Abstract
The blood–brain barrier (BBB) protects the brain and maintains neuronal homeostasis. BBB properties can vary between brain regions to support regional functions, yet how BBB heterogeneity occurs is poorly understood. Here, we used single-cell and spatial transcriptomics to compare the mouse median eminence, one of the circumventricular organs that has naturally leaky blood vessels, with the cortex. We identified hundreds of molecular differences in endothelial cells (ECs) and perivascular cells, including astrocytes, pericytes and fibroblasts. Using electron microscopy and an aqueous-based tissue-clearing method, we revealed distinct anatomical specializations and interaction patterns of ECs and perivascular cells in these regions. Finally, we identified candidate regionally enriched EC–perivascular cell ligand–receptor pairs. Our results indicate that both molecular specializations in ECs and unique EC–perivascular cell interactions contribute to BBB functional heterogeneity. This platform can be used to investigate BBB heterogeneity in other regions and may facilitate the development of central nervous system region-specific therapeutics.
Similar content being viewed by others
Main
The BBB is a physiological barrier between the blood and brain. Although BBB breakdown is involved in neurodegenerative diseases, an intact BBB is a major obstacle for central nervous system (CNS) drug delivery to treat neurological disorders1. Understanding the molecular mechanisms of BBB regulation will permit BBB manipulation for barrier repair or CNS drug delivery to improve disease treatment.
Different brain regions show different levels of blood vessel permeability. For example, the circumventricular organs (CVOs), specialized regions that include the median eminence (ME), are naturally leaky despite being adjacent to regions with a sealed BBB2. CVO neurons sense signaling compounds and secrete hormones into circulation to facilitate rapid communication with the periphery and regulate processes like feeding, cardiovascular function and thirst3,4. Moreover, BBB heterogeneity is observed in the hippocampus, basal ganglia and cerebellum; increased BBB permeability was reported in human aging and the early onset of neurodegenerative diseases1,5,6. Yet how these variations in BBB permeability occur is incompletely understood.
CNS capillary endothelial cells (cECs) constitute the BBB and have features like specialized tight junctions and low rates of transcytosis to regulate paracellular and transcellular trafficking, respectively7,8,9,10. To date, several studies have compared cECs from the CNS and peripheral tissue to identify molecular determinants of the BBB11. Yet BBB properties also require active induction and maintenance from the local environment12. Specifically, perivascular pericytes and astrocyte endfeet ensheath brain capillaries, forming the interface between ECs and neurons. Indeed, mice with reduced numbers of pericytes and astrocytes have a leaky BBB13,14,15,16. However, how local cues and cell interactions in the vascular microenvironment regulate regional brain barrier properties is largely unknown.
The major technical challenge to determining the mechanism underlying BBB heterogeneity is that ECs are rare in the brain, representing 4–6% of brain cells17. Some perivascular cells, including pericytes, are even less abundant18. Therefore, although typical unbiased single-cell transcriptomic studies of the brain often include vascular cells, they yield limited data about their transcriptomes owing to their relative scarcity following dissociation protocols optimized for neurons. To circumvent this problem, most studies of brain vascular and perivascular cells have relied on cell sorting from the entire brain19,20. This approach is not optimal for capturing BBB heterogeneity because it underrepresents smaller brain regions, which may contain transcriptionally diverse and specialized cells. Therefore, an investigation of regional vascular and perivascular cell heterogeneity necessitated the development of methods to enrich for brain ECs to discern differences in BBB-associated cells in small regions.
Here, we develop a platform to investigate how vascular and perivascular cells affect BBB functional heterogeneity in small, defined brain regions. We perform unbiased single-cell RNA sequencing (scRNA-seq) of a CVO, the ME, and a size-matched region (~0.05 × 0.2 × 1.2 mm3) of the somatosensory cortex (cortex) in the mouse brain. Comparison of these two small brain regions with distinct barrier properties revealed molecular differences in cECs and perivascular astrocytes and fibroblasts. Using spatial transcriptomics, we also identified molecular differences in pericytes. Correspondingly, we observed morphological differences in these cells and their interactions by electron microscopy and three-dimensional whole-brain imaging following tissue clearing by U.Clear. Finally, bioinformatics analysis identified regionally enriched ligand–receptor pairs, which may mediate the unique EC–perivascular cell interactions in these regions. Together, this work reveals both regional specializations of cECs and their unique interactions with surrounding perivascular cells, highlighting the importance of considering regional vascular and perivascular cell diversity to understand BBB heterogeneity and develop region-specific therapies.
Results
U.Clear reveals vascular differences in cortex and ME
We used U.Clear, an aqueous-based tissue-clearing protocol, to characterize ME and cortex blood vessels (Fig. 1a–e and Extended Data Fig. 1). U.Clear preserves endogenous fluorescence, permits the use of most antibodies to stain intact mouse tissues in their entirety and allows conventional confocal microscopy imaging. Consistent with previous reports in tissue sections21, after intravenous tracer injection, we observed tracer leak into the ME, but tracer remained confined to vessels in adjacent BBB-containing regions and the cortex in 3D (Fig. 1a, Extended Data Fig. 1a and Supplementary Video 1). As expected22,23, the ME vasculature lacks BBB markers glucose transporter 1 (GLUT1, encoded by Slc2a1) and claudin-5 (CLDN5) and expresses plasmalemma vesicle-associated protein (PLVAP) (Extended Data Fig. 1b–d). In addition, we found that the key BBB regulator, MFSD2 lysolipid transporter A (MFSD2A), was absent in the ME vasculature (Fig. 1b and Extended Data Fig. 1e).
a, Tracer leakage assay with tracer sulfo-NHS-biotin (magenta) and immunostaining for blood vessels (CD31, white) in cortex (upper panel) and ME (lower panel) following U.Clear. Tracer in circulation was washed out by perfusion before analysis. b, Co-immunostaining of CD31 (white) and MFSD2A (green) in cortex and ME. c, High-magnification images of capillaries (CD31) highlighting vessel morphology in cortex and ME (left) and quantification of vessel diameter (right) (n = 5 mice, three images per region in each mouse, with the same colors showing points from the same mice). Data presented as mean ± s.d., P = 3.604601 × 10−6, nested two-tailed t-test. d, High-magnification images of capillaries (CD31, white) and EC nuclei (ERG, red) in the cortex and ME (left). Quantification shows EC density, number of endothelial cell nuclei (ERG+) over the length of capillaries (n = 5 mice, three images per region in each mouse, with the same colors showing points from the same mice). Data presented as mean ± s.d., P = 4.569415 × 10−6, nested two-tailed t-test. e, Immunostaining and 3D reconstruction of three single Tomato+ ECs (red) within capillaries (CD31, white) in cortex and ME. Single ECs were labeled by a single low-dose injection of 4OH-tamoxifen in adult Cdh5-CreER:Ai14 mice 1 week before analysis. f, TEM images of a cortex capillary. Pseudocolors highlight different cells: cEC (E), pericyte (P), astrocyte endfoot (A), lumen (L) and neuropil. Insets show cEC tight junctions (white arrows), pericyte cells and astrocyte endfeet. g, TEM images of an ME blood vessel. Pseudocolors (as in f) highlight different cells: cECs, pericyte, fibroblast, lumen and neuropil. Insets show capillary fenestrations (white arrowheads), cEC tight junctions (white arrows), extracellular matrix-filled perivascular space (ECM), pericyte cells and fibroblast cells. h, Immunostaining for CD31 (red), mural cell marker PDGFRβ (white) and astrocyte endfoot marker aquaporin 4 (AQP4, green) in cortex (left) and ME (right).
Finally, with U.Clear, we observed distinct blood vessel morphologies in these regions. ME capillaries have a larger diameter and higher cEC density than cortex capillaries (Fig. 1c,d and Supplementary Video 2). Three-dimensional modeling after sparse labeling with an EC reporter (Cdh5-CreER; Ai14) revealed that ME capillary lumens are formed by more than one EC, while cortex capillary lumens comprised a single EC (Fig. 1e and Supplementary Video 3).
Vascular and perivascular cell organization in cortex and ME
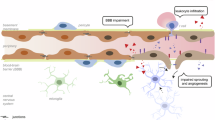
To understand how vascular and perivascular cells contribute to functional differences in BBB permeability, we first used transmission electron microscopy (TEM) to examine their interactions at the ultrastructural level. Cortex cECs are well known to interact closely with pericytes and astrocyte endfeet (Fig. 1f and Extended Data Fig. 2a). However, studies from several species indicated that ME perivascular cell interactions are quite different24. Indeed, we found that ME cECs are fenestrated and share a basement membrane with pericytes. Surprisingly, we did not see typical astrocyte endfeet surrounding ME capillaries. Rather, fibroblasts were present in a large perivascular space filled with extracellular matrix (Fig. 1g and Extended Data Fig. 2b,c). Tanycytes, specialized glial cells in CVOs2, were also not readily distinguishable in the ME parenchyma, which abuts the ME perivascular space on the dorsal side.
U.Clear revealed that cortex cECs, pericytes and astrocyte endfeet interact closely, and ME pericytes and fibroblasts (platelet-derived growth factor receptor beta (PDGFRβ)+) interact with cECs in the perivascular space (Fig. 1h). However, AQP4 is not polarized at ME astrocyte endfeet but rather is found throughout processes extending toward the ME vasculature (Extended Data Fig. 2d). These notable structural differences motivated us to identify their molecular underpinnings.
Regionally enriched cell types in cortex and ME by scRNA-seq
To identify molecular differences in ECs and perivascular cells, we performed inDrops scRNA-seq25,26 of the ME and a size-matched region of the cortex. We developed a tissue dissociation protocol to obtain efficient, unbiased recovery of vascular cells. All blood vessel-associated cell types are well represented in our dataset, with ECs comprising ~4% of cells, on par with estimates of their prevalence in the mouse brain. After quality control filtering (Methods), 58,117 high-quality cells were retained for further analysis; 35,879 from ME and 22,238 from the cortex. Unbiased cell clustering with Seurat identified 27 clusters corresponding to 11 cell types based on the expression of cell-type-specific transcripts (Fig. 2, Extended Data Fig. 3, Supplementary Figs. 1–3 and Supplementary Table 1). Notably, astrocytes from the ME and cortex clustered separately (Fig. 2 and Extended Data Fig. 3d,e).
a, Uniform manifold approximation and projection (UMAP) plot of 58,238 single-cell transcriptomes (35,934 from ME and 22,304 from cortex). Cell-type clusters were annotated post hoc based on their transcriptional profiles (Methods). The number of clusters identified for each cell type is indicated in the plot legend.
We first investigated EC regional differences by performing subclustering analysis, finding seven subtypes, including capillary ECs (cECs 1 and 2), arteriolar ECs (aECs 1 and 2) and venous ECs (vECs) (Fig. 3a, Extended Data Fig. 4 and Methods). ECs from the ME were found in all subclusters because ECs from ME-adjacent regions with BBB-containing blood vessels are inevitably included in our dissection. Despite this, an ME-specific EC subtype expressing Plvap emerged, which we confirmed by immunostaining (Fig. 3a, Extended Data Fig. 4a,b and Supplementary Table 1). Plvap-expressing ECs are cECs based on the expression of markers attributed to cECs20 (but not arteriolar or venous markers) and the absence of smooth muscle cells (which envelop arteries but not capillaries) (Extended Data Figs. 4a and 5a–c). Thus, we will refer to these cells as ‘ME cECs’. Finally, a small cluster of ECs was derived predominantly from the ME (36 out of 37 cells) that expressed markers characteristic of tip cells20, consistent with the characterization of this region as angiogenic27. Thus, we captured rare EC subtypes in small brain regions and ECs from all segments of the vascular tree, demonstrating that we can perform fine-grained molecular analysis and effectively investigate EC and perivascular cell heterogeneity with our method.
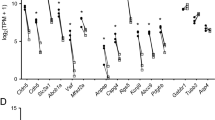
a, UMAP plot of 2,284 EC transcriptomes. Seven subtypes of ECs were identified with an unbiased analysis based on their transcriptional profiles (see Extended Data Fig. 4a). The number of each cell subtype profiled is indicated in the plot legend. b, Heatmap illustrating the average relative expression of regionally enriched genes in each subtype cluster identified in a that were validated by immunostaining. Regionally enriched genes show an average log2(fold change) > 0.6 (>1.5-fold change in expression) with an adjusted P < 0.05 by two-sided Wilcoxon test. c, Co-immunostaining for ME cEC-enriched EMCN (red) with CD31 (white) in cortex and ME. d, Co-immunostaining for ME cEC-enriched ESM1 (white), EMCN (red) and cortex cEC-enriched protein GLUT1 (green) in cortex and ME. e, Co-immunostaining for cortex cEC-enriched IGF1R (white), ME cEC-enriched protein EMCN (red) and cortex cEC-enriched protein GLUT1 (green) in cortex and ME. f, Co-immunostaining for cortex cEC-enriched BSG (white), GLUT1 (green) and EMCN (red) in cortex and ME.
ME and cortex cECs show transcriptional differences
We evaluated regional cEC differences, finding 445 differentially expressed genes (Extended Data Fig. 4b and Supplementary Table 1). We validated nine of these genes (summarized in Fig. 3b) by immunostaining: endomucin (EMCN) and endothelial cell-specific molecule 1 (ESM1) are expressed in ME cECs but not cortex cECs (Fig. 3c,d), whereas insulin-like growth factor 1 receptor (IGF1R), basigin (BSG) and SPARC/osteonectin, Cwcv and Kazal-like domains proteoglycan 2 (SPOCK2) (Fig. 3e,f and Extended Data Fig. 5d) are expressed in cortex cECs but are not detected in ME cECs (additional validation in Figs. 4f and 7d–f). These six cortex cEC-enriched genes are expressed in BBB-containing cECs throughout the brain, acting as common BBB-related genes. However, we anticipate that other cortex cEC-enriched genes may be expressed in a region-specific manner.
a, UMAP plot of 8,508 astrocyte transcriptomes. Astrocyte subtypes were identified with an unbiased analysis. The number of each cell subtype profiled is indicated in the plot legend. b, Violin plot showing the expression level of a subset of differentially expressed genes in each cluster identified in a. Differentially expressed genes show an average log2(fold change) of >0.6 (>1.5-fold change in expression) with an adjusted P value of <0.05 by two-sided Wilcoxon test. For comparison of ME astrocyte populations, coronal orientation is shown in each ME panel in c–f, with blood vessels on the bottom and the third ventricle toward the top. c, Left: Tomato in the cortex and ME of Slco1c1-CreER:Ai14 mice. Tomato (red) indicates Slco1c1 expression. Right: co-staining of CD31 (blood vessels, white). Note Tomato expression in cortex capillaries and astrocytes, but not ME capillaries and astrocytes. Single Tomato+ vessel in ME (green arrow) is arterial. Yellow arrows point at astrocytes in the cortex. d, Fluorescent labeling of astrocytes in cortex and ME using Glast-CreER:Ai14 mice after low-dose 4OH-tamoxifen to achieve sparse cell labeling. Top row shows immunostaining for Tomato+ astrocytes (red) and blood vessels (CD31, white). Yellow arrow indicates the ___location of the cell body, as determined by DAPI staining. Bottom row displays 3D reconstructions of astrocytes (red). Cells modeled in yellow are tanycytes. For a comparison of different astrocyte populations with the same scales, see Extended Data Fig. 7b. e, Left: GFP (green) in the cortex and ME of GFAP-GFP mice. Right: co-staining for CD31 (white, vessels). In the cortex, GFP only sparsely labeled peri-arterial astrocytes (yellow arrows). Scale bar, 50 µm in the top row and 20 µm in the middle and lower rows. f, Left: fluorescent labeling (green) of cortex astrocytes and ventricle-associated ME 2 astrocytes using Aldh1l1-GFP mice. Right: co-staining for CD31 (white) to label capillaries.
Gene set enrichment analysis of cEC differentially expressed genes corresponded to regional functional differences (Extended Data Fig. 5e and Supplementary Table 1). Cortex cEC genes were enriched for BBB-related pathways, including canonical Wnt signaling, in part owing to the expression of Lef1. We confirmed a lack of LEF1 activity in the ME28 (Extended Data Fig. 5f), consistent with reports of low Wnt activity in CVOs22,23. ME cEC-enriched genes were related to pathways like ghrelin signaling, in accordance with the role of ME in the hunger response29. Additionally, we observed enrichment for vascular endothelial growth factor (VEGF) signaling, in part owing to different expression patterns of VEGF receptors in ME and cortex cECs (Vegfr2 vs Vegfr1; Extended Data Fig. 4b and Supplementary Table 1).
Given that ME blood vessels share features with blood vessels in peripheral organs, we next compared gene expression patterns between ME or cortex cECs and cECs from peripheral tissues. We performed gene set enrichment for cell type signatures and determined the overlap of enriched genes in ME cECs, tip cells and cortex cECs with published datasets profiling cECs throughout the body20,30,31. Cortex cECs showed little similarity to peripheral ECs, whereas ME cECs showed overlap with ECs from the choroid plexus and the pancreas, kidney, colon and small intestine (Extended Data Fig. 6a–g), organs with fenestrated vessels. Moreover, a comparison of our data to ECs from the mouse pituitary gland23 and neurohypophysis32, which are adjacent to the ME, reveals some overlap (Extended Data Fig. 6h,i). Together, the ~400 molecular differences between cECs in the ME and cortex indicate that the differences in BBB permeability are at least in part a result of the molecular specialization of cECs.
ME astrocyte subtypes and their association with capillaries
Astrocyte endfeet ensheath brain capillaries with BBB properties; however, we found that ME astrocytes lack typical endfoot features (Fig. 1g,h and Extended Data Fig. 2), and astrocytes from each region clustered separately by scRNA-seq (Figs. 2 and 4a and Extended Data Fig. 7a–c). One gene, Slco1c1, previously known to be expressed in cortex astrocytes and cECs33, was absent in ME astrocytes and cECs (Figs. 3b and 4b). We validated this expression pattern with Slco1c1-CreER:Ai14 reporter mice (Fig. 4c and Extended Data Fig. 7d). Pathway analysis showed enrichment for ‘cell surface interactions at the vascular wall’ in cortex astrocytes and ‘GPCR signaling’ and ‘peptide-receptor interactions’ in ME astrocytes (Extended Data Fig. 7e).
Subclustering analysis identified four astrocyte subtypes, one from the cortex and three from the ME (Fig. 4a): ‘ME 1’, ‘ME 2’ and, consistent with previous reports in other CVOs, a subtype with high expression of Gfap predominantly (651 out of 678; 96%) from the ME (‘Gfap high’). Harmony analysis34 confirmed all subtypes are in a similar cell state (Extended Data Fig. 7f). We next compared these astrocyte subtypes to two published datasets that profiled multiple brain regions; our cortex astrocytes express markers of protoplasmic astrocytes from one of the datasets35 and Gfap-low frontal cortex astrocytes from the other36. Our ME astrocytes express markers of the diencephalon, and Gfap-high astrocytes express markers similar to ‘dorsal midbrain Myoc-expressing’ cells as reported in ref. 35 (Extended Data Fig. 7g,h). In ref. 36, ME subtypes showed similarity to substantia nigra and globus pallidus astrocytes (Extended Data Fig. 7i,j). Thus, ME astrocytes most likely represent novel subtypes.
We also found that ME and cortex astrocytes associate differently with blood vessels. We used reporters driven by Slc1a3 (Glast-CreER:Ai14) or Gfap (GFAP-EGFP37) to visualize individual astrocytes. Slc1a3 encodes GLAST and is expressed in both regions (Fig. 4b and Extended Data Fig. 7k). As expected, cortex GLAST+ astrocytes were stellate, with cell bodies situated away from the vasculature and extending numerous processes around blood vessels (Fig. 4d, Extended Data Fig. 71,m and Supplementary Video 4). GLAST+ ME astrocytes exhibited two morphologies (Fig. 4d, Extended Data Fig. 7l,m and Supplementary Video 4): one subtype was directly associated with ME blood vessels, nestled between the vessels with few, short processes, and the other subtype had cell bodies near the ventricle and long processes extending into the ME region but not associating with blood vessels. The third ME subtype, Gfap-EGFP+ astrocytes (‘Gfap high’) had cell bodies near the ventricle and extended numerous processes toward the vasculature (Fig. 4e, Extended Data Fig. 7n,o and Supplementary Video 4). To distinguish ME astrocyte subtypes 1 and 2, Aldh1l1-EGFP reporter mice were used. Aldh1l1 is expressed by ME 2 and Gfap-high astrocytes (Fig. 4b). Aldh1l1-expressing ME astrocytes have cell bodies near the ventricle and extend few processes toward the vasculature (Fig. 4f), indicating that ME 2 astrocytes correspond to the astrocytes interacting more distantly with the ME vasculature. Therefore, it is plausible that the ME 1 subtype represents the astrocytes nestled between ME blood vessels.
Thus, ME astrocytes are molecularly distinct from cortex astrocytes, lack endfeet typical of cortex astrocytes and show limited vascular association. In line with these differences, we found that most of the top 100 differentially expressed genes between ME and cortex astrocytes are predicted to be secreted or associated with the cell membrane (64% and 70%, respectively; Supplementary Table 1), suggesting that astrocyte molecular differences may be related to EC–astrocyte signaling (elaborated in Fig. 7).
Cortex pericyte–cEC interaction features by serial TEM
Given that cEC physical interaction with pericytes is important for the BBB, we first examined cortex cEC–pericyte interactions using serial TEM. We reconstructed pericytes and cECs in two capillaries from a published mouse visual cortex dataset38, as we expect features of this interaction to be present throughout the cortex. These reconstructions (Fig. 5a,b) show pericyte processes extending from the cell body along the length of blood vessels and wrapping around them (Supplementary Video 5). We quantified three features in four vessels (Fig. 5c and Extended Data Fig. 8a). First, we looked at close pericyte–cEC interactions, in which the extracellular matrix was not visible between the pericyte and EC, finding a close interaction in ~83% of sections on average. Next, we analyzed ‘peg and socket’ interactions, membrane invaginations between ECs and pericytes. These interactions were rare in vessel 2 (13.8% of sections), appeared more frequently in vessels 1 and 4 (46% and 50% of sections, respectively) and were prominent in vessel 3 (85% of sections). Finally, we quantified pericyte contact with EC tight junction clefts, finding this interaction more frequently in vessels 1 and 2 (64% and 40% of sections) than in vessel 3 (25% of sections). In vessel 4, tight junction clefts were not detectable in the ~6.5 μm analyzed (Extended Data Fig. 8b). Together, these findings detail pericyte–cEC interactions along the length of a cortex capillary. Consistent with a previous report39, we found frequent pericyte interactions with EC tight junctions, while ‘peg and socket’ interactions were concentrated in smaller domains.
a,b, Serial TEM reconstruction of EC and pericyte interactions in two cortex blood vessels. ECs are shown in green, pericytes in blue and the blood vessel lumen in red. Scale bars, 5 µm. c, Quantification of three features of pericyte–EC interaction in vessels in a and b and two additional vessels (n = 4 total vessels from one animal), displayed as a percentage of sections showing each feature. Each point represents a 50-section increment. Whiskers span the smallest and largest values, and the boxplot shows the median and first and third quantiles. d, Co-immunostaining for pan-pericyte marker PDGFRβ (red) and pan-EC marker ICAM2 (white) in cortex and ME. e, Immunostaining and 3D reconstruction of single Tomato+ pericytes (red) in touch with capillaries (CD31, white) in cortex and ME (two examples). Single pericytes labeled by single low-dose injection of 4OH-tamoxifen in adult Pdgfrb-CreER:Ai14 mice 1 week before analysis. f, Left: co-immunostaining for EC nuclei marker ERG (white) and pericytes labeled using Pdgfrb-CreER:Ai14; Pdgfra-GFP mice. Magenta arrowheads point at GFP−Tomato+ pericytes; cyan arrowheads point at ERG+ EC nuclei. Right: quantification of pericyte (GFP−Tomato+) to EC (ERG+) ratio using Pdgfra-H2B-GFP; Pdgfrb-CreER:Ai14 mice (n = 5 mice, for quantification three images per region and mouse were taken, same colors refer to same mice, data presented as mean ± s.d., P = 4.793057 × 10−6, nested two-tailed t-test). g, Co-immunostaining for pericyte marker desmin (DES, yellow), GFP (to visualize Pdgfra-H2B-EGFP in fibroblasts, green), EC marker CD31 (red) and nuclear marker Syto83 (blue) for GeoMX area of interest morphological identification. h, Volcano plot of differentially expressed genes between ME and cortex pericyte-enriched region of interest from GeoMX whole transcriptome profiling (also shown in Extended Data Fig. 8f). Differential expression was determined by linear mixed model analysis and significance assessed by FDR. Red points show log2(fold change) > |1| and FDR < 0.05 between cortex and ME pericyte-enriched regions of interest.
Distinct molecular and structural features of ME pericytes
Although brain pericytes have generally been viewed as homogenous across brain regions in rodents and humans40,41, we identified several regional differences. First, we observed striking morphological differences by immunostaining and sparse labeling in Pdgfrb-CreERT2:Ai14 reporter mice (Fig. 5d,e, Extended Data Fig. 8c,d and Supplementary Video 6). Cortex capillary-associated pericytes showed a characteristic ‘bump on a log’ morphology (a prominent cell body with long, thin processes extending along vessels), whereas ME pericytes had a more irregular shape (a less defined cell body found between blood vessels with processes of varying lengths). Additionally, although we observed cortex pericytes frequently interacting along a single vessel, some ME pericyte processes contacted several blood vessels. This interaction is reminiscent of pericyte–EC interactions in peripheral organs like the pancreas42.
Several studies have shown that a lower ratio of pericytes to ECs is associated with higher brain blood vessel permeability18,43,44. To quantify pericytes and ECs, we performed pericyte-specific labeling. Although Pdgfrb labels pericytes, it can also label fibroblasts, which surround ME cECs. Fibroblasts also express Pdgfra. Thus, we performed immunostaining for EC nuclei (Ets transcription factor (ERG)) in Pdgfra-H2B-GFP; Pdgfrb-CreERT2:Ai14 tdTomato reporter mice, quantifying tdTomato+GFP− pericytes and ERG+ ECs in each region. We found approximately half as many pericytes per EC in the ME than in the cortex (Fig. 5f), correlating with increased ME blood vessel permeability.
We next investigated molecular differences between ME and cortex pericytes using GeoMX whole transcriptome spatial profiling. As shown in Fig. 1a, leaky ME blood vessels are adjacent to non-leaky, BBB-containing blood vessels. Therefore, to gain higher resolution, we selected a spatial transcriptomic approach to unambiguously profile pericytes only from the ME region. As this method does not isolate single cells, we used Pdgfra-H2B-EGFP reporter mice and antibodies to distinguish fibroblasts (EGFP+), pericytes (DES+) and ECs (CD31+) (Fig. 5g). We selected pericyte-enriched areas around blood vessels in both regions and ME fibroblast-enriched areas as a control. We focused our analysis on genes expressed in pericytes (Methods and Supplementary Table 1). We confirmed pericyte enrichment based on expression of established markers (Extended Data Fig. 8e). We identified 137 differentially expressed genes between cortex and ME pericyte-enriched regions and 36 gene sets with differential enrichment by pathway analysis (Fig. 5h, Extended Data Fig. 8f and Supplementary Table 1). By immunostaining, we validated expression of one regionally enriched gene expressed in ME pericyte-enriched regions, Slc12a7 (encoding KCC4) (Extended Data Fig. 8g). Finally, we compared genes expressed in ME pericyte-enriched samples to human lung45, gut46 and kidney47 mural cells and the recently described T- and M-pericytes in the human brain40. In total, 20 out of 65 ME differentially expressed genes (30.8%) were found in mural cells from another peripheral tissue compared to 13 out of 77 (16.9%) of cortex differentially expressed genes (Extended Data Fig. 8h). Differentially expressed genes between ME and cortex pericyte-enriched regions and pericyte marker genes from our scRNA-seq dataset did not show enrichment for markers of human T-pericytes and M-pericytes (Extended Data Fig. 8i,j), and ME pericyte-enriched and fibroblast-enriched regions showed gene expression differences (Extended Data Fig. 9a,b and Supplementary Table 1).
In short, pericytes show different morphologies and capillary coverage in the ME and cortex. Although brain pericytes show transcriptomic differences from pericytes in the periphery19, our analysis revealed that brain pericytes may also show transcriptomic differences across regions. Together, these findings suggest that like astrocytes, pericytes probably contribute to BBB functional differences between the cortex and ME through their interactions with cECs (elaborated in Fig. 7).
Capillary-associated fibroblasts are present in the ME
Perivascular fibroblasts were observed previously in the cortex associated with large blood vessels19. Surprisingly, we found that only the ME contains numerous capillary-associated fibroblasts (Figs. 1g and 5f). To better characterize ME fibroblasts with TEM, we used a horseradish peroxidase (HRP) reporter driven by Pdgfra-CreERT2, finding fibroblasts in the ME extracellular space surrounding capillaries (Fig. 6a). We also used Pdgfra-CreERT2:Ai14 reporter mice to model fibroblast morphology with U.Clear. We found cortex fibroblasts along large vessels whereas ME fibroblasts were near capillaries, in accordance with our TEM data (Fig. 6b and Extended Data Fig. 9c,d).
a, TEM images of Pdgfra-CreER HRP reporter after DAB reaction in the ME. HRP is detected in the endoplasmic reticulum (white arrowheads) of Pdgfra-expressing fibroblast cells. b, Fluorescent labeling of fibroblasts in the cortex and ME using Pdgfra-CreER:Ai14 mice after low-dose 4OH-tamoxifen to achieve sparse cell labeling. Left: Tomato+ fibroblasts (red). Right column: merged with immunostaining for CD31 (white). Yellow arrow indicates artery. c, Co-immunostaining for fibroblasts with DECORIN (white) and collagen 1 (green) in cortex and ME.
Subclustering analysis of fibroblasts revealed three ME fibroblast subtypes (fibroblasts 1–3) and one subtype from the ME and cortex (fibroblast 4) (Extended Data Fig. 9e–h). Comparison of these subtypes to capillary-associated choroid plexus fibroblasts31 showed that ME-enriched subtypes exhibit similarity to third and fourth ventricle choroid plexus fibroblasts, whereas fibroblasts from subtype 4 are most similar to third ventricle meningeal fibroblasts (Extended Data Fig. 9i). Although all subtypes expressed Pdgfra, Dcn and Col1a1, we found that they were present only around ME capillaries (Fig. 6c and Extended Data Fig. 9j). Thus, the abundant fibroblasts near ME capillaries suggest that fibroblasts also have a role in regulating ME vascular permeability, perhaps by altering the composition of the extracellular matrix. Perivascular fibroblasts have also been observed near CNS capillaries in other CVOs and brain tumors48,49,50, indicating that fibroblasts may generally be associated with leaky CNS blood vessels in normal physiology and disease.
Bioinformatic method finds candidate ligand–receptor pairs
The proximity and direct physical interactions between cECs and perivascular cells suggested the feasibility of ligand–receptor interactions between these cells as a mechanism to regulate local blood vessel permeability. Indeed, a recent study showed such an interaction between pericytes and cECs at the BBB. Specifically, pericyte-secreted vitronectin interacts with receptor integrin alpha 5, expressed in neighboring cECs, to actively suppress transcytosis in cECs and thus maintain BBB integrity51. Similarly, recent studies indicate that the CVO environment contains cues to actively regulate leakiness22,23. To unbiasedly identify ligand–receptor pairs that may support intercellular signaling to regulate blood vessel permeability, we used CellChat52 and a co-expression method53. For the co-expression method, we sought to identify new interactions, supplementing a published database of experimentally validated interactions with predicted interactions of differentially expressed genes (Methods). By both methods, we identified known interactions important for the BBB, like PDGFβ–PDGFRβ between cECs and pericytes. Using CellChat, we found 25 and 33 enriched ligand–receptor pairs between cECs and pericytes in the cortex and ME, respectively, and 35 and 20 enriched ligand–receptor pairs between cECs and astrocytes in the cortex and ME, respectively (Extended Data Fig. 10a,b and Supplementary Table 1). Using the co-expression method, we found 37 and 62 enriched ligand–receptor pairs between cECs and pericytes in the cortex and ME, respectively, and 21 and 17 enriched ligand–receptor pairs between ECs and astrocytes in the cortex and ME (ME 1 astrocytes), respectively (Fig. 7a, Extended Data Fig. 10c,d and Supplementary Table 1). We confirmed the co-expression of one candidate cortex EC–astrocyte ligand–receptor pair: Bsg and Itga6. Immunostaining shows BSG expression in cortex cECs but not in ME cECs, and robust ITGA6 expression in cortex astrocyte endfeet—in addition to ECs—but decreased perivascular expression in the ME (Fig. 7b,c). Finally, extending our analysis to other ME cell types, we confirmed the expression of VEGFR2 (encoded by Kdr) in ME cECs and found co-expression of its ligand (VEGFA) in ME-specific tanycyte cells (Fig. 7d–f), in line with both methods.
a, Alluvial plot showing the number of significant (P < 0.05) co-expressed EC ligands and perivascular receptors. b, Co-immunostaining for BSG (white) and its receptor integrin α6 (ITGA6, red) in cortex and ME, validating elevated expression of ligand (BSG) and receptor (ITGA6) in cortex. c, Co-immunostaining for CD31 (white), AQP4 (green) and ITGA6 (red) in cortex. d, Co-immunostaining for VEGFR2 (red), EMCN (cyan, ME ECs) and GLUT1 (green, cortex ECs) in cortex and ME. EMCN and GLUT1 were used to label ECs instead of CD31 owing to antibody compatibility with VEGFR2. e, Co-immunostaining for VEGF (white), EMCN (cyan, ME ECs) and (GLUT1, green, cortex ECs) in cortex and ME. EMCN and GLUT1 were used to label ECs instead of CD31 owing to antibody compatibility with VEGF. f, Immunostaining illustrating complementary spatial distribution of ligand VEGF (white) and receptor VEGFR2 (red) in ME. Non-ME vessels are labeled in green (GLUT1).
This in silico analysis evaluated ligand–receptor expression patterns to identify candidate pericyte-derived and astrocyte-derived factors that may act upon ECs to maintain BBB integrity. Moreover, the co-expression method provides a molecular handle for future strategic investigation of EC–perivascular cell interactions. Future experimental characterization of this intercellular signaling, together with the fast-growing identification of additional signaling pathways in cECs, will provide a comprehensive understanding of local BBB regulation and heterogeneity.
Discussion
We combined scRNA-seq, spatial transcriptomic profiling, TEM and serial EM reconstruction, and U.Clear imaging to compare the vascular microenvironment of two brain regions showing BBB heterogeneity. In addition to molecular and morphological specializations of ECs and perivascular cells, we found distinct cell organization and identified putative ligand–receptor pairs that may mediate cell–cell signaling in the ME and cortex. These findings support the idea that intracellular signaling within ECs and intercellular signaling between ECs and perivascular cells control brain cEC permeability. This is in line with previous work showing that molecules like MFSD2A regulate permeability within ECs8,54, and that intercellular signaling with astrocytes through the Wnt pathway55 and pericytes through vitronectin–integrin regulate BBB formation51. This work serves as a foundation, revealing how alterations in cell interactions may control local blood vessel permeability and demonstrating the importance of performing both molecular and morphological characterizations to understand BBB properties.
The CVOs are key sites for body–brain communication. The leaky nature of CVO blood vessels permits rapid bidirectional communication between the circulation and the brain. Neurons that sense signals through the CVOs are being found to perform increasingly important and diverse functions related to body physiology regulation in health and disease. Multiple CVOs communicate with the hypothalamus56,57, a central regulator of temperature and sickness behavior during infection58,59, to coordinate humoral responses to environmental stimuli by affecting feeding behaviors, cardiovascular function and body temperature. Recent work also shows that CVOs may serve as an immune entry site in disease models60. This CVO vascular atlas can provide information to help understand how body–brain communication is achieved in this area to perform these essential functions. It has been also observed that blood vessels adjacent to the ME show increased leakiness during the physiological response to hunger3,61. Our molecular profiling of ME perivascular cells may facilitate future investigation of such plasticity by uncovering molecules and cell types involved in regulating CVO permeability.
Our transcriptomic analyses also lend further support to the idea that increased vascular permeability in the CNS is actively regulated and maintained by extrinsic factors in the local environment. Blood vessels in other CVOs, the choroid plexus and the choroid capillaries of the eye also show increased permeability2,62. Recent studies indicate that the permeability of these specialized vessels is not caused by a lack of barrier induction but is actively induced by the local microenvironment63. In choroid capillaries, VEGF secreted by the retinal pigment epithelium64 and inhibition of Wnt signaling regulate blood vessel permeability23. In the zebrafish pituitary, blood vessel permeability is induced by Cyp26b1, Tgfb and VEGF derived from pituicytes65. In the mouse area postrema, Wif1, which blocks Wnt activation, is expressed23. Although in all cases, permeability is locally induced in the perivascular environment, the cell source of these signals differs. By systematically analyzing ME cell–cell interactions, we found that VEGF is expressed in ME astrocytes and tanycytes and that ME cECs have enriched expression of VEGFR2. Additionally, in accordance with suppressed Wnt signaling in ME cECs (Extended Data Fig. 5f). The Wnt inhibitor Sfrp566 was expressed in ME astrocytes (Extended Data Fig. 7g). Moreover, the presence of ME capillary-associated fibroblasts, which are found in other leaky CNS regions, suggests that the extracellular matrix may also have a role in promoting vascular permeability in these regions.
Finally, this work revealed how performing scRNA-seq on small regions can uncover information about cell heterogeneity and the specialization of rare cell types within the brain. This platform can be applied in other brain regions to clarify how regional differences in cell organization and signaling affect BBB properties. Most CNS diseases affect specific brain regions. Therefore, alterations in cell signaling in disease could be driven by differences in the local composition or interactions of perivascular cells. Identifying the factors underlying BBB heterogeneity is an important step toward developing targeted therapies to make disease treatment as region-specific as possible.
Methods
Mice
All mouse experiments followed institutional and US National Institutes of Health guidelines and were approved by the Harvard University Institutional Animal Care and Use Committee. Mice were maintained on a 12 h light:12 h dark cycle at 22 °C and 55% humidity. Mice used in all experiments were 8–14 weeks old; both male and female mice were used unless otherwise indicated. The following mouse strains were used: wild type (C57BL/6N, Charles River Laboratories, strain 027), Ai14 (JAX, strain 007914)67, Aldh1l1-EGFP (JAX, strain 026033)68, GFAP-GFP (JAX, strain 003257)37, Glast-CreER (JAX, strain 012586)69, TCF/LEF-GFP (JAX, strain 032577)28, Cdh5-CreERT270, Slco1c1-CreERT233, Pdgfrb-CreERT2 (JAX, strain 029684)71, Pdgfra-H2B-EGFP (JAX, strain 007669)72, ROSA26LSL-ER-HRP (JAX, strain 034746) and Mfsd2ako (MMRRC, strain 032467-UCD)73.
Heterozygous Glast-CreER, Pdgfrb-CreERT2, Slco1c1-CreERT2 or Cdh5-CreERT2 mice were crossed with homozygous Ai14 mice to generate CreER-dependent reporter mice. Cell-type labeling was induced by five subsequent intraperitoneal injections of tamoxifen (1 mg per mouse). Brains were collected 2 weeks later. For sparse cell labeling, a single dose of 4OH-tamoxifen (0.4 mg per mouse) was injected 1 week before analysis. Given that the GLAST reporter can occasionally label tanycytes, which express Slc1a3 at a lower level than astrocytes, we distinguished tanycytes from ME astrocytes by morphology, expression of vimentin and cell body ___location relative to the third ventricle (Fig. 4d, Extended Data Fig. 7k,n,o and Supplementary Video 4).
U.Clear tissue clearing
U.Clear tissue clearing is a newly optimized protocol based on the Adipo-Clear framework74,75. In brief, mice were deeply anesthetized with ketamine and xylazine (100 mg kg−1) and subsequently intracardially perfused with cold 4% paraformaldehyde (PFA) in PBS. Brains were dissected and fixed overnight in 4% PFA at 4 °C. For claudin-5 staining, brains were perfused with cold PBS and drop-fixed in cold 100% methanol overnight before rehydrating brains in a series of 70% methanol–PBS, 30% methanol–PBS and PBS. After PBS washing, brains were dissected into a 5 × 5 × 5 mm cube of somatosensory cortex and a similarly sized cube of hypothalamus containing the ME. The resulting samples were delipidized by four washes (1 h, 2 h, 4 h, overnight) with SBiP buffer (200 µM Na2HPO4, 0.08% sodium dodecyl sulfate, 16% 2-methyl-2-butanol, 8% 2-propanol in H2O (pH 7.4)) at room temperature (~22 °C). Next, samples were transferred into B1n buffer (0.1% Triton X-100, 2% glycine, 0.01% 10 N sodium hydroxide, 0.008% sodium azide in H2O) for blocking under nutation at room temperature. On the next day, samples in B1n buffer were incubated at 37 °C for 1 h. For immunolabeling, samples were incubated at 37 °C for 2 days in primary antibodies diluted in PTxwH buffer (0.1% Triton X-100, 0.05% Tween-20, 0.002% heparin (w/v), 0.02% sodium azide in PBS) with gentle rocking. Samples were then washed four times with PTxwH (1 h, 2 h, 4 h, overnight). Samples were then incubated at 37 °C for 2 days in secondary antibodies diluted in PTxwH with gentle rocking and subsequently washed four times with PTxwH (1 h, 2 h, 4 h, overnight). For further delipidization, samples were immersed in SBiP buffer four times (1 h, 2 h, 4 h, overnight). Next, samples were washed twice in 0.5 mM Na2HPO4 (1 h, 2 h), twice in PB buffer (16 mM Na2HPO4, 4 mM NaH2PO4 in H2O) (1 h, 2 h) and finally twice in PTS solution (75% PB buffer, 25% 2,2’-thiodiethanol) (1 h, overnight), then equilibrated with histodenz gradient buffer with refractive index adjusted to 1.53 using thiodiethanol. Samples were stored at −20 °C until acquisition. To validate that BBB organization and morphology were intact following U.Clear, we performed a comparison to thick tissue sections (Extended Data Fig. 1f).
Immunohistochemistry
For anti-KCC4 (Slc12a7) and anti-CD31 co-immunostaining, wild-type brain tissue was perfused and fixed in 4% PFA and PBS as described above. The tissue was washed three times in PBS, equilibrated in 30% sucrose and PBS at 4 °C and flash-frozen in NEG-50 for cryosectioning. Then, 25 μm coronal sections were blocked for 1 h at room temperature in PBS plus 10% normal donkey serum and 0.1% Triton X-100, then incubated in blocking buffer with primary antibodies overnight at 4 °C. Samples were washed three times in PBS with 0.1% Triton X-100, then incubated in blocking buffer with secondary antibodies for 1 h at room temperature. Samples were washed three times in PBS with 0.1% Triton X-100 and stained with DAPI.
Antibodies
Primary and secondary antibodies used in this study are detailed in Supplementary Table 2.
A polyclonal antibody to the carboxyl terminus of mouse Mfsd2a was generated by New England Peptide using Institutional Animal Care and Use Committee-approved protocols. Rabbits were immunized with a KLH-conjugated peptide (Ac-CSDTDSTELASIL-OH). Antiserum was purified by peptide affinity column. Antibody specificity was validated in Mfsd2ako mice (Extended Data Fig. 1e).
Light microscopy
Cleared and stained brains and tissue sections were analyzed at high resolution with a Leica TCS SP8 confocal microscope. U.Clear Z-stacks were processed and 3D-reconstructed with Imaris software (v.9.3.1; Oxford Instruments). Immunostained sections were processed with FIJI (v.2.1.0). Photoshop CC and Illustrator CC (Adobe) were used for image formatting.
Image analysis
Capillary diameter, EC density and EC pericyte coverage were quantified from three ~50 µm-thick 40× confocal stacks of capillaries in cortex and ME per mouse. To measure capillary thickness, the area covered by three different capillaries in each image, labeled by CD31 immunostaining, was measured and divided by their respective vessel length. The average of these three diameters was used as the average capillary length for an image, represented as a single data point on a graph. To measure EC nuclei per vessel length, ERG+ EC nuclei were counted and total capillary length was measured. All analysis was performed blinded. Each data point in the graph represents an individual image. To measure pericyte coverage, EC nuclei were labeled with ERG antibody. Pericytes (GFP−, Tomato+) were identified using Pdgfra-H2B-GFP; Pdgfrb-CreERT2:Ai14 mice. All analysis was performed blinded. Each data point in the graph represents an individual image. In all analyses, data points from the same mice are depicted in the same color, values are shown as mean ± s.d. and significance was determined using a nested two-tailed t-test in GraphPad Prism (v.8). Data distribution was assumed to be normal, but this was not formally tested.
Tracer permeability assays
EZ-Link sulfo-NHS-LC-biotin tracer was injected retro-orbitally under short isoflurane anesthesia (0.2 mg g−1 bodyweight). Brains were dissected 30 min after injection after perfusion with 4% PFA as described above.
scRNA-seq and analysis
Sample isolation and dissociation
For each experimental replicate, cortex and ME were isolated from five 9-week-old male mice, pooled and processed together. Male mice were used to limit variations in the ME by the estrous cycle, as the ME is responsive to reproductive hormones2. Mice were killed at 08:00 h to avoid circadian cycle variation. Dissociation into single cells was performed using a protocol adapted from a previous publication76. Brains were dissected in ice-cold dissociation medium (DM; 1× Hank's balanced salt solution without calcium and magnesium, 0.01 M HEPES, 9 mM MgCl2, 35 mM d-glucose pH 7.35). First, the ME was removed then the brain was cut into 1 µm thick sections by sectioning matrix (Ted Pella). Cortex samples were obtained with a 1 mm biopsy puncher (Harris Uni-Core). Samples were dissociated using the papain dissociation system (Worthington) according to the manufacturer’s instructions with the following modifications. Dissociation was performed at 37 °C for 45 min with gentle agitation in DM plus papain (20 U ml−1) and DNase (200 U ml−1). After centrifugation with ovomucoid inhibitor in DM, cells were washed in DM containing 0.04% BSA and resuspended in DM with 0.04% BSA and 15% Optiprep (Sigma-Aldrich) for inDrops cell encapsulation, performed by the Single Cell Core at Harvard Medical School.
inDrops library preparation, sequencing and data processing
Two replicates of approximately 3,000 cells were collected from each experimental sample. inDrops was performed as described previously25,26 using v3 barcodes, generating 22 and 30 libraries from cortex and ME samples, respectively. Libraries were pooled and sequenced in 21 runs with the NextSeq 500 using the high output flow cell (Illumina), pooling 3,000 to 12,000 cells per sequencing run. Transcripts were processed with bcbio-nextgen inDrops3 data pipeline (v.1.2.8) using the default parameters.
Quality control and filtering
Analysis was performed with R77 (v.4.0.2) in RStudio78 using the Seurat analysis package (v.4.0.2)79,80,81. In each library, empty droplets were predicted using the EmptyDrops function82 and doublets were predicted with scrublet83. The levels of ambient RNA in ME and cortex were estimated separately using SoupX84. In brief, before filtering, all samples from each region were merged into a single dataset. Each dataset was clustered using the default Seurat analysis parameters to assign tentative cluster identities. Using the assumption that the background profile is the same as the average expression across all cells, we performed a custom estimation of the soup profile using the ‘setSoupProfile’ command. The corrected count matrices were then merged to generate the combined dataset.
To select for high-quality cells, we filtered based on number of genes expressed (at least 500), number of reads per cell (at least 750), percentage of mitochondrial genes (≤15%) and, by novelty index, the ratio of the number of genes detected to the number of reads for each cell (≥0.4). Clustering analysis was performed using the default Seurat analysis pipeline. In brief, the dataset was log-normalized and scaled to 10,000 transcripts per cell. Highly variable genes were determined with the Vst selection method using the default 2,000 features. All genes were then scaled across all cells so the mean expression of each gene is 0 and the variance of each gene is 1. Next, principal component analysis was performed using the calculated variable features.
The top 30 principal components were used in downstream steps based on the output of the ElbowPlot function. Clustering was performed at resolution 0.5 to identify broad cell types, resulting in 35 clusters. After clustering, cells predicted to be empty droplets (EmptyDrops output false discovery rate (FDR) > 0.01) and doublets (scrublet output score of >0.25) were removed from the dataset. Additionally, one cluster in which more than 70% of the cells had a doublet score of >0.25 and showed expression of marker genes of both neurons and oligodendrocytes was removed from the dataset. The dataset was then re-analyzed as above without these cells, resulting in 104,091 cells and 33 clusters. After subclustering analyses (described below), the dataset was re-analyzed to generate the final dataset of 58,117 cells in 30 clusters with an average number of unique molecular identifiers per cell of 4,283 and an average number of genes per cell of 2,197. We defined clusters as regionally enriched by the scProportionTest algorithm85 (v.0.0.0.9000).
Cell type assignment and subclustering analyses
Marker genes for each cluster were determined with the FindAllMarkers function using the default Wilcoxon test and the following parameters: only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25. To assign cell types, known marker genes were used (Supplementary Table 3).
Classification of cell subtypes
For iterative subclustering, analysis was performed as for the complete dataset described above unless indicated. Clusters that expressed marker genes characteristic of multiple cell types or that were isolated from only one sample were removed (described below), and the data were reclustered until all subtype clusters showed expression of known marker genes for the given cell type. ECs, astrocytes, neurons, oligodendrocytes, mural cells, fibroblasts and pars tuberalis cells were subclustered individually; tanycytes and ependymal cells, and microglia, PVMs and T cells were subclustered together.
ECs
ECs were subclustered four times to remove contamination from SMCs and pericytes and to remove clusters with ribosomal marker genes. This analysis resulted in seven subclusters that correspond to cECs, vECs, aECs, Plvap+ ECs and tip cells (Fig. 3 and Extended Data Fig. 4). aECs express the marker genes Bmx and Vegfc; vECs express marker genes Lcn2 and Nr2f2; and cECs lacked expression of arteriolar and venous marker genes and expressed marker genes Mfsd2a and Tfrc. cECs 1 and 2 differ in their expression of immediate early genes, probably because of EC activity-induced transcription76 or activation following tissue dissociation86. aECs 1 express arteriolar markers at a lower level than aECs 2, suggesting that they represent ECs at the capillary-to-arteriolar transition.
Mural cells
To identify mural cell subtypes, mural cells were subclustered three times (first at resolution 0.5, then at resolution 2) to remove clusters with EC marker genes, resulting in four clusters that include two pericyte subclusters and two smooth muscle cell clusters. One cluster was removed that expressed both EC and pericyte marker genes, considered doublets. The remaining clusters express pericyte or SMC marker genes, with or without activation markers.
Astrocytes
Subclustering analysis was performed first at resolution 2, then at resolution 0.4 to remove clusters expressing neuron and oligodendrocyte marker genes and ribosomal marker genes, resulting in four subclusters.
Fibroblasts
Initial subclustering analysis of fibroblasts (resolution 2, 20 principal components) revealed 12 subclusters. Subclusters expressing EC marker genes were removed and the dataset was reclustered (resolution 0.5, 20 principal components), resulting in four subclusters. The smallest subcluster contained cells from only one experimental replicate. These cells were removed and the data were reclustered (resolution 0.5, 20 principal components) to reveal four subclusters.
Microglia, PVMs and T cells
Subclustering analysis removed subclusters with vEC, oligodendrocyte and astrocyte marker genes, resulting in eight immune cell subtypes87,88.
Tanycytes and ependymal cells
Subclustering of tanycyte and ependymal cells (20 principal components, resolution 0.5) removed clusters with oligodendrocyte or PVM marker genes or ribosomal marker genes, revealing eight ME-derived subclusters, with subtypes consistent with previous reports89.
Oligodendrocytes
Subclustering analysis of oligodendrocytes removed astrocyte and immune cell contamination, revealing 12 subclusters, consistent with previous reports36,90.
Neurons
Subclustering analysis removed astrocyte and oligodendrocyte contamination and low-quality clusters with Malat1 or ribosomal genes as marker genes, resulting in 23 subclusters; 10 from the cortex and 13 from the ME.
Pars tuberalis
Subclustering analysis removed clusters with neuron, astrocyte and keratinocyte contamination, resulting in four subclusters.
Pathway enrichment analysis
Pathway enrichment analysis was performed as previously described91. In brief, curated gene sets (C2) and cell type signature gene sets (C8) were downloaded from the MSigdB (v.7.5.1)92,93; mouse gene IDs were converted to human homologs using SynGO94. Differentially expressed genes were calculated in Seurat using the two-sided Wilcoxon test as indicated. Pathway enrichment was determined using bc3net95 (v.1.0.4) with default parameters and plotted with the pheatmap function (v.1.0.12) as −log of the adjusted FDR.
Differential gene expression analysis and comparison to pituitary gland, neurohypophysis and peripheral ECs
Differentially expressed genes between cortex and ME cECs were calculated with the FindMarkers function (Wilcoxon test, min.pct > 0.25, avg_log2FC > 0.6). The intersection of the top 100 enriched genes in ME cECs, the top 100 marker genes in vascular ECs from the mouse neurohypophysis32 and the top 100 genes in pituitary ECs23 was displayed using ggvenn (v.0.1.9). For ECs from peripheral tissues, the top 50 enriched genes were compared to the top 50 marker genes from ME cECs, tip cells and cortex-derived cECs 1 subtypes. Barplots show the per cent overlap of the top 50 genes. Finally, the overlap between all enriched genes in kidney cECs, pancreas cECs30 and choroid plexus ECs31, and ME cECs-enriched differentially expressed genes was displayed using ggvenn (v.0.1.10).
Integration analysis of astrocytes and comparison to astrocyte subtypes
Integration analysis was performed with Harmony34 in Seurat using the RunHarmony function. Genes differentially enriched in astrocyte subtypes in a previous publication35 were visualized with VlnPlot and FeaturePlot features in Seurat. For comparison to aggregate metacell astrocyte subtypes from another publication36, astrocyte subtype aggregated expression was calculated with the AggregateExpression function in Seurat. Differentially expressed genes in astrocyte subtypes from this study were determined with the FindAllMarkers function (Wilcoxon test, min.pct > 0.25, avg_log2FC > 0.6), and samples from both studies were clustered based on the expression of these genes with pheatmap. Expression of the top 15 genes in the most similar astrocyte subtypes in a previous publication36 was also visualized with pheatmap.
GeoMX spatial whole transcriptomic profiling
Tissue preparation
Pdgfra-H2B-EGFP mice were anesthetized with ketamine and xylazine, then transcardially perfused with 15 ml of ice-cold PBS followed by 50 ml of ice-cold fixative solution (4% PFA in PBS). Brains were extracted and post-fixed for 3 × 20 min, then 4 h in fixative solution on ice. Brains were further fixed in fixative solution at 4 °C overnight, then for a final 4 h the next day. Brains were washed three times for 5 min in PBS before paraffin embedding. Then, 5 μm sections containing the ME and/or cortex were mounted on SuperFrost slides (Fisherbrand).
GeoMX digital spatial profiling
For digital spatial profiling (DSP), spatial transcriptomics was performed in the NanoString GeoMx DSP platform using the mouse whole transcriptome atlas for >19,000 transcripts of protein-encoding genes. The DSP workflow was carried out by NanoString Technologies through the Technology Access Program.
Slides were baked, deparaffinized and rehydrated using graded ethanol. Target epitope retrieval was performed with Tris-EDTA (pH 9.0), then proteinase K to expose RNA targets. In situ hybridization with the whole transcriptome probes was performed overnight. The next day, slides were washed to remove off-target probes and samples were stained with morphology markers, to distinguish ECs, pericytes and fibroblasts (Supplementary Table 2), and Syto83 (1:10; Invitrogen) to visualize cell nuclei. Fluorescent images were collected by a GeoMx DSP instrument for region of interest selection.
ME and cortex DES+GFP− areas (pericyte segments) and ME DES−GFP+ areas (fibroblast segments) were collected for transcriptional profiling. Labeled cells associated with large blood vessels were excluded as much as possible to reduce the collection of vascular smooth muscle cells. Additionally, samples were selected from the middle of the ME region to avoid smooth muscle cells (Extended Data Fig. 5a–c). In total, samples from eight animals of both sexes were profiled over three separate days for a total of 79 segments. Ultraviolet light was used to photocleave antibodies and release oligodendrocytes from areas of interest. Oligodendrocytes were collected and quantified by next-generation sequencing, and reads were processed into digital counts for data analysis.
DSP analysis
Data were analyzed in R (v.4.1.2) using GeomxTools (v.3.1.1).
Quality control
First, all segments passed a sequencing quality control assessment. Next, negative control probes were used to estimate background and downstream gene detection and to remove outliers. The limit of quantification of negative control probes in each region of interest was calculated to identify genes detected above background.
Several segments were removed because of low gene detection (<10% of the probes detected). Samples from six animals remained for analysis. Gene filtering was performed, resulting in 7,844 targets detected above the limit of quantification in 30% or more segments. Owing to differential distribution of cell types in the ME and cortex (Supplementary Table 1), genes attributed to astrocyte, L5.PT.CTX and EC profiles from a published study96 and genes from ME cECs, cortex cECs and tip cell subtypes were removed from the analysis (unless they overlapped with the annotated pericyte profile). A total of 2,921 genes remained from 53 of the 79 segments from four animals. Quartile three normalization was performed for genes in each segment. Principal component analysis followed by dimensional reduction showed that ME segments cluster separately from cortex segments and that ME pericytes and fibroblasts cluster separately. Coefficient of variation analysis of the top 292 genes (90th quantile) showed that these genes cluster by region and cell type.
Differential gene expression and pathway analysis
Differential gene expression analysis was performed on a per-gene basis, modeling normalized gene expression using a linear mixed-effect model to account for the sampling of multiple segments from each tissue. To compare ME and cortex pericyte segments, the following formula was used: gene ~ pericyte region segment + (1 + pericyte region segment per animal). To compare ME pericyte and fibroblast segments, the following formula was used: gene ~ cell type segment + (1 + cell type segment per animal). Differentially expressed genes were considered significant at a FDR < 0.05 and |log2(fold change)| > 1. Differentially expressed genes were compared to enriched genes from human mural cells from the lung45, gut46 kidney47 and brain40 and visualized by UpSetR (v.1.4.0)97. Differentially expressed genes were also compared to mouse choroid plexus pericytes31 and lung pericyte-enriched genes reported previously19. Finally, pathway analysis was performed with GSVA (v.1.46.0)98 using the KEGG BRITE database. A total of 337 gene sets were scored, with each gene set containing 5 and 500 genes. Enriched pathways were significant at FDR < 0.05. Plots were generated with the EnhancedVolcano (v.1.6.0) and UpsetR packages. We also found enrichment of eight gene sets and 22 genes in ME fibroblasts relative to ME pericytes (Extended Data Fig. 9a,b and Supplementary Table 1), including EGFP and cortex fibroblast markers Islr and Ptgds.
TEM
Mice were anesthetized with ketamine and xylazine, then transcardially perfused with cold, 5% glutaraldehyde, 4% PFA and 0.1 M sodium cacodylate. Brains were dissected and post-fixed overnight at 4 °C in fixative solution. Brains were then washed overnight in 0.1 M sodium cacodylate. Brains were washed three times for 15 min in 0.1 M sodium cacodylate, then sectioned into 100 μm coronal sections by vibratome. HRP reporter strains were processed with diaminobenzidine as described previously99. Regions of interest were microdissected, post-fixed in 1% osmium tetroxide and 1.5% potassium ferrocyanide, dehydrated and embedded in epoxy resin. Ultrathin sections of 80 nm were cut from the block surface, collected on copper grids and counter-stained with Reynold’s lead citrate before examination under a 1200EX electron microscope (JEOL) equipped with a 2k CCD digital camera (AMT).
Serial TEM reconstruction
Serial TEM data of the visual cortex was generated previously (450 μm × 450 μm × 50 μm volume)38. Capillaries were reconstructed in FIJI100 using TrakEM2 (ref. 101). ECs and pericytes were manually traced in each section in the dataset, then the images were rendered together to create a 3D reconstruction. Vessel 1 was reconstructed through 794 serial 40 nm sections or 31.8 μm, with 66 sections excluded. Vessel 2 was reconstructed in 490 serial 40 nm sections or 19.6 μm, with 28 sections excluded. Renderings were processed in blender (blender.org). Two additional vessels were analyzed for interaction features: vessel 3 over 22 μm and vessel 4 over 6.5 μm.
Ligand–receptor analysis
We analyzed our dataset with CellChat52 (v.1.6.0) and performed an analysis similar to that in a previous publication53. In brief, a ligand–receptor database was assembled. To facilitate the discovery of novel ligand–receptor interactions, the reference database was supplemented with differentially expressed genes in cECs and astrocytes (Supplementary Table 1). The predicted subcellular localization of each gene was determined using the Uniprot database102. For those genes known or predicted to be localized to the plasma membrane, secreted proteins or extracellular matrix components, the STRING database103 was queried to identify candidate interaction partners. Interaction partners with experimental validation were added to our ligand–receptor database. Established interactions53 are displayed in uppercase (for example, ‘PDGFB–PDGFRB’) and candidate ligand–receptor pairs are displayed in lowercase (for example, ‘Bsg–Itga6’). An interaction score was calculated for each ligand–receptor pair for two candidate interacting cell subtypes of interest by multiplying the average expression of the ligand gene in the candidate signaling cell and the average expression of the receptor gene in the candidate receiving cell. ME pericyte average expression data was generated by coercing the GeoMX data into a Seurat object (R v.4.1.3). An interaction score cutoff was determined by bootstrapping. In brief, the average expression of all genes in each cell subtype was calculated. For each iteration, the dataset was randomized with replacement, and interaction scores were calculated between the ligand-expressing cell subtype of interest and 2,192 random genes (the size of the supplemented database). This iteration was repeated 1,000 times to generate a null distribution of interaction scores. We focused on interaction scores >40, as these values were observed with a one-sided P value of <0.01 after Bonferroni correction for multiple comparisons (Supplementary Table 1). For the determination of unique ligand–receptor pairs in pericytes, pairs from the ME that were above this threshold in all cell types except ME pericytes were excluded, as we expect that they are probably a result of methodological differences.
Statistics and reproducibility
Data collection and analysis were performed blind to the conditions of the experiments where indicated. All representative immunofluorescence and TEM images were performed in three or more mice and repeated in at least three independent experiments. For confocal and electron microscopy data, we performed preliminary experiments to identify the variation. We then performed a power test to identify appropriate sample sizes of images per mouse.
A total of 52 total inDrops scRNA-seq samples were collected on 15 separate days, with two technical replicates from ME and cortex samples on each day (except for days 12–15, which were ME only). Sequencing libraries were generated over 9 days to minimize variation owing to library preparation. For GeoMX DSP, samples from eight animals of both sexes were profiled over three separate days. For transcriptomic experiments, sample sizes were chosen based on the yield of high-quality vascular cells. For scRNA-seq, we aimed to profile at least 100 cells per cluster from each region of our cell types of interest. For GeoMX, we based our sample size on the reproducible clustering of samples from multiple animals on separate experiment days both by sample region and enriched cell type.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The scRNA-seq data and GeoMX spatial profiling generated during this study are available for download at the Gene Expression Omnibus (GEO) (accession GSE241206). The analyzed scRNA-seq dataset has been uploaded to the Single Cell Portal (singlecell.broadinstitute.org/single_cell/study/SCP2553).
Code availability
The source code to run ligand–receptor analysis is available at GitHub (https://github.com/gulabneuro/scRNAseq-ligand_receptor).
References
Sweeney, M. D., Sagare, A. P. & Zlokovic, B. V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 14, 133–150 (2018).
Kaur, C. & Ling, E.-A. The circumventricular organs. Histol. Histopathol. 32, 879–892 (2017).
Jiang, H. et al. MCH neurons regulate permeability of the median eminence barrier. Neuron 107, 306–319.e9 (2020).
Hicks, A.-I., Kobrinsky, S., Zhou, S., Yang, J. & Prager-Khoutorsky, M. Anatomical organization of the rat subfornical organ. Front. Cell. Neurosci. 15, 691711 (2021).
Montagne, A. et al. Blood–brain barrier breakdown in the aging human hippocampus. Neuron 85, 296–302 (2015).
Al-Bachari, S., Naish, J. H., Parker, G. J. M., Emsley, H. C. A. & Parkes, L. M. Blood–brain barrier leakage is increased in Parkinson’s disease. Front. Physiol. 11, 593026 (2020).
Reese, T. S. & Karnovsky, M. J. Fine structural localization of a blood–brain barrier to exogenous peroxidase. J. Cell Biol. 34, 207–217 (1967).
Ben-Zvi, A. et al. Mfsd2a is critical for the formation and function of the blood–brain barrier. Nature 509, 507–511 (2014).
O’Brown, N. M., Pfau, S. J. & Gu, C. Bridging barriers: a comparative look at the blood–brain barrier across organisms. Genes Dev. 32, 466–478 (2018).
Langen, U. H., Ayloo, S. & Gu, C. Development and cell biology of the blood–brain barrier. Annu. Rev. Cell Dev. Biol. 35, 591–613 (2019).
Kaplan, L., Chow, B. W. & Gu, C. Neuronal regulation of the blood–brain barrier and neurovascular coupling. Nat. Rev. Neurosci. 21, 416–432 (2020).
Stewart, P. A. & Wiley, M. J. Developing nervous tissue induces formation of blood–brain barrier characteristics in invading endothelial cells: a study using quail-chick transplantation chimeras. Dev. Biol. 84, 183–192 (1981).
Armulik, A. et al. Pericytes regulate the blood–brain barrier. Nature 468, 557–561 (2010).
Bell, R. D. et al. Pericytes control key neurovascular functions and neuronal phenotype in the adult brain and during brain aging. Neuron 68, 409–427 (2010).
Daneman, R., Zhou, L., Kebede, A. A. & Barres, B. A. Pericytes are required for blood–brain barrier integrity during embryogenesis. Nature 468, 562–566 (2010).
Heithoff, B. P. et al. Astrocytes are necessary for blood–brain barrier maintenance in the adult mouse brain. Glia 69, 436–472 (2021).
Motoike, T. et al. Universal GFP reporter for the study of vascular development. Genesis 28, 75–81 (2000).
Armulik, A., Genové, G. & Betsholtz, C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell 21, 193–215 (2011).
Vanlandewijck, M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018).
Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779.e20 (2020).
Wang, Y., Smallwood, P. M., Williams, J. & Nathans, J. A mouse model for kinesin family member 11 (Kif11)-associated familial exudative vitreoretinopathy. Hum. Mol. Genet. 29, 1121–1131 (2020).
Benz, F. et al. Low Wnt/β-catenin signaling determines leaky vessels in the subfornical organ and affects water homeostasis in mice. eLife 8, 204 (2019).
Wang, Y. et al. Beta-catenin signaling regulates barrier-specific gene expression in circumventricular organ and ocular vasculatures. eLife 8, 3221 (2019).
Kobayashi, H., Matsui, T. & Ishii, S. Functional electron microscopy of the hypothalamic median eminence. Int. Rev. Cytol. 29, 281–381 (1970).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Zilionis, R. et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12, 44–73 (2016).
Morita, S., Ukai, S. & Miyata, S. VEGF-dependent continuous angiogenesis in the median eminence of adult mice. Eur. J. Neurosci. 37, 508–518 (2012).
Cho, C., Smallwood, P. M. & Nathans, J. Reck and Gpr124 are essential receptor cofactors for Wnt7a/Wnt7b-specific signaling in mammalian CNS angiogenesis and blood–brain barrier regulation. Neuron 95, 1056–1073.e5 (2017).
Schaeffer, M. et al. Rapid sensing of circulating ghrelin by hypothalamic appetite-modifying neurons. Proc. Natl Acad. Sci. USA 110, 1512–1517 (2013).
Feng, W., Chen, L., Nguyen, P. K., Wu, S. M. & Li, G. Single cell analysis of endothelial cells identified organ-specific molecular signatures and heart-specific cell populations and molecular features. Front. Cardiovasc. Med. 6, 165 (2019).
Dani, N. et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell 184, 3056–3074.e21 (2021).
Chen, Q., Leshkowitz, D., Blechman, J. & Levkowitz, G. Single-cell molecular and cellular architecture of the mouse neurohypophysis. eNeuro 7, 1–14 (2020).
Ridder, D. A. et al. TAK1 in brain endothelial cells mediates fever and lethargy. J. Exp. Med. 208, 2615–2623 (2011).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Zeisel, A. et al. Molecular architecture of the mouse nervous system. Cell 174, 999–1014.e22 (2018).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030.e16 (2018).
Zhuo, L. et al. Live astrocytes visualized by green fluorescent protein in transgenic mice. Dev. Biol. 187, 36–42 (1997).
Lee, W.-C. A. et al. Anatomy and function of an excitatory network in the visual cortex. Nature 532, 370–374 (2016).
Ornelas, S. et al. Three-dimensional ultrastructure of the brain pericyte-endothelial interface. J. Cereb. Blood Flow Metab. 41, 2185–2200 (2021).
Yang, A. C. et al. A human brain vascular atlas reveals diverse mediators of Alzheimer’s risk. Nature 603, 885–892 (2022).
Xia, M. et al. Single-cell RNA sequencing reveals a unique pericyte type associated with capillary dysfunction. Theranostics 13, 2515–2530 (2023).
Almaça, J., Weitz, J., Rodriguez-Diaz, R., Pereira, E. & Caicedo, A. The pericyte of the pancreatic islet regulates capillary diameter and local blood flow. Cell Metab. 27, 630–644.e4 (2018).
Mäe, M. A. et al. Single-cell analysis of blood–brain barrier response to pericyte loss. Circ. Res. 128, e46–e62 (2021).
Nikolakopoulou, A. M. et al. Pericyte loss leads to circulatory failure and pleiotrophin depletion causing neuron loss. Nat. Neurosci. 22, 1089–1098 (2019).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Elmentaite, R. et al. Cells of the human intestinal tract mapped across space and time. Nature 597, 250–255 (2021).
He, B. et al. Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes. Nat. Commun. 12, 2141 (2021).
Bouchaud, C., Le Bert, M. & Dupouey, P. Are close contacts between astrocytes and endothelial cells a prerequisite condition of a blood–brain barrier? The rat subfornical organ as an example. Biol. Cell 67, 159–165 (1989).
Duda, D. G. et al. Malignant cells facilitate lung metastasis by bringing their own soil. Proc. Natl Acad. Sci. USA 107, 21677–21682 (2010).
Schaffenrath, J. et al. Blood–brain barrier alterations in human brain tumors revealed by genome-wide transcriptomic profiling. Neuro. Oncol. 23, 2095–2106 (2021).
Ayloo, S. et al. Pericyte-to-endothelial cell signaling via vitronectin–integrin regulates blood–CNS barrier. Neuron 110, 1641–1655.e6 (2022).
Jin, S. et al. Inference and analysis of cell–cell communication using CellChat. Nat. Commun. 12, 1088 (2021).
Kumar, M. P. et al. Analysis of single-cell RNA-seq identifies cell–cell communication associated with tumor characteristics. Cell Rep. 25, 1458–1468.e4 (2018).
Andreone, B. J. et al. Blood–brain barrier permeability is regulated by lipid transport-dependent suppression of caveolae-mediated transcytosis. Neuron 94, 581–594.e5 (2017).
Guérit, S. et al. Astrocyte-derived Wnt growth factors are required for endothelial blood–brain barrier maintenance. Prog. Neurobiol. 199, 101937 (2021).
Yu, S., François, M., Huesing, C. & Münzberg, H. The hypothalamic preoptic area and body weight control. Neuroendocrinology 106, 187–194 (2018).
Saper, C. B. Neurobiological basis of fever. Ann. N Y Acad. Sci. 856, 90–94 (1998).
Hrvatin, S. et al. Neurons that regulate mouse torpor. Nature 583, 115–121 (2020).
Osterhout, J. A. et al. A preoptic neuronal population controls fever and appetite during sickness. Nature 606, 937–944 (2022).
Schulz, M. & Engelhardt, B. The circumventricular organs participate in the immunopathogenesis of experimental autoimmune encephalomyelitis. Cerebrospinal Fluid Res. 2, 8–8 (2005).
Langlet, F. et al. Tanycytic VEGF-A boosts blood–hypothalamus barrier plasticity and access of metabolic signals to the arcuate nucleus in response to fasting. Cell Metab. 17, 607–617 (2013).
Brinks, J. et al. Exploring the choroidal vascular labyrinth and its molecular and structural roles in health and disease. Prog. Retin. Eye Res. 87, 100994 (2021).
Ben-Zvi, A. & Liebner, S. Developmental regulation of barrier- and non-barrier blood vessels in the CNS. J. Int. Med. 292, 31–46 (2022).
Marneros, A. G. et al. Vascular endothelial growth factor expression in the retinal pigment epithelium is essential for choriocapillaris development and visual function. Am. J. Pathol. 167, 1451–1459 (2010).
Anbalagan, S. et al. Pituicyte cues regulate the development of permeable neuro-vascular interfaces. Dev. Cell 47, 711–726.e5 (2018).
Tong, S. et al. Sfrp5/Wnt pathway: a protective regulatory system in atherosclerotic cardiovascular disease. J. Interferon Cytokine Res. 39, 472–482 (2019).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Tsai, H.-H. et al. Regional astrocyte allocation regulates CNS synaptogenesis and repair. Science 337, 358–362 (2012).
de Melo, J. et al. Injury-independent induction of reactive gliosis in retina by loss of function of the LIM homeodomain transcription factor Lhx2. Proc. Natl Acad. Sci. USA 109, 4657–4662 (2012).
Wang, Y. et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010).
Gerl, K. et al. Inducible glomerular erythropoietin production in the adult kidney. Kidney Int. 88, 1345–1355 (2015).
Hamilton, T. G., Klinghoffer, R. A., Corrin, P. D. & Soriano, P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 23, 4013–4025 (2003).
Tang, T. et al. A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28, 749–755 (2010).
Chi, J. et al. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226–236.e3 (2018).
Chi, J., Crane, A., Wu, Z. & Cohen, P. Adipo-clear: a tissue clearing method for three-dimensional imaging of adipose tissue. J. Vis. Exp. 137, 58271 (2018).
Hrvatin, S. et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci. 21, 120–129 (2018).
R Core Team. R: a language and environment for statistical computing (R Foundation for Statistical Computing, 2020).
RStudio Team. RStudio: Integrated Development for R. R Studio, PBC.
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291.e9 (2019).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience 9, giaa151 (2020).
Miller, S. A. et al. LSD1 and aberrant DNA methylation mediate persistence of enteroendocrine progenitors that support BRAF-mutant colorectal cancer. Cancer Res. 81, 3791–3805 (2021).
Lacar, B. et al. Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun. 7, 11022 (2016).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019).
Li, Q. et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10 (2019).
Campbell, J. N. et al. A molecular census of arcuate hypothalamus and median eminence cell types. Nat. Neurosci. 20, 484–496 (2017).
Chen, R., Wu, X., Jiang, L. & Zhang, Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. 18, 3227–3241 (2017).
Habib, N. et al. Disease-associated astrocytes in Alzheimer’s disease and aging. Nat. Neurosci. 23, 701–706 (2020).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Koopmans, F. et al. SynGO: an evidence-based, expert-curated knowledge base for the synapse. Neuron 103, 217–234.e4 (2019).
de Matos Simoes, R. & Emmert-Streib, F. Bagging statistical network inference from large-scale gene expression data. PLoS ONE 7, e33624 (2012).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e26 (2021).
Conway, J. R., Lex, A. & Gehlenborg, N. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33, 2938–2940 (2017).
Hänzelmann, S., Castelo, R. & Guinney, J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 14, 7 (2013).
Zhang, Q., Lee, W.-C. A., Paul, D. L. & Ginty, D. D. Multiplexed peroxidase-based electron microscopy labeling enables simultaneous visualization of multiple cell types. Nat. Neurosci. 22, 828–839 (2019).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Cardona, A. et al. TrakEM2 software for neural circuit reconstruction. PLoS ONE 7, e38011 (2012).
The UniProt Consortium. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 47, D506–D515 (2018).
Szklarczyk, D. et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2018).
Acknowledgements
We would like to thank members of the Gu laboratory for comments on this manuscript; the Single Cell Core at Harvard Medical School (HMS) for performing the scRNA-seq sample preparation; the Nanostring Technology Access Program; E. Raviola for advice and discussion; S. Hrvatin, M. A. Nagy and R. Kirschner for advice and technical assistance; Q. Zhang and D. Ginty for providing R26-LSL-HRP mice; M. Schwanniger for providing Slco1c1-CreER mice; and R. Adams for providing Cdh5-CreERT2 mice. Imaging, consultation and/or services were in part performed in the Neurobiology Imaging Facility. This facility is supported in part by the HMS and Boston Children's Hospital (BCH) Center for Neuroscience Research as part of a National Institute of Neurological Disorders and Stroke (NINDS) P30 Core Center grant (NS072030). Electron microscopy imaging, consultation and services were performed in the HMS Electron Microscopy Facility. S.J.P was supported by the Damon Runyon Cancer Research Foundation (DRG-2289-17). U.H.L. was supported by an EMBO (European Molecular Biology Organization) long-term fellowship (ALTF 42-2017). R.A.L. was supported by the Howard Hughes Medical Institute Exceptional Research Opportunities Program. Large-scale serial EM dataset generation and resource availability was supported by the National Institutes of Health (NIH) (R21NS085320 and R01NS108410) to W-C.A.L. C.G. is a Howard Hughes Medical Institute Investigator. The research of C.G. was supported in part by a Faculty Scholar grant from the Howard Hughes Medical Institute. This work was also supported by the Fidelity Biosciences Research Initiative (C.G.), an Allen Distinguished Investigator Award, a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Family Foundation (C.G.), the NINDS (R35NS116820 and DP1NS092473 to C.G.) and the National Cancer Institute (DP1NS092473 to C.G.). Research reported in this publication was also supported by the National Institute On Drug Abuse of the NIH under award number RF1DA048786 to C.G. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
S.J.P, U.H.L and C.G. conceived the project, designed the experiments and wrote the manuscript with input from all authors. S.J.P. and U.H.L. performed experiments and data analysis. T.F., I.P. and F.N. performed 3D reconstruction of serial TEM data. R.A.L. performed immunostainings in Fig. 7f. W.-C.A.L. provided technical guidance for serial TEM analysis. Z.W. provided technical guidance and experimental resources for U.Clear experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Dritan Agalliu and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Morphological, molecular and functional differences of the vasculature between the ME and cortex.
(a) Tracer leakage assay with Sulfo-NHS-Biotin (magenta) and immunostaining for blood vessels (CD31, white) in cortex (upper panel) and ME (lower panel). Tracer in circulation was washed out by perfusion prior to analysis. Scale bar 10 µm. (b) Co-immunostaining of GLUT1 (green) and PLVAP (red) in ME and cortex. Scale bar 100 µm. (c) High magnification images of capillaries showing distinct Glut1 (green) and Plvap (red) expression pattern and vessel morphology in the cortex and ME. Scale bar 20 µm. (d) Co-immunostaining of CD31 (white) and tight junction protein Cldn5 (green) in cortex and ME. Scale bar 10 µm. (e) Validation of specificity of newly generated polyclonal antibody against MFSD2A (green) by immunostaining of cortex from wild type and Mfsd2ako mice. Scale bar 100 µm. (f) Immunostaining of CD31 (white) in the ME in thick tissue section. Scale bar 100 µm.
Extended Data Fig. 2 TEM reveals differences in organization of the vasculature and cellular environment in the ME and cortex.
(a) TEM images of a cortical capillary. As outlined in the legend, pseudocolors highlight different cells: cEC (purple), pericyte (teal), astrocyte endfoot (cyan), lumen (L, white) and neuropil (yellow). Insets show cEC tight junctions (white arrows), pericyte cells (P, teal) and astrocyte endfeet (A, cyan). Scale bar represents 1 µm (left). (b) TEM images of two blood vessels in the ME, (i) and (ii). As outlined in the legend, pseudocolors highlight different cells: cEC (purple), pericyte (teal), fibroblast (red), lumen (L, white) and neuropil (yellow). Insets show capillary fenestrations (white arrowheads), cEC tight junctions (white arrows), extracellular matrix-filled perivascular space (ECM), pericyte cells (P, teal) and fibroblast cells (F, red). Scale bar represents 1 µm. (c) TEM images of two groups of ME blood vessels. Scale bar represents 4 µm. (d) Co-immunostaining for astrocyte endfoot marker Aqp4 (green) and CD31 (white) in cortex and ME. Scale bar 20 µm.
Extended Data Fig. 3 Single cell RNA sequencing of median eminence and a size-matched region of somatosensory cortex reveals unique cell types in each brain region.
(a) Bar plot showing distribution of cells in each cell type for each experimental replicate. Replicates 12–15, in bold, are from the ME region only. (b) Bar plot showing distribution of cells in each cell type for each library batch preparation. Batch 9, highlighted in bold, is comprised of libraries from the ME region only. (c) Dot plot showing average expression of one cell type-specific transcript used to annotate cluster cell types in Fig. 2. Additional transcripts used for annotation are detailed in Methods. (d) Tukey box and whisker plot depicting the number of genes detected per cell in all identified clusters in Fig. 2. Box shows the median and first and third quartiles, whiskers represent 1.5 times the interquartile range. The cell number in each cluster per sample region is indicated at the right of each plot, with cortex-enriched clusters highlighted in red and ME-enriched clusters highlighted in blue (as determined in (e)). Data was collected on 15 separate days, with two technical replicates from ME and cortex samples in each replicate (except for replicates 12–15, which were ME only). ME and cortex regions were isolated from the same 5 mice in each replicate and pooled by region prior to dissociation. (e) Point-range plot showing the relative differences in cell proportions for each cluster in (d) between the ME and cortex. Regional enrichment was determined by permutation test, with significance assessed by false discovery rate following bootstrapping (implemented by the scProportionTest R package). Clusters showing regional enrichment (log2 fold-change greater than |4.5| and FDR < 0.05) are labeled and shown in red.
Extended Data Fig. 4 Novel regionally enriched genes identified in cECs in the ME and cortex.
(a) Dot plot of average expression of vascular zonation markers and several transcripts used to annotate cluster cell types in Fig. 3a. Actb and Cdh5 are expressed in all populations, and Cldn5 and Slc2a1 are cortex-enriched transcripts, as illustrated in Extended Data Fig. b-d. Lcn2, Vcam1, Lrg1, and Slc38a5 are enriched in vECs; Nr4a1, Jun and Fos are enriched in cECs 2; Apln, Aplr and Trp53i11 are enriched in tip cells; Vegfc, Sema3g and Gkn3 are enriched in aECs 1 and 2; Tfrc and Mfsd2a are enriched in cECs 1 and 2; Emcn, Esm1, and Plvap are enriched in ME cECs; Unc5b is enriched in aECs 1; Rgcc and Kdr are enriched in capillary ECs (cECs 1 and 2 and ME cECs). (b) Heatmap illustrating the top 75 differentially expressed genes from each group when comparing cortex-derived cECs 1 and cECs 2 with ME cECs. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat (minimum percentage = 25%, log2-fold change > 0.6, adjusted p-value < 0.05). (c) Heatmap illustrating endothelial activity-induced transcripts (reported in Hrvatin, et al.76) that are differentially expressed between cECs 1 and cECs 2. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat (minimum percentage = 10%, log2-fold change > 0.25, adjusted p-value < 0.05). (d) Percentage of EC subtypes sequenced by experimental replicate. Experimental replicates with only ME samples are highlighted in blue. (e) Percentage of EC subtypes sequenced by library batch. Library batches with only ME samples are highlighted in blue.
Extended Data Fig. 5 Immunostaining reveals ME capillary boundary and validates differential gene expression across regions.
(a) Co-immunostaining with anti-CD31 (white) and anti-SMA (red) antibodies to visualize arteries in the cortex and ME. Scale bar 200 µm. (b) High magnification images of blood vessels (CD31, white), highlighting vessels interacting with SMA-positive smooth muscle cells (red) in cortex (top) and ME (bottom). Scale bar 20 µm. (c) Co-immunostaining with anti-CD31 (white) and anti-PLVAP (red) antibodies in the ME. Yellow arrows indicate arteries in this region. Scale bar 50 µm. (d) Co-immunostaining of SPOCK2 (white), GLUT1 (green) and EMCN (red) in the cortex and ME of P5 wild type mouse. Scale bar 10 µm. (e) Heatmap showing significantly upregulated Reactome pathways for the top 115 differentially expressed genes in cortex-derived cECs 1 and 2 and ME cECs. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat comparing cortex-derived cECs 1 and 2 with ME cECs (minimum percentage = 25%, log2-fold change > 0.6, adjusted p-value < 0.05). p-value was calculated with a one-sided Fisher’s exact test, and -log(FDR) values are shown. Upregulated pathways from other pathway databases can be found in Supplementary Table 1. (f) Co-immunostaining for CD31 (white), endothelial nuclei marker ERG (red), and GFP in cortex and ME of TCF/LEF-GFP Wnt-signaling reporter mice. GFP expression (green) indicates activation of the Wnt-signaling pathway. Arrows indicate ERG and GFP double positive nuclei. Scale bar 50 µm.
Extended Data Fig. 6 Comparison of scRNAseq of ME-derived Plvap + ECs to published ECs from the mouse neurohypophysis, pituitary gland and peripheral organs.
(a) Heatmap showing the top 10 most similar cell types when analyzing the top 115 differentially expressed genes for each cEC subtype. Differentially expressed genes were determined by two-sided test in Seurat comparing each subcluster to all other astrocyte cell subclusters with min.pct=0.25 and logFC>0.6. Hypergeometric p-value was calculated and -log(FDR) values are shown. Brain cell types are highlighted in blue, while peripheral cell types are highlighted in green. (b) Table showing genes common between the top 50 differentially expressed genes in Plvap ECs, tip cells and cECs1 (calculated by two-sided Wilcoxon test in Seurat comparing each EC subtype to all other ECs with min.pct=0.25 and logFC>0.6) and marker genes of ECs from the choroid plexus from Dani et al.31. (c) Bar plot showing the percentage overlap of the top 50 enriched genes from Plvap ECs, tip cells and cECs 1 EC subtypes (calculated by two-sided Wilcoxon test in Seurat comparing each EC subtype to all other ECs with min.pct=0.25 and logFC>0.6) and each of the top 50 enriched genes in ECs reported from Kalucka et al.20. (d) Table showing genes common between Plvap+ ECs, tip cells and cECs 2 in the organs with the highest level of overlap in (c). (e) Bar plot showing the percentage overlap of the top 50 enriched genes from Plvap ECs, tip cells and cECs 1 EC subtypes (calculated by two-sided Wilcoxon test in Seurat comparing each EC subtype to all other ECs with min.pct=0.25 and logFC>0.6) and each of the top 50 enriched genes in ECs reported from Feng et al.30. (f) Table showing genes common between Plvap+ ECs, tip cells and cECs 1 in the organs with the highest level of overlap in (e). (g) Venn diagram showing the overlap between EC marker genes from the kidney and pancreas from Feng et al.30 and the choroid plexus from Dani et al.31 with marker genes of Plvap ECs (calculated by two-sided Wilcoxon test in Seurat comparing Plvap ECs to all other ECs with min.pct=0.25 and logFC>0.6). The 9 genes common to all samples are listed in the center. (h) The top 100 genes enriched in ME-derived Plvap+ ECs when compared to cortex-derived cECs (as in (a), blue) were compared to the top 100 genes enriched in vascular ECs from the mouse neurohypophysis32 (gray) and the top 100 genes enriched in ECs isolated from the mouse pituitary gland23 (red). Plvap+ EC enriched genes were calculated by two-sided Wilcoxon test in Seurat comparing each EC subtype to all other ECs. (i) Table containing the identities of the 8 genes expressed by all samples compared in (h).
Extended Data Fig. 7 Astrocyte subtypes and their interactions with blood vessels are distinct between the ME and cortex and from published datasets.
(a) Heatmap illustrating the top 5 genes differentiating each astrocyte cell subcluster. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat comparing each subcluster to all other astrocyte cell subclusters. (b) Percentage of astrocyte subtypes sequenced by experimental replicate. Experimental replicates with only ME samples are highlighted in bold (12–15). (c) Percentage of astrocyte subtypes sequenced by library batch. Library batches with only ME samples are highlighted in bold (9). (d) Fluorescent labeling of cortex blood vessels and astrocyte populations (Tomato, red) in cortex and ME using Slco1c1-CreER:Ai14 mice. Co-staining for blood vessels (CD31, white). Scale bar 100 µm. (e) Heatmap showing the significance of upregulated pathways for the top 50 differentially expressed genes for each astrocyte subtype. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat comparing each subcluster to all other astrocyte cell subclusters with min.pct=0.25 and logFC>0.6. p-value was calculated with a one-sided Fisher’s exact test, and -log(FDR) values are shown. (f) UMAP projections of astrocyte transcriptomes following Harmony cross-correlation analysis. (g) Violin plots showing the expression of GLAST (Slc1a3), Sfrp5, and of markers reported previously35 of telencephalon astrocytes, expressed in cortex astrocytes (Lhx2, Foxg1, Mfge8) and diencephalon astrocytes expressed in ME astrocytes (Gfap, Aqp4, Slc6a11, Agt, Slc7a10, Fgfr3, Cldn10, Igsf1, Itih3, Ntsr2). (h) UMAP plots highlighting the expression of markers (purple) reported previously35 of Myoc-expressing astrocytes in Gfap high population (outlined in black). (i) Clustergram showing similarity of ME and cortex astrocyte subtypes to those reported previously36. The expression patterns of the top 25 differentially expressed genes in the 4 astrocyte subtypes (calculated by two-sided Wilcoxon test in Seurat comparing each astrocyte subtype to all other astrocytes, with min.pct = 0.25 and logFC > 0.6) were clustered in aggregate metacells using the pheatmap R package with the default parameters. (j) Heatmap (scaled by row) illustrating the top 15 genes distinguishing the FC_8-1_Gfap- astrocytes (FC), GP_5-1.Astrocyte.Gja1.Gfap (GP), and SN_7-2_Astrocyte.Gja1.Cst3 (SN) subtypes previously reported36. Genes expressed in at least 2 of the above astrocyte subtypes are labeled as ‘multiple.’ (k) Violin plot showing expression of Slc1a3, which encodes GLAST, in astrocyte and tanycyte populations. (l) Fluorescent labeling of astrocyte populations (Tomato, red) in cortex and ME using Glast-CreER:Ai14 mice with a high dose of tamoxifen. Co-staining for blood vessels (CD31, white). Scale bar 50 µm. (m) Fluorescent labeling of astrocyte populations in the cortex and ME using Glast-CreER:Ai14 mice after a low dose of tamoxifen to achieve sparse cell labeling. Upper row: immunostaining for Tomato-positive astrocytes (red) and blood vessels (CD31, white). Lower row displays 3D reconstructions of astrocytes (red). Scale bar 20 µm (upper) and 10 µm (lower). (n) Co-immunostaining for GFAP-enriched astrocyte population (GFAP, red) and tanycyte marker Vimentin (yellow). Scale bar 30 µm. (o) Co-immunostaining for tanycyte marker VIMENTIN (yellow) and pan EC marker CD31 (white) in ME. Ventral and coronal view of ME shown, note tanycyte protrusions in touch with ME vessels. Scale bar 30 µm (upper panel) and 15um (lower panel).
Extended Data Fig. 8 Mural cells associated with cortex and ME blood vessels show distinct morphology and transcriptomic differences.
(a) Representative cross section images of (i) vessel 1 and (ii) vessel 2 reconstruction by serial TEM. Pseudocolors show reconstructed regions: blood vessel lumen (L, red); EC (green); and pericyte cell (blue). White arrows indicate EC tight junctions (purple), and white boxes highlight ‘peg and socket’ pericyte-EC interactions. Scale bar represents 1 µm. (b) Representative image of cortex vessel 4 from serial TEM dataset. Scale bar represents 1 µm. (c) Immunostaining for PDGFRβ (white, left panels) and Imaris 3D reconstruction of pericytes (white, middle and right panels) in cortex and ME. Co-staining for Glut1 (green) and Emcn (red) to mark capillaries in the cortex and ME, respectively. Scale bar 10 µm. (d) High magnification images of reconstructed pericytes (PDGFRβ, white) in contact with capillaries (Glut1, green and Emcn, red) in cortex and ME. Arrows point at ME pericyte protrusions not in contact with capillaries. Scale bar 5 µm. (e) Violin plots showing expression of published markers of brain pericytes in both ME and cortex areas of interest from GeoMX whole transcriptome profiling. (f) Heatmap of differentially expressed genes (log2 fold-change greater than |1| and FDR < 0.05) between cortex and ME pericyte-enriched regions of interest from GeoMX whole transcriptome profiling (also shown in Fig. 5h). Differential expression was determined by linear mixed model analysis and significance assessed by false discovery rate (FDR). (g) Immunostaining for CD31 (green) and SLC12A7 (red) in ME and cortex in coronal tissue sections. Scale bar 10 µm. (h) Upset plot showing overlap between human pericyte cell type signatures and differentially expressed genes in ME and cortex pericyte-enriched regions. (i) Upset plot showing overlap between human brain pericyte cell type signatures and ME and cortex pericyte-enriched differentially expressed genes (Fig. 5h). (j) Upset plot showing overlap between human pericyte cell type signatures and pericyte marker genes from our scRNAseq dataset (calculated by two-sided Wilcoxon test in Seurat comparing pericytes to other mural cells, with min.pct=0.25 and logFC>0.6).
Extended Data Fig. 9 Fibroblasts in the ME.
(a) Heatmap of differentially expressed genes between ME pericyte- and fibroblast-enriched areas of interest from GeoMX whole transcriptome profiling. (log2 fold-change greater than |1| and FDR < 0.05). Differential expression was determined by linear mixed model analysis and significance assessed by false discovery rate (FDR). (b) Upset plot showing overlap between ME fibroblast-enriched regions from GeoMx whole transcriptome profiling and fibroblasts from choroid plexus in Dani et al.31. (c) 3D reconstructions of fibroblasts in the ME and cortex are shown in red (based on Pdgra-CreER:Ai14 Tomato expression). Immunostaining for CD31 is shown in white. Scale bar 15 µm. (d) Another 3D view of (g) showing the ___location of fibroblasts below ME blood vessels. Scale bar 5 µm. (e) UMAP plot of 714 fibroblast transcriptomes. Fibroblast subtype clusters were identified with an unbiased analysis. The number of cells identified for each subtype is indicated in the plot legend. (f) UMAP plot in (a) colored by sample region. (g) VlnPlot showing the expression of Cola1a, Dcn and Pdgfra in fibroblast subclusters. (h) Heatmap illustrating the top 5 genes differentiating each fibroblast subcluster. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat comparing each subcluster to all other mural cell subclusters with min.pct=0.25 and log2_FC > 0.6 thresholds. (i) Table showing overlap of fibroblasts 1 and 4 subtypes with fibroblast topics from Dani et al.31. Differentially expressed genes were determined by two-sided Wilcoxon test in Seurat comparing the fibroblast 1 subcluster to the fibroblast 4 subcluster, with min.pct =0.25 and log2_FC > 0.6 thresholds. (j) Co-immunostaining for fibroblast-enriched protein, DECORIN (white) and Tomato in cortex and ME. Blood vessels labelled with Tomato using Cdh5-CreERT2:Ai14 mice. Scale bar 20 µm.
Extended Data Fig. 10 Differences in perivascular cell signaling capacity in the ME and cortex.
(a, b) Bubble plots from CellChat analysis showing ligand-receptor pairs between (b) cortex ECs and pericytes and astrocytes and (c) ME Plvap ECs and astrocytes, tanycytes, pericytes and fibroblasts. ME pericyte data is from the scRNAseq dataset. p-values were calculated by permutation test in the CellChat R package. (c) Overview of ligand-receptor analysis methodology and workflow. (d) EC ligand-receptor interaction scores with pericyte, astrocyte, tanycyte and fibroblast receptors in the ME or cortex with values > 40 and p-values < 0.01. Candidate, ME-enriched ligand-receptor interaction investigated in Fig. 7 is highlighted in blue. Candidate, cortex-enriched ligand-receptor interaction investigated in Fig. 7 is highlighted in red. Uppercase ligand-receptor pairs are from Kumar et al. database. ME pericyte data is from GeoMX spatial transcriptomic profiling.
Supplementary information
Supplementary Information
Supplementary Tables 1–3, Supplementary Videos 1–6, Supplementary Figs. 1–3
Supplementary Table 1
scRNAseq cell type- and subtype-enriched genes, pathway analysis, spatial transcriptomics data and ligand–receptor interaction scores
Supplementary Video 1
Sulfo-NHS-biotin leakage in cortex and ME. Immunostaining for EC marker CD31 (white) and BBB leakage tracer (sulfo-NHS-biotin, magenta) showing no leakage of tracer in cortex and tracer leaking out of vessels in ME.
Supplementary Video 2
Morphology of capillaries in cortex and ME. High magnification images of capillaries immunostained for CD31 (white) show morphology of cortex and ME capillaries.
Supplementary Video 3
Morphology of ECs in cortex and ME. Imaris 3D reconstruction of single ECs (red) in cortex and ME labeled with tdTomato in Cdh5-CreERT2:Ai14 mouse. Co-staining for EC marker CD31 (white).
Supplementary Video 4
Morphology of astrocytes in cortex and ME. Imaris 3D reconstruction of single astrocytes (red) and tanycytes (yellow, in ME) in cortex and ME labeled with Tomato in Slc1a3-CreERT2:Ai14 mouse. Co-staining for EC marker CD31 (white) and Imaris 3D reconstruction of single GFAP+ astrocyte (green) in ME labeled with GFP in GFAP-EGFP mouse. Co-staining for EC marker CD31 (white).
Supplementary Video 5
Serial TEM blood vessel reconstructions. 3D reconstruction of two blood vessel-pericyte interactions from a serial TEM dataset of the visual cortex, highlighting pericytes (blue), an endothelial cell (green) and the blood vessel lumen (red).
Supplementary Video 6
Morphology of pericytes in cortex and ME. Imaris 3D reconstruction of single pericytes (red) in cortex and ME labeled with Tomato in Pdgfrb-CreERT2:Ai14 mouse. Co-staining for EC marker CD31 (white).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pfau, S.J., Langen, U.H., Fisher, T.M. et al. Characteristics of blood–brain barrier heterogeneity between brain regions revealed by profiling vascular and perivascular cells. Nat Neurosci 27, 1892–1903 (2024). https://doi.org/10.1038/s41593-024-01743-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-024-01743-y
This article is cited by
-
Molecular and cellular characteristics of cerebrovascular cell types and their contribution to neurodegenerative diseases
Molecular Neurodegeneration (2025)
-
Optimized enrichment of murine blood–brain barrier vessels with a critical focus on network hierarchy in post-collection analysis
Scientific Reports (2025)
-
Immune control of brain physiology
Nature Reviews Immunology (2025)
-
PD-L1/PD-1 checkpoint pathway regulates astrocyte morphogenesis and myelination during brain development
Molecular Psychiatry (2025)