Abstract
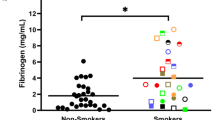
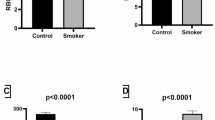
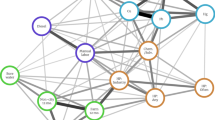
Exposure to fire smoke has become a global health concern and is associated with increased morbidity and mortality. There is a lack of understanding of the specific immune mechanisms involved in smoke exposure, with preventive and targeted interventions needed. After exposure to fire smoke, which includes PM2.5, toxic metals and perfluoroalkyl and polyfluoroalkyl substances, epidemiology-based studies have demonstrated increases in respiratory (for example, asthma exacerbation), cardiac (for example, myocardial infarction, arrhythmias), neurological (for example, stroke) and pregnancy-related (for example, low birthweight, premature birth) outcomes. However, mechanistic studies exploring how smoke exposure disrupts cellular homeostasis are lacking. Therefore, we collected blood from smoke-exposed individuals (n = 31) and age-matched and sex-matched non-smoke-exposed controls (n = 29), and investigated these complex interactions using a single-cell exposomic approach based on both methylation and mass cytometry. Overall, our data demonstrated a strong association between smoke exposure and methylation at 133 disease-relevant gene loci, while immunophenotyping showed increased homing and activation biomarkers. We developed an application of mass cytometry to analyze single-cell/metal binding and found, for example, increased levels of mercury in dead cells and cadmium in the live and dead cell populations. Moreover, mercury levels were associated with years of smoke exposure. Several epigenetic sites across multiple chromosomes were associated with individual toxic metal isotopes in single immune cells. Our methods for detecting the effect of smoke exposure at the single-cell level and the study results may help to determine the timing of exposure and identify specific molecular targets that could be modified to prevent and manage exposure to smoke.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The DNA methylation and mass cytometry datasets (including manually gated live single cells and cell subsets) generated during the current study are available via the Harvard Dataverse data repository at https://doi.org/10.7910/DVN/5PWZJM. Source data are provided with this paper.
Code availability
The R code needed to reproduce the results of the study is available at https://github.com/knadeaulab/Smoke_Exposure_Study.
References
Wu, Y. et al. Synergistic aircraft and ground observations of transported wildfire smoke and its impact on air quality in New York City during the summer 2018 LISTOS campaign. Sci. Total Environ. 773, 145030 (2021).
Childs, M. L. et al. Daily local-level estimates of ambient wildfire smoke PM2.5 for the contiguous US. Environ. Sci. Technol. 56, 13607–13621 (2022).
Chen, G. et al. Mortality risk attributable to wildfire-related PM2.5 pollution: a global time series study in 749 locations. Lancet Planet Health 5, e579–e587 (2021).
Demers, P. A. et al. Carcinogenicity of occupational exposure as a firefighter. Lancet Oncol. 23, 985–986 (2022).
Gu, W. et al. Particulate polycyclic aromatic hydrocarbons and metals, DNA methylation and DNA methyltransferase among middle-school students in China. Sci. Total Environ. 926, 172087 (2024).
Martin, E. M. & Fry, R. C. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu. Rev. Public Health 39, 309–333 (2018).
Wallace, D. R. et al. Toxic-metal-induced alteration in miRNA expression profile as a proposed mechanism for disease development. Cells 9, 901 (2020).
Guo, X. et al. Associations of blood levels of trace elements and heavy metals with metabolic syndrome in Chinese male adults with microRNA as mediators involved. Environ. Pollut. 248, 66–73 (2019).
Manić, L. et al. Epigenetic mechanisms in metal carcinogenesis. Toxicol. Rep. 9, 778–787 (2022).
Liu, J. C. & Peng, R. D. The impact of wildfire smoke on compositions of fine particulate matter by ecoregion in the Western US. J. Expo. Sci. Environ. Epidemiol. 29, 765–776 (2019).
Lopez, A. M., Pacheco, J. L. & Fendorf, S. Metal toxin threat in wildland fires determined by geology and fire severity. Nat. Commun. 14, 8007 (2023).
Burke, M. et al. The contribution of wildfire to PM2.5 trends in the USA. Nature 622, 761–766 (2023).
From Wildfire Smoke to PFAS: Innovative EPA Scientists Address Longstanding Research Gaps https://www.epa.gov/sciencematters/wildfire-smoke-pfas-innovative-epa-scientists-address-longstanding-research-gaps (United States Environmental Protection Agency, 2022).
Wee, S. Y. & Aris, A. Z. Revisiting the “forever chemicals”, PFOA and PFOS exposure in drinking water. NPJ Clean Water 6, 57 (2023).
van den Dungen, M. W., Murk, A. J., Kampman, E., Steegenga, W. T. & Kok, D. E. Association between DNA methylation profiles in leukocytes and serum levels of persistent organic pollutants in Dutch men. Environ. Epigenet. 3, dvx001 (2017).
Kishi, R. et al. Importance of two birth cohorts (n = 20,926 and n = 514): 15 years’ experience of the Hokkaido Study on Environment and Children’s Health: malformation, development and allergy. Nihon Eiseigaku Zasshi 73, 164–177 (2018).
Liu, P., Yang, F., Wang, Y. & Yuan, Z. Perfluorooctanoic acid (PFOA) exposure in early life increases risk of childhood adiposity: a meta-analysis of prospective cohort studies. Int. J. Environ. Res. Public Health 15, 2070 (2018).
Morin, A. et al. A functional genomics pipeline to identify high-value asthma and allergy CpGs in the human methylome. J. Allergy Clin. Immunol. 151, 1609–1621 (2023).
Goodrich, J. M. et al. Repeat measures of DNA methylation in an inception cohort of firefighters. Occup. Environ. Med. 79, 656–663 (2022).
Liu, Y. et al. Gestational perfluoroalkyl substance exposure and DNA methylation at birth and 12 years of age: a longitudinal epigenome-wide association study. Environ. Health Perspect. 130, 37005 (2022).
Slütter, B., Pewe, L. L., Kaech, S. M. & Harty, J. T. Lung airway-surveilling CXCR3hi memory CD8+ T cells are critical for protection against influenza A virus. Immunity 39, 939–948 (2013).
Deiuliis, J. A. et al. Pulmonary T cell activation in response to chronic particulate air pollution. Am. J. Physiol. Lung Cell. Mol. Physiol. 302, L399–L409 (2012).
Townley, R. G. & Agrawal, S. CRTH2 antagonists in the treatment of allergic responses involving TH2 cells, basophils, and eosinophils. Ann. Allergy Asthma Immunol. 109, 365–374 (2012).
Chen, G. et al. Inhibition of CRTH2-mediated Th2 activation attenuates pulmonary hypertension in mice. J. Exp. Med. 215, 2175–2195 (2018).
Oyesola, O. O. et al. The prostaglandin D2 receptor CRTH2 promotes IL-33-induced ILC2 accumulation in the lung. J. Immunol. 204, 1001–1011 (2020).
Spits, H. & Mjösberg, J. Heterogeneity of type 2 innate lymphoid cells. Nat. Rev. Immunol. 22, 701–712 (2022).
Hilvering, B. et al. Synergistic activation of pro-inflammatory type-2 CD8+ T lymphocytes by lipid mediators in severe eosinophilic asthma. Mucosal Immunol. 11, 1408–1419 (2018).
Hinks, T. S. C., Hoyle, R. D. & Gelfand, E. W. CD8+ Tc2 cells: underappreciated contributors to severe asthma. Eur. Respir. Rev. 28, 190092 (2019).
Esensten, J. H., Helou, Y. A., Chopra, G., Weiss, A. & Bluestone, J. A. CD28 costimulation: from mechanism to therapy. Immunity 44, 973–988 (2016).
Glinos, D. A. et al. Genomic profiling of T-cell activation suggests increased sensitivity of memory T cells to CD28 costimulation. Genes Immun. 21, 390–408 (2020).
Collier, J. L., Weiss, S. A., Pauken, K. E., Sen, D. R. & Sharpe, A. H. Not-so-opposite ends of the spectrum: CD8+ T cell dysfunction across chronic infection, cancer and autoimmunity. Nat. Immunol. 22, 809–819 (2021).
Radziszewska, A., Moulder, Z., Jury, E. C. & Ciurtin, C. CD8+ T cell phenotype and function in childhood and adult-onset connective tissue disease. Int. J. Mol. Sci. 23, 11431 (2022).
Mulliken, J. S. et al. Risk of systemic fungal infections after exposure to wildfires: a population-based, retrospective study in California. Lancet Planet Health 7, e381–e386 (2023).
Zhou, X. et al. Excess of COVID-19 cases and deaths due to fine particulate matter exposure during the 2020 wildfires in the United States. Sci. Adv. 7, eabi8789 (2021).
Landguth, E. L. et al. The delayed effect of wildfire season particulate matter on subsequent influenza season in a mountain west region of the USA. Environ. Int. 139, 105668 (2020).
Singh, S. P., Zhang, H. H., Foley, J. F., Hedrick, M. N. & Farber, J. M. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J. Immunol. 180, 214–221 (2008).
Gómez-Melero, S. & Caballero-Villarraso, J. CCR6 as a potential target for therapeutic antibodies for the treatment of inflammatory diseases. Antibodies 12, 30 (2023).
Liu, H. & Rohowsky-Kochan, C. Regulation of IL-17 in human CCR6+ effector memory T cells. J. Immunol. 180, 7948–7957 (2008).
Starner, T. D., Barker, C. K., Jia, H. P., Kang, Y. & McCray, P. B. Jr. CCL20 is an inducible product of human airway epithelia with innate immune properties. Am. J. Respir. Cell Mol. Biol. 29, 627–633 (2003).
Li, Y.-J. et al. Correction: progress of CCL20-CCR6 in the airways: a promising new therapeutic target. J. Inflamm. 22, 2 (2025).
Zuniga, E. I. & Harker, J. A. T-cell exhaustion due to persistent antigen: quantity not quality? Eur. J. Immunol. 42, 2285–2289 (2012).
Webster, J. P., Kane, T. J., Obrist, D., Ryan, J. N. & Aiken, G. R. Estimating mercury emissions resulting from wildfire in forests of the Western United States. Sci. Total Environ. 568, 578–586 (2016).
McKee, A. S. & Fontenot, A. P. Interplay of innate and adaptive immunity in metal-induced hypersensitivity. Curr. Opin. Immunol. 42, 25–30 (2016).
Mou, Y. et al. Environmental pollutants induce NLRP3 inflammasome activation and pyroptosis: roles and mechanisms in various diseases. Sci. Total Environ. 900, 165851 (2023).
Vergilio, C. S., Carvalho, C. E. V. & Melo, E. J. T. Mercury-induced dysfunctions in multiple organelles leading to cell death. Toxicol. In Vitro 29, 63–71 (2015).
Isley, C. F. & Taylor, M. P. Atmospheric remobilization of natural and anthropogenic contaminants during wildfires. Environ. Pollut. 267, 115400 (2020).
Cui, Z.-G., Ahmed, K., Zaidi, S. F. & Muhammad, J. S. Ins and outs of cadmium-induced carcinogenesis: mechanism and prevention. Cancer Treat. Res. Commun. 27, 100372 (2021).
Faroon, O. et al. Toxicological Profile for Cadmium (Agency for Toxic Substances and Disease Registry, 2012).
Driller, R. et al. Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype. J. Biol. Chem. 294, 13269–13279 (2019).
Chiossone, L., Dumas, P.-Y., Vienne, M. & Vivier, E. Natural killer cells and other innate lymphoid cells in cancer. Nat. Rev. Immunol. 18, 671–688 (2018).
Baliu-Piqué, M. et al. Short lifespans of memory T-cells in bone marrow, blood, and lymph nodes suggest that T-cell memory is maintained by continuous self-renewal of recirculating cells. Front. Immunol. 9, 2054 (2018).
Choi, J. E. et al. Heavy metal exposure linked to metabolic syndrome in Korean male firefighters: FRESH cohort cross-sectional analysis. Sci. Rep. 13, 14016 (2023).
Allonneau, A. et al. Lead contamination among Paris Fire Brigade firefighters who fought the Notre Dame Cathedral fire in Paris. Int. J. Hyg. Environ. Health 233, 113707 (2021).
Leary, D. B., Takazawa, M., Kannan, K. & Khalil, N. Perfluoroalkyl substances and metabolic syndrome in firefighters: a pilot study. J. Occup. Environ. Med. 62, 52–57 (2020).
Liang, S. et al. Sensei: how many samples to tell a change in cell type abundance? BMC Bioinformatics 23, 2 (2022).
Thomas, M. F. et al. Single-cell transcriptomic analyses reveal distinct immune cell contributions to epithelial barrier dysfunction in checkpoint inhibitor colitis. Nat. Med. 30, 1349–1362 (2024).
De Mel, S. et al. Single cell multi-omic profiling of multiple myeloma with t(4;14) finds an immune microenvironment gene signature that correlates with clinical outcomes. Blood 138, 2653 (2021).
Li, J. et al. Integrated multi-omics single cell atlas of the human retina. Preprint at Res. Sq. https://doi.org/10.21203/rs.3.rs-3471275/v1 (2023).
Rodriguez-Meira, A. et al. Single-cell multi-omics identifies chronic inflammation as a driver of TP53-mutant leukemic evolution. Nat. Genet. 55, 1531–1541 (2023).
Aguilera, J. et al. Granzymes, IL-16, and poly(ADP-ribose) polymerase 1 increase during wildfire smoke exposure. J. Allergy Clin. Immunol. Glob. 2, 100093 (2023).
Barros, B., Oliveira, M. & Morais, S. Firefighters’ occupational exposure: contribution from biomarkers of effect to assess health risks. Environ. Int. 156, 106704 (2021).
Kaushik, A. et al. CD8+ T cell differentiation status correlates with the feasibility of sustained unresponsiveness following oral immunotherapy. Nat. Commun. 13, 6646 (2022).
Guidelines on Drawing Blood: Best Practices in Phlebotomy (World Health Organization, 2010).
Fuss, I. J., Kanof, M. E., Smith, P. D. & Zola, H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr. Protoc. Immunol. https://doi.org/10.1002/0471142735.im0701s85 (2009).
Leipold, M. D. & Maecker, H. T. Phenotyping of live human PBMC using CyTOFTM mass cytometry. Bio Protoc. 5, e1382 (2015).
Van Gassen, S. et al. FlowSOM: using self-organizing maps for visualization and interpretation of cytometry data. Cytometry A 87, 636–645 (2015).
Hahne, F., Gopalakrishnan, N., Khodabakhshi, A.H., Wong, C. & Lee, K. flowStats: Statistical methods for the analysis of flow cytometry data, R package version 4.16.0 https://bioconductor.org/packages/flowStats (2024).
McInnes, L., Healy, J. & Melville, J. UMAP: uniform manifold approximation and projection for dimension reduction. Preprint at https://arxiv.org/abs/1802.03426 (2018).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Scrucca, L., Fraley, C., Murphy, T. B. & Raftery, A. E. Model-Based Clustering, Classification, and Density Estimation Using mclust in R (CRC Press, 2023).
Hansen, K. D. IlluminaHumanMethylationAllergymanifest. GitHub https://github.com/hansenlab/IlluminaHumanMethylationAllergymanifest (2023).
Xu, Z., Niu, L. & Taylor, J. A. The ENmix DNA methylation analysis pipeline for Illumina BeadChip and comparisons with seven other preprocessing pipelines. Clin. Epigenetics 13, 216 (2021).
Du, P., Kibbe, W. A. & Lin, S. M. lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019).
Makowski, D., Ben-Shachar, M. S. & Lüdecke, D. bayestestR: describing effects and their uncertainty, existence and significance within the Bayesian framework. J. Open Source Softw. 4, 1541 (2019).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Use R!) (Springer, 2009).
Blighe. K, Rana, S. & Lewis, M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling, R package version 1.22.0 https://bioconductor.org/packages/EnhancedVolcano (2024).
Kolde, R. pheatmap: Pretty Heatmaps, R package version 1.0.12 https://cran.r-project.org/web/packages/pheatmap/index.html (2019).
Acknowledgements
We thank the firefighters for their service and commitment to health research and the San Francisco Cancer Prevention Foundation for their support. The following sources of funding were used: AIRHEALTH PPG P01HL152953 (K.C.N.); the San Francisco Cancer Prevention Foundation (K.C.N.); Pregnancy R01 ES032253 (K.C.N.); the Asthma and Allergic Diseases Cooperative Research Center (K.C.N.); grant no. U19AI167903 (K.C.N.); T32 (E. Simonin); and the Keck Foundation (M. Burke). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
M.M.J. conceived the study design, managed all aspects of the study, performed the data collection and developed and wrote the manuscript. A.K. designed and executed the data analysis workflow and interpretation, created the figures and wrote the manuscript. O.A.K. contributed to sample collection and processing, and performed the mass cytometry, data analysis and manuscript development. E.Smith assisted with the study design, firefighter recruitment and performed the data collection. Y.P. and L.B. performed the mass cytometry. S.A. and J.A. performed the data collection. X.Z. contributed to data analysis and results interpretation. E. Simonin, J.A., A.F., M.C., O.B., R.S.C., E.P., M.S., C.A.A., M. Burke and M. Bondy reviewed the manuscript. M.A. and C.A.A. supervised the mass cytometry and provided critical revisions of the manuscript. K.C.N. conceived the study design, supervised all aspects of the study and wrote and finalized the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Liam Messin, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Figs. 1–19 and Tables 1–4.
Supplementary Table 1
Supplementary Tables 1–4 as a single Excel sheet.
Supplementary Data 1
Source data for Supplementary Figs. 1–19.
Supplementary Data 1
Computational data.
Supplementary Data 2
Computational data.
Supplementary Data 3
Computational data.
Supplementary Data 4
Computational data.
Supplementary Data 5
Computational data.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Johnson, M.M., Kaushik, A., Kline, O.A. et al. Immune impacts of fire smoke exposure. Nat Med (2025). https://doi.org/10.1038/s41591-025-03777-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41591-025-03777-6