. N. & Schreiber, R. D. Neoantigens in cancer immunotherapy. Science 348, 69–74 (2015).
Verdegaal, E. M. et al. Neoantigen landscape dynamics during human melanoma–T cell interactions. Nature 536, 91–95 (2016).
Khong, H. T. & Restifo, N. P. Natural selection of tumor variants in the generation of ‘tumor escape’ phenotypes. Nat. Immunol. 3, 999–1005 (2002).
Spiotto, M. T. et al. Increasing tumor antigen expression overcomes ‘ignorance’ to solid tumors via crosspresentation by bone marrow-derived stromal cells. Immunity 17, 737–747 (2002).
Ott, P. A. et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221 (2017).
Sahin, U. et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547, 222–226 (2017).
Hilf, N. et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature 565, 240–245 (2019).
Keskin, D. B. et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239 (2019).
Qi, L. S. et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013).
Gilbert, L. A. et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451 (2013).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR–Cas9 complex. Nature 517, 583–588 (2015).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Tanenbaum, M. E., Gilbert, L. A., Qi, L. S., Weissman, J. S. & Vale, R. D. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159, 635–646 (2014).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1–9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Mingozzi, F. & High, K. A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 12, 341–355 (2011).
Kirsch, I., Vignali, M. & Robins, H. T-cell receptor profiling in cancer. Mol. Oncol. 9, 2063–2070 (2015).
Ciucci, T. & Bosselut, R. Gimap and T cells: a matter of life or death. Eur. J. Immunol. 44, 348–351 (2014).
Martincorena, I. & Campbell, P. J. Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015).
Liu, X. S. & Mardis, E. R. Applications of Immunogenomics to Cancer. Cell 168, 600–612 (2017).
van den Boorn, J. G. & Hartmann, G. Turning tumors into vaccines: co-opting the innate immune system. Immunity 39, 27–37 (2013).
Marabelle, A., Tselikas, L., de Baere, T. & Houot, R. Intratumoral immunotherapy: using the tumor as the remedy. Ann. Oncol. 28, 33–43 (2017).
Townsend, S. E. & Allison, J. P. Tumor rejection after direct costimulation of CD8+ T cells by B7-transfected melanoma cells. Science 259, 368–370 (1993).
Chen, L. et al. Costimulation of antitumor immunity by the B7 counterreceptor for the T lymphocyte molecules CD28 and CTLA-4. Cell 71, 1093–1102 (1992).
Ribas, A. et al. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell 170, 1109–1119 (2017).
Zamarin, D. et al. Localized oncolytic virotherapy overcomes systemic tumor resistance to immune checkpoint blockade immunotherapy. Sci. Transl. Med. 6, 226ra232 (2014).
Suva, M. L. et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell 157, 580–594 (2014).
Roulois, D. et al. DNA-Demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162, 961–973 (2015).
Shalem, O. et al. Genome-scale CRISPR–Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Wang, T., Wei, J. J., Sabatini, D. M. & Lander, E. S. Genetic screens in human cells using the CRISPR–Cas9 system. Science 343, 80–84 (2014).
Robbins, P. F. et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat. Med. 19, 747–752 (2013).
Russell, S. et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860 (2017).
Bainbridge, J. W. et al. Long-term effect of gene therapy on Leber’s congenital amaurosis. N. Engl. J. Med. 372, 1887–1897 (2015).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Broz, M. L. et al. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell 26, 638–652 (2014).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Linderman, G. C., Zhao, J. & Kluger, Y. Zero-preserving imputation of scRNA-seq data using low-rank approximation. bioRxiv https://www.biorxiv.org/content/10.1101/397588v1 (2018).
Maaten, Lvd & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Maaten, Lvd Accelerating t-SNE using tree-based algorithms. J. Mach. Learn. Res. 15, 3221–3245 (2014).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Bolotin, D. A. et al. MiXCR: software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015).
Nazarov, V. I. et al. tcR: an R package for T cell receptor repertoire advanced data analysis. BMC Bioinforma. 16, 175 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Acknowledgements
We thank all members of the Chen laboratory, as well as various colleagues in the Department of Genetics, Systems Biology Institute, Immunobiology Program, BBS Program, Comprehensive Cancer Center, Stem Cell Center and the School of Medicine at Yale, for their assistance and/or scientific discussion. We thank the Center for Genome Analysis, Center for Molecular Discovery, Pathology Tissue Services, Histology Services, High Performance Computing Center, West Campus Analytical Chemistry Core, and West Campus Imaging Core and Keck Biotechnology Resource Laboratory at Yale for technical support. We thank P. Cresswell for assistance with ELISpot. S.C. was supported by Yale SBI/Genetics Startup Fund, National Institutes of Health/National Cancer Institute (NIH/NCI) (grant nos DP2CA238295, R01CA231112, R33CA225498, U54CA209992-8697, RF1DA048811, P50CA196530-A10805, P50CA121974-A08306), Damon Runyon Dale Frey Award (grant no. DFS-13-15), Melanoma Research Alliance (grant nos 412806, 16-003524), St Baldrick’s Foundation (grant no. 426685), Breast Cancer Alliance, Cancer Research Institute, American Association for Cancer Research (grant nos 499395, 17−20-01-CHEN), the Mary Kay Foundation (grant no. 017−81), the V Foundation (grant no. V2017-022), Ludwig Family Foundation, DoD (grant no. W81XWH-17-1-0235), Sontag Foundation and Chenevert Family Foundation. C.S.F. was supported by NIH/NCI Cancer Center Support Grant (no. 3P30CA016359). G.W. was supported by CRI Irvington and RJ Anderson Postdoctoral Fellowships. R.D.C., J.P. and M.B.D. were supported by the Yale MSTP training grant from the NIH (grant no. T32GM007205). P.R. was supported by a Yale PhD training grant from NIH (grant no. T32GM007499). X.D. was supported by a C Revson Postdoctoral Fellowship.
Author information
Authors and Affiliations
Contributions
G.W., R.D.C. and S.C. designed the study. G.W. performed most of the experiments in the present study. R.D.C. designed the libraries and analyzed most of the high-throughput data. Z.B., L.Z., Y.E., X.D., M.B.D., L.Y., X.Z., L.S. and H.Y. assisted with the experiments. P.A.R. and J.J.P. assisted with the data analysis. C.S.F. provided clinical insights. S.C. conceived the study, secured funding and supervised the work. G.W., R.D.C. and S.C. prepared the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
A patent application has been filed by Yale University related to data in the present study. C.S.F. serves a consulting role for Agios, Bain Capital, Bayer, Celgene, Dicerna, Five Prime Therapeutics, Gilead Sciences, Eli Lilly, Entrinsic Health, Genentech, KEW, Merck, Merrimack Pharmaceuticals, Pfizer, Roche, Sanofi, Taiho and Unum Therapeutics, and also serves as a director for CytomX Therapeutics and owns unexercised stock options for CytomX and Entrinsic Health.
Additional information
Peer review information Zoltan Fehervari was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Integrated supplementary information
Supplementary Figure 1 CRISPRa in cancer cells induces stronger effector function of T cells by boosting the presentation of targeted antigens.
a, The expression of dCas9 protein in E0771-dCas9-VP64 and E0771-dCas9-VP64-MPH cell lines was confirmed by western blot using a monoclonal antibody against Cas9. Anti-GAPDH was used as an internal control. Representative data out of two independent experiments. b-c, OT-I CD8+ T effector cells after co-culture with E0771-OVA cancer cells. In each treatment group, n = 11 independent cell co-cultures, pooled from three independent experiments. b, Quantification of the geometric mean fluorescent intensity (MFI) of IFNγ in OT-I CD8+ T effector cells after co-culture with E0771-OVA cancer cells for 3h. Two sided Welch’s unpaired t-test: CRISPRa sgRNAs vs. vector in E:T = 1:1, p = 0.0713; CRISPRa sgRNAs vs. vector in E:T = 0.5:1, p = 0.0368. c, The percentage of cells undergoing apoptosis among all non-dead cancer cells after co-culture with OT-I CD8+ T effector cells for 24h. Two sided Welch’s unpaired t-test: CRISPRa sgRNAs vs. vector in E:T = 1:1, p = 0.001; CRISPRa sgRNAs vs. vector in E:T = 0.5:1, p = 0.002. d, Tukey boxplots (interquartile range (IQR) boxes with 1.5 × IQR whiskers) showing library representation of genome-scale mouse CRISPRa (SAM) library-transduced E0771 cells. n = 6 infection replicates from two independent experiments. Out of the 66,749 sgRNAs in the library, the six replicates cover 99.44% to 99.69% of the total sgRNAs. e, Tumor growth curves of E0771- Vector tumors that were treated with PBS (n = 5 independent mice), α-CD8 (n = 3), or c-MAEGI + α -CD8 (n = 3). Blue arrows, c-MAEGI treatment; brown arrows, α -CD8 treatment. Two-way ANOVA: PBS vs α-CD8, p = 0.9744; PBS vs. c-MAEGI + α-CD8, p = 0.6037; α-CD8 vs. c-MAEGI + α-CD8, p = 0.705. f, Representative peripheral blood flow cytometry analysis of CD8+ T cells after PBS or α-CD8 treatment, gated on CD45+CD3+ cells. Top row, PBS treatment; bottom row, α-CD8 treatment. Left column, DPI = 14; right column, DPI = 21. X-axis of scatterplots, CD4; Y-axis of scatterplots, CD8a. Percentages are noted in each quadrant. Over 50,000 PBMCs were analyzed for each sample. g, Bar plots detailing the % of CD8+ T cells among CD45+ PBMCs, 14 days after PBS or α-CD8 treatment. Data from PBS treated mice (n = 3), and α-CD8 treated mice (n = 4). Two-tailed unpaired t-test: PBS vs. α-CD8, p < 0.0001. h, Bar plots detailing the % of CD8+ T cells among CD45+ PBMCs, 21 days after PBS or α-CD8 treatment. Data from PBS treated mice (n = 4), and α-CD8 treated mice (n = 4). Two-tailed unpaired t-test: PBS vs. α-CD8, p < 0.0001. Error bars: Data points in this figure are presented as mean ± s.e.m. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Figure 2 AAV-delivered CRISPRa enhances the expression and presentation of targeted antigens.
a, AAV-CRISPRa activation of multiple endogenous genes in a pooled manner. The transcript levels of Cd70, Cd80, Cd86 were quantified by qRT-PCR after transducing E0771-dCas9-VP64 cells with either AAV-Vector or AAV-sgRNA-miniLib (n = 3 cell infection replicates from one representative experiment). Two-sided unpaired t-test for sgRNAs vs. vector: Cd70, p < 0.0001; Cd80, p = 0.0005; Cd86, p < 0.0001. b, AAV-CRISPRa enhances the cell surface presentation of a model antigen in the form of peptide-MHC-I complexes. In cells transduced with lentivirus to express ovalbumin (Ova) under a PGK promoter, AAV-CRISPRa targeted to the PGK promoter augments the presentation of Ova-derived SIINFEKL-MHC-I complex on the cell surface. Quantitative analysis of geometric mean fluorescence intensity (MFI) of PE-SIINFEKL-H-2Kb on cells treated with AAV-Vector or AAV-sgRNAs. n = 6 cell infection replicates (SIINFEKL-H-2Kb staining in AAV-Vector), n = 6 (isotype in AAV-Vector), n = 6 (SIINFEKL-H-2Kb staining in AAV-sgRNAs), or n = 6 (isotype in AAV-sgRNAs) from four independent experiments. Two sided Mann-Whitney test: SIINFEKL-H-2Kb staining vs. isotype in AAV-Vector, p = 0.091; SIINFEKL-H-2Kb staining vs. isotype in AAV-sgRNAs, p = 0.0022; SIINFEKL-H-2Kb staining in AAV-sgRNAs vs. AAV-Vector, p = 0.0411. Error bars: All data in this figure are presented as mean ± s.e.m., with individual data points shown. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Figure 3 Exome-guided design of precision MAEGI to activate endogenous mutant genes.
a, Exome sequencing identified genic mutations in E0771 cells, indicated over their positions in the mouse genome. Genes were colored by the presence or absence of SNPs and/or indels. b, Scatter plot showing the number of mutations as SNPs or indels per gene. Genes were filtered by a minimum number of total mutations in E0771 cells compared to wildtype C57BL/6J mammary fat pad. Examples of the most highly mutated genes are Tekt2, Hmmr, Acer2, G3bp1, Rsc1a1, Slu7, Fat2 and Skint3. A total of 1,116 genes were used in custom library design. c, Violin density plot of sgRNA representation in the E0771 exome-guided AAV-p-MAEGI plasmid library (n = 3,839 sgRNAs), showing 100% representation of the library. d, Representative images of mice at experimental end-point from PBS, AAV-Vector, and AAV-p-MAEGI groups in the E0771 syngeneic tumor model. e, Growth curves of E0771 syngeneic tumors in Rag1–/– mice treated by PBS (n = 6), AAV-Vector (n = 5), or AAV-p-MAEGI (n = 6) by intratumoral administration at indicated times (blue arrows). n = biologically independent mice. Two-way ANOVA: AAV-Vector vs. PBS, p = 0.0653; AAV-p-MAEGI vs. PBS, p = 0.6982; AAV-p-MAEGI vs. AAV-Vector, p = 0.0634. Error bars: All data in this figure are presented as mean ± s.e.m., with individual data points shown. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Figure 4 Histological characterization of E0771 tumors treated with AAV-p-MAEGI.
a-c, Representative pictures of H&E (a), CD8 (b), or Ki67 (c), staining of E0771 tumors collected from mice treated with PBS, AAV-Vector, or AAV-p-MAEGI. (Tumor harvest dpi = 36) (n = 2 out of 6 mice from two independent experiments with similar results). d, Quantification of % Ki67 positivity in E0771 tumors collected from mice treated with PBS, AAV-Vector, or AAV-p-MAEGI. Scale bars of zoomed-out images and insets = 200 μm. e-g, AAV infection efficiency assessed by intratumoral delivery of GFP-expressing AAVs and flow cytometry analysis. e, Percentage of GFP+ cells (mean ± s.e.m.) within tumors from mice 4 days after intratumoral injection of PBS (n = 3 mice) or AAV-GFP (n = 12), grouped by CD45+ and CD45- cells. f, Percentage of GFP+ cells (mean ± s.e.m.) within tumors from mice 12 days after intratumoral injection of PBS (n = 2 mice) or AAV-GFP (n = 8), grouped by CD45+ and CD45- cells. g, Percentage of GFP+ cells in various organs 4 days after intratumoral injection of AAV-GFP (n = 6 mice).
Supplementary Figure 5 Characterization of the dual-AAV CRISPRa system, and representative flow cytometry gating.
a, Schematics of using a dual AAV vector system to deliver CRISPRa components. For in vitro testing, Pmel was targeted with the dual AAV CRISPRa system (AAV-dCas9 and AAV-U6-sgPmel-EF1a-MPH). 5-7 days post-infection, expression levels of Pmel were determined by quantitative RT-PCR. n = 4 infection replicates, pooled from two independent experiments (unpaired two-sided Mann-Whitney test, p = 0.0286). b, Representative flow cytometry gating pictures of CD3+, CD4+, and CD8+ T cell populations within tumors from mice treated with PBS, AAV-Vector or AAV-p-MAEGI. c, Representative flow cytometry gating of monocytes, neutrophils, MHCII+ cells, dendritic cells, and macrophages in tumors from mice treated with PBS, AAV-Vector, or AAV-p-MAEGI, 19 days after tumor induction, using a previously described gating strategy. d, Percentage of dendritic cells in tumors from mice treated with PBS (n = 6 mice), AAV-Vector (n = 10), or AAV-p-MAEGI (n = 12). Two-tailed Mann-Whitney test: AAV-Vector vs. PBS, p = 0.0075; AAV-p-MAEGI vs. PBS, p = 0.0008; AAV-p-MAEGI vs. AAV-Vector, p = 0.0692. e, Percentage of macrophages in tumors from mice treated with PBS (n = 6 mice), AAV-Vector (n = 10), or AAV-p-MAEGI (n = 12). Two-tailed Mann-Whitney test: AAV-Vector vs. PBS, p = 0.6353; AAV-p-MAEGI vs. PBS, p = 0.5532; AAV-p-MAEGI vs. AAV-Vector, p = 0.9229. f, Percentage of monocytes in tumors from mice treated with PBS (n = 6 mice), AAV-Vector (n = 10), or AAV-p-MAEGI (n = 12). Two-tailed Mann-Whitney test: AAV-Vector vs. PBS, p = 0.1471; AAV-p-MAEGI vs. PBS, p = 0.5533; AAV-p-MAEGI vs. AAV-Vector, p = 0.0426. g, Percentage of neutrophils in tumors from mice treated with PBS (n = 6 mice), AAV-Vector (n = 10), or AAV-p-MAEGI (n = 12). Two-tailed Mann-Whitney test: AAV-Vector vs. PBS, p = 0.0559; AAV-p-MAEGI vs. PBS, p = 0.6165; AAV-p-MAEGI vs. AAV-Vector, p = 0.4176. Error bars: All data in this figure are presented as mean ± s.e.m., with individual data points shown. Asterisks: * p < 0.05, ** p < 0.01, *** p < 0.001.
Supplementary Figure 6 TCR richness and CDR3 amino acid sequences from major tumor-infiltrating T cell clones.
CDR3 amino acid sequences from major (top 3) clones in three example TIL samples from PBS, AAV-Vector, AAV-g-MAEGI, and AAV-p-MAEGI treated mice.
Supplementary Figure 7 Processing of scRNA-seq data to exclude CD45- cells and characteristic markers of each scRNA-seq cluster.
a, t-SNE of all single cells passing the filtering criteria, colored by k-means clustering (AAV-p-MAEGI, n = 4381 cells; AAV-Vector, n = 3482). b-f, Pooled analysis of all CD45+ cells, grouped by cluster (k1, n = 124 cells; k2, n = 409; k3, n = 383; k4, n = 1056; k5, n = 317; k6, n = 734; k8, n = 1781; k9, n = 2060). b, Violin density plots of Ptprc (encoding CD45) expression, grouped by k-means cluster. Cluster 7 is largely negative for Ptprc expression. c, Violin density plots of Cd3e, Cd4, CD8a, Il7r, and Gzmb expression, defining different T cells. d, Violin density plots of Cd19, Cd20, Pax5, Jchain, and Ly6d expression, defining different B cells. e, Violin density plots of Ly75, Batf3, Clec10a, and CD209a expression, defining different dendritic cells. f, Violin density plots of Cd14, Fcgr1, and Adgre1 (encoding F4/80) expression, defining macrophages and monocytes.
Supplementary information
Supplementary Information
Supplementary Figs. 1–7.
Supplementary Table 1
Raw data and statistics of mouse and cellular experiments
Supplementary Table 2
Processed data and analysis of high-throughput sequencing experiments
Rights and permissions
About this article
Cite this article
Wang, G., Chow, R.D., Bai, Z. et al. Multiplexed activation of endogenous genes by CRISPRa elicits potent antitumor immunity. Nat Immunol 20, 1494–1505 (2019). https://doi.org/10.1038/s41590-019-0500-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-019-0500-4
This article is cited by
-
The present and future of the Cancer Dependency Map
Nature Reviews Cancer (2025)
-
Sensitizing solid tumors to CAR-mediated cytotoxicity by lipid nanoparticle delivery of synthetic antigens
Nature Cancer (2025)
-
Comprehensive review of CRISPR-based gene editing: mechanisms, challenges, and applications in cancer therapy
Molecular Cancer (2024)
-
Epigenome editing technologies for discovery and medicine
Nature Biotechnology (2024)
-
Epigenetic therapy potentiates transposable element transcription to create tumor-enriched antigens in glioblastoma cells
Nature Genetics (2024)