Abstract
Plant metabolism faces a challenge of investing enough enzymatic capacity to a pathway without overinvestment. As it takes energy and resources to build, operate, and maintain enzymes, there are benefits and drawbacks to accurately matching capacity to the pathway influx. The relationship between functional capacity and physiological load could be explained through symmorphosis, which would quantitatively match enzymatic capacity to pathway influx. Alternatively, plants could maintain excess enzymatic capacity to manage unpredictable pathway influx. In this study, we use photorespiration as a case study to investigate these two hypotheses in Betula papyrifera. This involves altering photorespiratory influx by manipulating the growth environment, via changes in CO2 concentration and temperature, to determine how photorespiratory capacity acclimates to environmental treatments. Surprisingly, the results from these measurements indicate that there is no plasticity in photorespiratory capacity in B. papyrifera, and that a fixed capacity is maintained under each growth condition. The fixed capacity is likely due to the existence of reserve capacity in the pathway that manages unpredictable photorespiratory influx in dynamic environments. Additionally, we found that B. papyrifera had a constant net carbon assimilation under each growth condition due to an adjustment of functional rubisco activity driven by changes in activation state. These results provide insight into the acclimation ability and limitations of B. papyrifera to future climate scenarios currently predicted in the next century.
Similar content being viewed by others
Introduction
Anthropogenic activities are rapidly changing the composition and thermal conditions of the global atmosphere, leading to fundamental trade-offs in photosynthetic carbon metabolism. Burning fossil fuels has and continues to liberate enormous quantities of CO2 and other greenhouse gases into the atmosphere, which has increased global temperatures due to their role in radiative heat transfer1. The most recent Intergovernmental Panel on Climate Change (IPCC) report indicates that global surface temperatures have increased faster in the last 50 years due to anthropogenic greenhouse gas release than in any other 50-year period in the previous 2000 years2. The influx of CO2 into the atmosphere will directly alter photosynthetic carbon metabolism, in addition to operating at elevated temperatures, as atmospheric CO2 and O2 are substrates for the initial carbon-fixing enzyme of photosynthesis, rubisco.
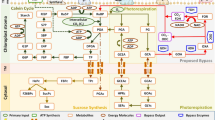
Rubisco has dual substrate affinity for both CO2 and O2, with each acting as a competitive inhibitor to the other3,4,5,6,7. The fixation of CO2 through rubisco carboxylation (vc) initiates net carbon assimilation (A) through the Calvin-Benson cycle, while the fixation of O2 through rubisco oxygenation (vo) initiates photorespiration (Fig. 1)3,6,8. The enrichment of atmospheric CO2 concentrations and warming of the atmosphere exert opposite effects on rates of rubisco vc and vo. Considering just an increase in CO2 concentration, rubisco catalyzes more carboxylation reactions due to the heightened partial pressure of CO2 surrounding the enzyme9. Conversely, a warmer atmosphere enhances rates of rubisco oxygenation through temperature-dependent shifts in rubisco specificity for CO2 relative to O2 and a decrease in CO2/O2 gas solubility10,11,12. Additionally, prior to rubisco vc or vo, rubisco must be carbamylated to prime the catalytic sites of the enzyme to accept CO2 or O213,14. The generation of misfire products which prevent further reactions in the active site of rubisco can reduce the efficiency of the enzyme, and conformational changes are required by rubisco activase to restore functionality15,16,17. Rubisco activase, the chaperone protein that promotes these conformational changes, regulates functional rubisco activity17,18,19,20,21,22,23. Unlike rubisco, the activity of rubisco activase is influenced only by temperature (the rate of rubisco activation by rubisco activase decreases with rising temperatures), but not by the CO2 concentration surrounding the enzyme20. Although past research has elucidated the mechanisms of rubisco under elevated CO2 concentrations and increased temperatures, our understanding of how/if photorespiration, downstream of rubisco, acclimates to these environmental changes is limited.
Following the oxygenation of RuBP by rubisco in the chloroplast and the formation of phosphoglycolate (2-PG), phosphoglycolate phosphatase (PGP) converts 2-PG into glycolate. Glycolate is transported to the peroxisome where glycolate oxidase (GO) catalyzes the conversion of glycolate and O2 to glyoxylate and hydrogen peroxide (H2O2). H2O2 is decomposed in the peroxisome into H2O and O2 by catalase (CAT), while glyoxylate is animated with glutamate or alanine to produce glycine via aminotransferase (GGAT or AGAT). Glycine in transported into the mitochondrion and decarboxylated to produce serine by the glycine decarboxylase complex and serine hydroxymethyltransferase. Serine is transported back to the peroxisome and converted to hydroxypyruvate by serine glyoxylate aminotransferase (SGAT). Hydroxypyruvate is reduced by hydroxypyruvate reductase (HPR) to form glycerate. Glycerate is transported back to the chloroplast and converted by glycerate kinase (GLYK) to 3-PGA, which re-enter the C3 cycle. Image reproduced from: Catalase protects against nonenzymatic decarboxylations during photorespiration in Arabidopsis thaliana/Bao et al./Plant Direct Volume 5/Issue 12. Copyright (c) [2021] authors hold copyright and have given permission to reproduce.
Specifically, it is unknown whether photorespiratory capacity downstream of rubisco acclimates to changes in growth CO2 concentration and temperature to maintain vo or whether excess capacity exists in photorespiration constitutively. This capacity of photorespiration is set by the maximal reaction velocity (Vmax) for the enzymes downstream of rubisco. While rubisco sets the rate of 2-phosphoglycolate (2-PG) production through vo (photorespiratory influx), the enzymes downstream of rubisco must process the subsequent photorespiratory intermediates that are produced. Some of these reactions process biologically inert photorespiratory intermediates, like glycine, but others degrade biologically active intermediates, such as 2-phosphoglycolate (2-PG), glycolate, and H2O2. If there is mismatch between photorespiratory influx and the capacities of downstream photorespiratory reactions, photorespiratory intermediates are likely to accumulate due to insufficient conversion rates. Accumulation of either 2-PG or glycolate under photorespiratory pressure can lead to a decrease in A through inhibition of triose phosphate isomerase and sedoheptulose-1,7-bisphosphatase, or through interference with rubisco activity and RuBP regeneration, respectively24,25,26,27,28,29,30. Accumulation of H2O2, a signaling molecule involved in both stress and developmental processes, leads to cell death when catalase-deficient Nicotiana tabacum mutant plants were exposed to high photorespiratory pressure31. Managing these intermediates is thus important to maintain plant vigor, especially in changing environments where photorespiratory influx is highly dynamic.
Photorespiratory influx can change in short- (seconds to days) or long-term (weeks-years) time scales depending on the growth environment. Daily photorespiratory influx can increase due to heat wave anomalies, causing a greater carbon efflux associated with photorespiration compared to historic averages32. While at the opposite end, C3 plants adapted to hot-arid environments, like Rhazya stricta, have double the rates of vo compared to Nicotiana tabacum when grown together under identical growth temperatures in a glasshouse33. Whether there are transient or permanent changes to photorespiratory influx, it is unclear if photorespiratory enzymes, downstream of rubisco, acclimate their capacities to manage changes in vo.
There are two main hypotheses that could explain the relationship between photorespiratory influx and downstream metabolic capacity. One concept to describe the relationship between functional capacity and physiological load is symmorphosis. Symmorphosis means that the capacity optimally matches the demand placed on the system34,35,36. Symmorphosis in the case of photorespiration would exist if the enzymatic capacities (Vmax) quantitatively match the vo pressure tied to growth conditions, such that photorespiratory capacity would be optimized to photorespiratory influx. A benefit to symmorphosis for photorespiratory capacity is optimal energy expenditure. As it takes energy and resources to build, operate, and maintain enzymes, optimally matching capacity to influx would alleviate wasted energy that could go into growth or fitness. Alternatively, another hypothesis to explain the structure–function relationship would be the presence of a reserve capacity embedded into the pathway37,38. We will refer to this reserve or excess capacity as a “safety factor”. The presence of safety factors in the photorespiratory pathway would appear as higher capacities (Vmax) of the downstream enzymes than appears to be needed, but that could manage dynamic photorespiratory influx in fluctuating conditions, while maintaining A. The benefit of overinvestment in photorespiratory capacity would be efficient degradation of photorespiratory intermediates so that minimal to no accumulation of these intermediates would occur. A downside to this overinvestment would be that the excess energy needed for photorespiratory enzyme synthesis, operation, regulation, and maintenance to keep capacity high could otherwise be devoted elsewhere in the plant.
To explore these hypotheses, we determined the plasticity of enzyme capacity across the photorespiratory pathway over six different CO2 concentration and temperature growth environments in Betula papyrifera. B. papyrifera is ideal for studying plasticity of photorespiration under future conditions as it is native to the boreal forest, a biome predicted to experience the most significant increase in temperature compared to all forest biomes by 2100, concurrent with rising atmospheric CO2 concentrations39,40,41. Since vo in absolute terms and the ratio of vo/vc are sensitive to both CO2 concentration and temperature, these environmental shifts should influence photorespiratory influx during growth. The growth environments were designed using a factorial approach which aimed to mimic current, moderate, and extreme climate change scenarios for the boreal region40,42. To test the validity of these hypotheses in B. papyrifera, biochemical and physiological mechanisms were probed using biochemical assays with gas-exchange data from a previous study to determine if photorespiratory capacity acclimates to environmental treatments.
In the present study, we concluded that there is no plasticity in photorespiratory capacity in B. papyrifera grown under different CO2 concentrations and temperatures. Instead, a fixed photorespiratory capacity is maintained, likely due to the existence of safety factors embedded in the pathway. Interestingly, the initial reactions in the photorespiratory pathway have lower safety factors than later ones, highlighting potential enzymatic bottlenecks that may limit the rate of reaction due to low enzyme activities. Additionally, we found that B. papyrifera had a constant A under each growth condition due to an adjustment of functional rubisco activity driven by changes in activation state. These results provide insight into the acclimation ability and limitations of B. papyrifera to future climate scenarios currently predicted in the next century.
Results
B. papyrifera photorespiratory enzyme activities across six growth environments
To assess the acclimation potential of photorespiratory capacity in B. papyrifera, we measured chloroplastic and peroxisomal enzyme activities under saturating substrate concentration to determine an in vitro Vmax (Figs. 2, 3; Supplemental Figures 2, 3; Supplemental Tables 1, 2). These enzyme activities were measured in leaves of B. papyrifera grown under 6 different environmental conditions at 25 °C and 35 °C using crude protein extract. The 6 different growth conditions were designed to represent ambient, moderate, and extreme future climate scenarios, where CO2 concentrations (either ambient or elevated (AC or EC)) and air temperature (ambient, ambient + 4 °C and ambient + 8 °C (T0, T4, T8)) differed. The enzyme assayed were rubisco (RBC), phosphoglycolate phosphatase (PGP), glycolate oxidase (GO), catalase (CAT), glutamate glyoxylate aminotransferase (GGAT), alanine glyoxylate aminotransferase (AGAT), serine glyoxylate aminotransferase (SGAT), hydroxypyruvate reductase (HPR), and glycerate kinase (GLYK). B. papyrifera had similar RUB, PGP, GO, CAT, GGAT, AGAT, SGAT, HPR, and GLYK activities per leaf area (and on chlorophyll and protein basis) when grown under any of the six growth environments at 25 °C and 35 °C assay temperatures (Figs. 2, 3; Supplemental Figures 2, 3). GO was the only enzyme to have a significant growth temperature effect at 35 °C assay temperature. Thus, the maximal activities of the photorespiratory enzymes do not acclimate in parallel to vo and appears to have the same enzymatic photorespiratory capacity regardless of environmental growth conditions.
Chloroplastic photorespiratory enzyme activities in B. papyrifera at 25 °C and 35 °C. Specific activities per m2 leaf area were measured in B. papyrifera using crude protein extract for rubisco (total activity), phosphoglycolate phosphatase, and glycerate kinase. Colors represent temperature treatments, with ambient temperature in yellow, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. No hatching pattern denotes ambient CO2 concentration, while hatching represents elevated CO2. Shown are the boxplots as well as the points indicating the biological replicates (n = 6). Significant difference between treatment types is indicated by letters as determined by Two-way ANOVA and Tukey–Kramer ad hoc test with p < 0.05.
Peroxisomal photorespiratory enzyme activities in B. papyrifera at 25 °C and 35 °C. Specific activities per m2 leaf area were measured in B. papyrifera using crude protein extract for peroxisomal enzymes glycolate oxidase, catalase, hydroxypyruvate reductase, glutamate glyoxylate aminotransferase, alanine glyoxylate aminotransferase, and serine glyoxylate aminotransferase. Colors represent temperature treatments, with ambient temperature in yellow, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. No hatching pattern denotes ambient CO2 concentration, while hatching represents elevated CO2. Shown are the boxplots as well as the points indicating the biological replicates (n = 6). Significant difference between treatment types is indicated by letters as determined by Two-way ANOVA and Tukey–Kramer ad hoc test with p < 0.05.
To determine whether downstream enzyme activities scale with rubisco activity, the initial enzyme of the photorespiratory pathway, downstream enzyme’s activities were plotted against total rubisco activity using the entire kinetic dataset measured for each enzyme (Fig. 4). Linear regressions were fitted to establish correlation to total rubisco activity. The activity of the enzymes PGP, GO, CAT, SGAT, HPR, and GLYK all had significant (p-value < 0.01) positive correlations to rubisco activity, while the activity of GGAT, and AGAT were not correlated with rubisco activity.
Downstream photorespiratory enzyme activities correlation to rubisco activity in B. papyrifera. A correlation of the specific activities per m2 leaf area of the eight downstream photorespiratory enzymes (phosphoglycolate phosphatase, glycolate oxidase, catalase, glutamate glyoxylate aminotransferase, alanine glyoxylate aminotransferase, serine glyoxylate aminotransferase, hydroxypyruvate reductase, and glycerate kinase) versus rubisco are visualized in B. papyrifera using crude protein extract. Colors represent temperature treatments, with ambient temperature in yellow, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. Shape represents CO2 concentration, with ambient CO2 being circles and elevated CO2 being triangles. Linear regressions are fitted with the corresponding p-values (p < 0.01) and adjusted R2.
The temperature response ratio (or Q10) of the enzyme activities were calculated by dividing the activity at 35 °C by the activity at 25 °C (Supplemental Fig. 4). The Q10 of RBC, PGP, GO, CAT, GGAT, AGAT, SGAT, HPR, and GLYK were not affected by growth CO2 concentration or temperature. RBC, PGP, GO, CAT, HPR, and GLYK had Q10 above 1, indicating a thermal dependence of enzyme activity. However, the GGAT, AGAT, and SGAT had Q10 equal to or below 1, indicating aminotransferases activity is likely temperature independent in B. papyrifera in the range assayed (25 °C and 35 °C).
B. papyrifera rubisco deactivates under elevated CO2 conditions and high temperature
In vivo net carbon fixation is driven by functional rubisco activity rather than total rubisco activity, which is heavily regulated and only considers the activity of rubisco with open active sites. To evaluate the response of in vivo rubisco activity to different growth environments in B. papyrifera, rubisco activation state was measured (Fig. 5). B. papyrifera grown under T0AC and T4AC had a higher rubisco activation state than trees grown under T4EC and T8EC conditions. When compared against the active rubisco at current climate conditions (T0AC), rubisco deactivates by 3.3% (T4AC), 13.1% (T8AC), 15.6% (T0EC), 24.7% (T4EC), and 31.4% (T8EC). The two-way ANOVA reveals a clear CO2 concentration effect (p-value < 0.001) and a temperature effect (p-value < 0.05), but no interactive effect (Supplemental Table 4).
Rubisco activation state. Initial and chemically activated Specific activities per m2 leaf area were measured in B. papyrifera using crude protein extract for rubisco and divided to give activation state on a percentage basis. Colors represent temperature treatments, with ambient temperature in yellow, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. No hatching pattern denotes ambient CO2 concentration, while hatching pattern represents elevated CO2. Significant difference between treatment types is indicated by letters as determined by Two-way ANOVA and Tukey–Kramer ad hoc test with p < 0.05.
B. papyrifera rubisco carboxylation is not affected by CO2 concentration or temperature, but rubisco oxygenation activity and the ratio v c/v o varies significantly
To assess the ability of B. papyrifera to fix carbon under the six different growth environments, A, vo, vc, vo/vc were resolved at the time of leaf development using temperature response curves (Fig. 6, 7). Changing CO2 and temperature during growth did not cause significant differences in calculated A, and vc, but did cause a significant difference in vo, and vo/vc at the time of leaf harvest. B. papyrifera vo and vo/vc are sensitive to CO2 concentration (p-value < 0.001), but not to temperature (Supplemental Table 5). Thus, these environmental conditions altered photorespiratory influx in B. papyrifera within CO2 conditions. However, the change in photorespiratory influx did not alter A across growth conditions.
Net carbon fixation in B. papyrifera. The temperature response of net carbon assimilation [A; (A)], and the A rate at the time of leaf harvest (B). Colors represent temperature treatments, with ambient temperature in yellow, orange, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. Solid line and no hatching pattern denotes ambient CO2 concentration, while dotted line and hatching pattern represents elevated CO2. Shown are (A) non-linear temperature response model of A at 10 °C, 20 °C, 30 °C, and 40 °C (interpolation and extrapolation are indicated with color and gray scales, respectively), and (B) bar plot with ± SE bars representing A rates from 3 to 5 biological replicates. Significant difference between the harvest A rates treatment types is indicated by letters as determined by Two-way ANOVA and Tukey–Kramer ad hoc test with p < 0.05.
Rubisco carboxylation and oxygenation in B. papyrifera. The temperature response of rubisco oxygenation rate [vo, A)], rubisco carboxylation rate [vc, (C)], and rubisco oxygenation per carboxylation [vo/vc, E)]. The vc (B), vo (D), and vo/vc (F) rate at the time of leaf harvest. Colors represent temperature treatments, with ambient temperature in yellow, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. Solid line and no hatching pattern denotes ambient CO2 concentration, while dotted line and hatching pattern represents elevated CO2. Shown are (A) non-linear temperature response models of vo, vc and vo/vc at 10 °C, 20 °C, 30 °C, and 40 °C (interpolation and extrapolation are indicated with color and gray scales, respectively), and (B) bar plot with ± SE bars representing vo, vc and vo/vc rates from 3 to 5 biological replicates. Significant difference between the harvest A rates treatment types is indicated by an asterisk as determined by Two-way ANOVA and Tukey–Kramer ad hoc test with p < 0.05.
Three output parameters (i.e., the maximum rate on the temperature response curve, Vmax; the temperature optimum, Topt, and the maximum temperature greater than the optimum at which the biological rate drops to zero, Tmax) were additionally solved from the temperature response curves for A, vo, and vc and compared between the B. papyrifera growth environments (Supplemental Figure 5A–I). No significant differences were revealed across growth conditions for any of the parameters.
B. papyrifera photorespiratory safety factors across six growth environments
To evaluate the relationship between photorespiratory influx and downstream metabolic capacities, safety factors were calculated under each growth environment in B. papyrifera at 25 °C to quantify the excess capacity (Fig. 8). To calculate “safety factors”, downstream photorespiratory enzyme activities m-2 s-1 at 25 °C were divided by vo estimated at 25 °C using the temperature response curve. If a safety factor is below 1, then photorespiratory influx is greater than enzymatic capacity. If a safety factor is equal to 1, then photorespiratory influx is accurately matched to the enzymatic capacity and indicates symmorphosis. If a safety factor is above 1, then photorespiratory influx is less than the enzymatic capacity and indicates a reserve capacity. Broadly, plants grown under ambient CO2 concentrations had lower safety factors across the downstream enzymes than plants grown under elevated CO2 conditions due to the decrease in vo in plants from the high CO2 concentrations. In particular, PGP, GO, and GGAT had safety factors of ~ 1 under T0AC, T4AC, and T8AC growth conditions, which suggest symmorphosis, but safety factors above 1 under T0EC, T4EC, and T8EC growth conditions. CAT, AGAT, SGAT, HPR, and GLYK had safety factors above 1 for all growth conditions, revealing the reserve capacity. The two-way ANOVA reveals a clear CO2 concentration effect (p-value = < 0.005) in all enzyme safety factors, and a temperature effect in PGP (p-value < 0.05), but no interactive effect (Supplemental Table 6).
Safety Factors of downstream photorespiratory enzymes at 25 °C in B. papyrifera. Safety factors (specific activities per m2 leaf area measured at 25 °C divided by corresponding mean vo at 25 °C) of downstream photorespiratory enzymes (PGP, GO, CAT, GGAT, AGAT, SGAT, HPR, and GK) were calculated in B. papyrifera. Colors represent temperature treatments, with ambient temperature in yellow, ambient temperature + 4 °C in orange, and ambient temperature + 8 °C in red. No hatching pattern denotes ambient CO2 concentration, while hatching pattern represents elevated CO2. Shown are the boxplots as well as the points indicating the biological replicates (n = 6). Significant difference between treatment types is indicated by letters as determined by Two-way ANOVA and Tukey–Kramer ad hoc test with p < 0.05.
Discussion
This study demonstrates that photorespiratory enzyme activity has predominately a fixed capacity, rather than an acclimation response, to changing CO2 and temperature in B. papyrifera (Figs. 2, 3; Supplemental Figures 2, 3). Changing CO2 and temperature growth conditions did not cause a significant difference in photorespiratory influx between the six growth conditions but did significantly alter the vo/vc as estimated from previously measured gas exchange during this experiment (Fig. 7D, F). However, significant CO2 concentration effect was identified in the absolute rates of vo and vo/vc (p-value < 0.005), but no temperature effect was found (Supplemental Table 5) Trends in this experiment are consistent with decreasing rates of rubisco oxygenation due to elevated CO2, and a slight decrease with temperature caused by decreases in rubisco activation state (Figs. 5, 7). This indicates that the regulation of photorespiratory enzymes is not tied simply to temperature during development. The inability of photorespiratory enzymes, downstream of rubisco, to acclimate to environmental conditions raises the question: does photorespiratory enzymatic capacity need to acclimate to vo, or does excess photorespiratory capacity already exist?
If photorespiratory capacity acclimates to maintain a change in vo to prevent an over-investment in cellular resources or minimize photorespiratory intermediate accumulation, then enzyme activities would adjust to changing vo in B. papyrifera. An optimized photorespiratory pathway would adhere to the symmorphosis hypothesis, where the enzyme activities would quantitatively adjust to vo34,35,36. Historically, B. papyrifera trees inhabit high latitude regions and are frequently found in boreal forests biomes that experience cold-moderate temperatures, with freezing temperatures for over half the year41. Under a boreal forest environment, we would expect B. papyrifera to be adapted to low photorespiratory pressure, therefore a minimal photorespiratory capacity should be maintained. In this study, where B. papyrifera is exposed to different CO2 and temperature conditions during growth, we would expect changes in vo to adjust photorespiratory capacity in parallel. Based on the mean vo at 25 °C, which are 5.96 μmol m-2 s-1 (T0AC), 4.63 μmol m-2 s-1 (T4AC), 3.94 μmol m-2 s-1 (T8AC), 2.64 μmol m-2 s-1 (T0EC), 2.58 μmol m-2 s-1 (T4EC), 1.75 μmol m-2 s-1 (T8EC), the downstream enzyme activities at 25 °C would need to be ~ 6 μmol m-2 s-1 (T0AC), ~ 5 μmol m-2 s-1 (T4AC), ~ 4 μmol m-2 s-1 (T8AC), ~ 3 μmol m-2 s-1 (T0EC & T4EC), ~ 2 μmol m-2 s-1 (T8EC) to manage this carbon influx following rubisco oxygenation. Under ambient CO2 conditions, the activities of PGP, GO, GGAT are near 5 μmol m-2 s-1 and broadly match the photorespiratory influx, but the other downstream enzymes have activities greater than this. In contrast, under elevated CO2 conditions we would expect enzyme activities closer to 3 μmol m-2 s-1, to match the vo, but we do not see this. Instead, the enzyme activities have no significant changes to what is measured under ambient conditions (T0AC). Since photorespiratory enzyme activities do not acclimate to vo in B. papyrifera, adjustment of photorespiratory capacity through symmorphosis is not supported in B. papyrifera at the tested growth conditions.
An alternative hypothesis to describe the fixed capacity of the photorespiratory enzymes is the existence of “safety factors” embedded into the pathway43. With this hypothesis, the photorespiratory pathway has a reserve capacity to handle photorespiratory influx under dynamic conditions. Therefore, we would expect greater photorespiratory enzyme capacities in B. papyrifera than are required by “typical” physiological demand to manage rapid changes in vo without compromising A. In all environmental conditions B. papyrifera were grown under, photorespiratory enzymes’ activities matched or exceeded vo and were not significantly altered by differences in photorespiratory influx brought by changes in CO2 or temperature. The safety factor metric, quantified by dividing the enzyme activity at saturating substrate concentrations (Vmax) by the flux of the substrate it needs to process, in this case the production of 2-phosphoglycolate (vo), reveals presence of a safety factor for each photorespiratory enzyme downstream of rubisco. Additionally, we find that A is consistent between the six growth conditions, suggesting that the accumulation of bioactive photorespiratory intermediates, that would inhibit photosynthetic carbon fixation, are minimized (Fig. 6B).
Interestingly, the initial reactions of photorespiration have lower safety factors (Enzyme activity / vo) than the reactions later in the pathway (Fig. 8). This is primarily due to the larger photorespiratory influx in the ambient CO2 conditions (T0AC, T4AC, T8AC), compared to elevated CO2 conditions (T0EC, T4EC, T8EC), in tandem with the fixed absolute activities of the photorespiratory enzymes (Fig. 2, 3 and Supplemental Table 6). Since B. papyrifera had lower absolute activities of PGP, GO, and GGAT compared to CAT, AGAT, SGAT, HPR, and GLYK across growth conditions, the safety factors calculated for these initial enzymes were lower. In particular, when B. papyrifera was grown under T0AC, T4AC, and T8AC, the safety factor ranged from 0.787 to 1.267 in PGP, GO, and GGAT, but ranged from 1.751 to 3.314 under T0EC, T4EC, and T8EC.
Low safety factors in the initial photorespiratory enzymes suggest that these reactions may be rate limiting in B. papyrifera and potential enzymatic bottlenecks in the pathway. Enzymatic bottlenecks are defined here as steps in a metabolic pathway where the rate of the reaction is significantly limited by the activity of the enzyme. The pattern of underinvestment in enzyme activity in the initial reactions of photorespiration in B. papyrifera may apply universally to C3 species and increasing the activity of these enzymes may improve photosynthetic efficiency when photorespiratory influx is unpredictable. Past work supports this hypothesis generally, for example, in Rhazya stricta, a C3 desert extremophile, the early photorespiratory enzymes PGP and CAT had elevated activities when compared to Nicotiana tabacum33. R. stricta is adapted to hot-arid environments, where temperatures range from 26 °C to 43 °C, that promote an increase in photorespiratory flux44. The increased activity of PGP and CAT adapted in R. stricta may provide an increase in safety factor in these initial enzymes to reduce enzymatic bottlenecks and maintain minimal inhibitor or H2O2 accumulation during periods of high vo in this species. Interestingly, species that do not show this constitutive safety factor, may have increased photosynthetic resilience by increasing enzyme capacity. For example, PGP overexpression in Arabidopsis thaliana improved A likely by maintaining lower steady-state pools of 2-phosphoglycolate after short- and long-term exposure to elevated temperatures25,45. Another photorespiratory enzyme, glycine decarboxylate complex (GDC), is expected to have a low safety factor, although it was not measured in this study. Overexpression of one of the four proteins that compose the complex, H-protein, enhance GDC activity and is associated with an increase in A in Arabidopsis thaliana46. Other researchers, have overexpressed H-protein in Nicotiana tabacum and have similarly shown an increase in plant biomass47.
It is also worth noting the difference in the safety factor between GGAT and AGAT in B. papyrifera, as it might be suggestive of the amino donor B. papyrifera uses to facilitate the transamination reaction (Fig. 8D, E). Peroxisomal aminotransferases in the photorespiratory pathway utilize glycolate as an amino acceptor, yet the specific amino donor across species remains unknown due to promiscuity48,49,50,51. In various plant species, glutamate and alanine are supported to be the main donor for glycine production from glycolate52. In Arabidopsis thaliana, glutamate is supported to be the main donor as GGAT-KO mutants reveal photorespiratory phenotype48. In our work, the larger safety factor in AGAT is caused by the higher absolute activity of AGAT (~ 15 μmol Alanine m-2 s-1), than GGAT (~ 4 μmol Glutamate m-2 s-1) across growth environments. The larger activity may indicate the preference for alanine as the amino doner during periods of high photorespiratory influx (Fig. 3D, E). In agreement with this, past work in R. stricta and N. tabacum have greater absolute activities of AGAT (~ 20 & ~ 9 μmol Alanine m-2 s-1, respectively), than GGAT (~ 2 & ~ 2 μmol Glutamate m-2 s-1, respectively), suggesting that alanine may be the amino donor in these species as well33.
Although outside of the scope of this study, it is also likely that some photorespiratory enzymes in B. papyrifera are subject to posttranslational modifications (PTM) that further regulate enzyme activity in vivo. We recognize that photorespiratory capacity measured in vitro in this study is different than the enzymatic rates under physiologically-relevant substrate concentrations in vivo. PTMs might provide fine-tune metabolic control of the photorespiratory system by transient inactivation of a particular enzyme, or positively or negatively alter enzyme activity in relation to environmental pressure53,54. Recently, regulation of the photorespiratory pathway has been found to include protein phosphorylation, ubiquitination, acetylation, and redox modifications54,55,56,57.
Although many of the enzymes downstream of rubisco do not adjust their activities to environmental pressure, we identified that the activities of many downstream photorespiratory enzymes scaling to total rubisco activity (Fig. 4). Our results demonstrate a significant correlation of PGP, GO, CAT, SGAT, HPR, and GLYK to total rubisco activity, however no correlation of GGAT, and AGAT to rubisco activity. The coordinated change of PGP, GO, CAT, SGAT, HPR, and GLYK activities associated with rubisco may be an effective strategy to alter pathway capacity. Especially when considering the safety factor of PGP and GO, tight coordination may circumvent the underinvestment in enzyme capacity early in the photorespiratory pathway. The mechanism that causes this coordinated regulation may be at the transcript-level via co-expression of multiple photorespiratory genes58. The activities of the aminotransferase enzymes (GGAT, AGAT, and SGAT) are not well explained by the variation seen in rubisco activity.
The results from this work suggest B. papyrifera acclimates functional rubisco capacity (total activity * activation state) to growth conditions through adjustments in rubisco activation state (Figs. 2, 4). When compared to active rubisco at current climate conditions (T0AC), rubisco deactivates by 3.3% (T4AC), 13.1% (T8AC), 15.6% (T0EC), 24.7% (T4EC), and 31.4% (T8EC). The deactivation of rubisco is largest when compared within the CO2 concentrations. Both a CO2 effect (p-value < 0.001) and temperature effect (p-value < 0.05) were identified (Supplemental Table 4) Our activation state data in B. papyrifera agree with what is universally observed when rubisco is exposed to increasing CO2 and/or temperature treatments19,20,22,59,60. The deactivation of rubisco associated with elevated CO2 is likely driven by a combination of a regulated response in RuBP regeneration, and possibly rubisco activase sensitivity to high temperature in B. papyrifera (Cen et al., 2005). Under elevated CO2 conditions, rubisco consumes RuBP faster than it is regenerated compared to ambient CO2 conditions. This greater consumption reduces ATP, influencing the ATP:ADP ratio, which is tied to the regulation of rubisco activase. A lower ATP:ADP ratio in elevated CO2 conditions compared to ambient CO2 conditions reduces the activity of rubisco activase, therefore lowering the catalytic capacity of rubisco to rebalance the RuBP regeneration to consumption capacity17,61,62. The decrease in activation state with temperature coincides with a decrease in rubisco activase activity under elevated temperatures17,21,22,63,64. It is also possible, although not measured in this study, that total rubisco content changes among the 6 growth conditions. A decline in rubisco enzyme content would effectively reduce total activity (Vmax = [rubisco] * kcat), thereby decreasing in vivo functional rubisco capacity. In vitro total rubisco carboxylation activity in B. papyrifera, although not significant between growth conditions, appears to decrease with increasing temperatures on a protein basis at both 25 °C and 35 °C assay temperatures (Supplemental Figure 3A). This apparent decrease in total activity of rubisco could be biologically meaningful. In vitro rubisco oxygenation activity (vo,max), although not measured, is anticipated to be lower, with similar trends as rubisco carboxylation activity, since both reactions are catalyzed by the same enzyme.
With the differences in active rubisco between the six climate scenarios, in vivo A, estimated from the average growth temperatures at the time of harvest, remains constant (Fig. 6B). A can be dissected using Farquhar Berry von Caemmerer (FBvC) model of C3 leaf photosynthesis (A = vc − 0.5vo − RL) to explore the underlying biochemical mechanisms65. In vivo vc, estimated from the average growth temperatures at the time of harvest, were not different between the six growth conditions (Fig. 7A). In vivo vo were greater in ambient CO2 conditions compared to elevated CO2 conditions (Fig. 7B). This lower vo in B. papyrifera under elevated CO2 results from a higher CO2 partial pressure around rubisco that competitively inhibits vo. Therefore, the growth environment shapes the frequency of rubisco catalyzing the oxygenation of RuBP. In vivo estimates of RL were greater at moderate (T4), and extreme (T8) growth temperatures compared to ambient (T0) growth temperatures (Supplemental Figure 6). A significant temperature effect was identified, but no CO2 concentration or temperature:CO2 interactive effect was found (Supplemental Table 5). The temperature sensitivity of RL aligns with the Arrhenius equation used to model the temperature response at the time of leaf harvest. The greater CO2 release from RL at moderate (0.57 μmol m-2 s-1 (T4AC) & 0.71 μmol m-2 s-1 (T4EC)) and extreme (0.64 μmol m-2 s-1 (T8AC) & 0.61 μmol m-2 s-1 (T8EC)) temperature conditions decrease A more than at ambient temperatures (0.36 μmol m-2 s-1 (T0AC) & 0.42 μmol m-2 s-1 (T0EC)) due to the greater CO2 release.
Acclimation in B. papyrifera reveals differences in the fresh weight per leaf area, but is not widespread to the photosynthetic pigments, nor total soluble protein (Supplemental Figure 7). This indicates that the photosynthetic capacity (total chlorophyll) and metabolic activity (soluble protein) likely remains relatively constant per leaf area, but the physical properties of the leaves are acclimating to their growth environment. B. papyrifera leaves had a greater fresh weight per leaf area in T4EC and T8EC than the other growth conditions. Both a CO2 effect (p-value < 0.001), temperature effect (p-value < 0.05), and interaction effect of CO2:temperature (p-value < 0.001) were identified in the leaf weight per leaf area (Supplemental Table 7). A related study using this same experimental material identified significant CO2 and temperature effects in dry mass per leaf area, nitrogen on a leaf area basis, and carbon to nitrogen ratio (see Fig. 4 in Schmiege et al., 2023)66. This provides further evidence that plant leaf morphology acclimates to environmental growth condition.
Conclusion
In summary, these results suggest that photorespiration, downstream of rubisco, does not acclimate distinct enzyme activities to environmental pressures in parallel to photorespiratory influx, but instead has a fixed capacity that scales relative to total rubisco activity. This fixed capacity is due to safety factors embedded into the photorespiratory pathway, although initial reactions have a lower safety factor than later ones, potentially revealing enzymatic bottlenecks. A majority of downstream photorespiratory enzyme activities correlated to total rubisco activity indicating that rubisco, instead of the environmental factors, sets the capacity for photorespiration. Additionally, A remained consistent between environmental pressures as a consequence of acclimation of rubisco activation state in B. papyrifera. This work provides physiological and biochemical mechanisms for the acclimation ability and limitations in B. papyrifera to future climate scenarios.
Methods
Plant material and growth conditions
Paper birch (B. papyrifera [Marshall]) seeds were sourced from the Canadian National Tree Seed Center and grown under six different climate treatments within the Biotron Experimental Climate Change Research Centre of Western University (Supplemental Fig. 1)66 (Hammer et al., unpublished). Seeds were sown in 11.6 L pots filled with Pro-Mix BX Mycorrhizal growth medium (Premier Tech Home and Garden) and slow-release fertilizer (Slow-Release Plant Food, 12–4-8, Miracle Gro®, The Scotts Company). Growth conditions for each glasshouse were set to the following 6 climate treatments: Ambient CO2 (398 ± 54 ppm) or Elevated CO2 (739 ± 48 ppm; elevated CO2 treatments were maintained by adding pure CO2 to the chambers until the elevated setpoint was reached), with 1 of 3 temperature treatments: ambient temperature, ambient + 4 °C, or ambient + 8 °C. Ambient temperature conditions were set to a 5-year day/night average for Algonquin Park, ON (45°58′N, 78°48′W) (Supplemental Fig. 1). Moreover, day (6:00–20:00)/night (12:00–5:00) ambient growth temperatures ranged from 28.0–8.08 °C/24.3–7.89 °C (T0AC), 28.8–9.76 °C/25.2–8.91 °C (T0EC), 32.6–8.73 °C/29.2–11.0 °C (T4AC), 32.9–10.3 °C/29.3–11.3 °C (T4EC), 36.9–10.8 °C/32.5–11.6 °C (T8AC), 36.0–6.46 °C/32.2–11.6 °C (T8EC) across the growth season. Plants grew in the glasshouses for 5 months before the youngest, fully expanded leaves of B. papyrifera were non-destructively measured (beginning of September) to assess temperature response of net carbon assimilation (A) for each growth condition. Different leaves, from different plants from each growth treatment, were harvested under ambient illumination (400–600 μmol m-2 s-1) in the glasshouses afterwards (mid-September) for enzyme activity assays. Specifically, ~ 4 g of leaf material were harvested from each plant, frozen in liquid N2, and stored at − 80 °C. A total of five biological replicates were measured for in vivo gas exchange, and six biological replicates were collected for enzyme activity assays. Since these seeds were sourced from the Canadian National Tree Seed Center and reared under controlled conditions no voucher specimens were created, no additional permissions to collect or sample were required, and all methods were carried out in accordance with relevant guidelines and regulations.
Preparing crude protein extract for protein quantification, total chlorophyll content, enzyme activity assays
All photorespiratory enzyme activities, total chlorophyll content, and protein quantification were measured from the same biological replicate (or homogenate) for each growth condition in efforts to control day-to-day variation and pipetting error. Six biological replicates (one biological replicate from each growth condition) were measured at a single time and took 3 days to complete. On day 1, crude protein was extracted from frozen leaf tissue from each growth treatment, immediately flash frozen in liquid N2, and stored at −80 °C to maximize enzyme stability (described below). On days 2 and 3, nine enzyme activity assays, total chlorophyll content, and protein quantification were measured. The order of enzyme activities and assay temperatures were randomized between the days for the six biological replicates to control for the effects of extraction thaw time (from − 80 °C to 0 °C on ice) on enzyme activity, total chlorophyll extraction, or protein quantification.
Clarified crude protein extract was prepared by grinding approximately 50–75 mg of B. papyrifera leaf material by mortar and pester over liquid N2 and homogenized on ice with 1.5 mL of the extraction buffer (50 mM EPPS buffer, pH 8.0, containing 1 mM EDTA, 10 mM DTT, 0.1% Triton X-100 [v/v], 0.5% polyvinylpyrrolidone (PVPP), and 20 μL 1X SigmaFAST Protease Inhibitor Cocktail, EDTA Free (Sigma, St. Louis, MO, USA)), using a 2 mL glass-to-glass homogenizer (Kontes Glass Co., Vineland, NJ, USA). The homogenate was transferred into a 2 mL plastic Eppendorf tube and clarified by centrifugation for 15 min at 15,000 g and 4℃ (Eppendorf Centrifuge 5424R, Eppendorf, Enfield, CT, USA). The supernatant, containing the clarified crude protein extract was split into two 1.5 mL Eppendorf tubes (each containing roughly 750 μL), and immediately flash frozen in liquid N2 after centrifugation and stored at − 80 °C. The clarified crude protein extract was split to avoid freeze–thaw and preserve enzyme activity during days 2 and 3. The addition of the protease inhibitor in the extraction buffer preserved enzyme activity as well.
Enzyme activity assays
All enzyme activities (except for RBC activation state and CAT) were measured by spectrophotometric assays with the use of SpectraMax M2 Plate reader and SoftMax Pro7 software (Molecular Devices, San Jose, CA, USA). The maximum enzyme activity under saturating substrate concentrations (Vmax; corresponding to enzyme capacity) of rubisco (RBC), phosphoglycolate phosphatase (PGP), glycolate oxidase (GO), glutamate:glyoxylate aminotransferase (GGAT), alanine:glyoxylate aminotransferase (AGAT), serine:glyoxylate aminotransferase (SGAT), hydroxypyruvate reductase (HPR), and glycerate kinase (GLYK) activity assays were performed in a 200 μL total reaction mix using polystyrene or acrylic UV transparent 96-well microplates (Corning, Kennebunk, ME, USA) as described in33. While the RBC activation state and catalase (CAT) activity assay was performed in 1 mL reaction mix using an Cary60 UV–Vis Spectrophotometer (Agilent Technologies, Santa Clara, California, USA) and Oxygraph + Oxygen Monitoring System and Oxytrace+ software (Hansatech Instruments, Penteny, UK). The pH of reactions was selected based on the organellar pH where the reaction occurs67,68,69,70. All enzyme assays were performed with 2–3 technical replicates on 5–6 independent biological replicates using leaf tissue from different plants (Fig. 2, 3; Supplemental Figures 2, 3).
Rubisco activity and activation state assays
The activity and activation state of rubisco was determined by coupling the formation of 3-phosphoglcerate to NADH oxidation in initial and chemically activated rubisco from crude protein extracted of B. papyrifera leaves as described in71. For this assay, a 1 mL cuvette was prepared containing 908 μL rubisco reaction buffer (50 mM HEPES, pH 7.8, 20 mM MgCl2, 1 mM ETDA, 1 mM ATP, 5 mM Creatine Phosphate, 20 mM NaHCO3, 0.2 mM NADH), 20 μL of coupling enzyme, 0.5 mM RuBP. The cuvette was mixed with a stir paddle and placed into the spectrophotometer. In the meantime, fresh crude protein extract was prepared from the same leaf as the other enzyme activity assays with CO2-free extraction buffer. Approximately 50 mL of extraction buffer was prepared as described above (without the addition of Triton X-100) and sparged using 100% N2 at 0.25 L per minute for 1 h. After adding the Triton X-100, 50–75 mg of ground B. papyrifera leaf material was homogenized using 1.5 mL of extraction buffer. The homogenate was transferred into a 2 mL plastic Eppendorf tube and clarified by centrifugation for 60 s at 15,000 g and 4℃ (Eppendorf Centrifuge 5424R, Eppendorf, Enfield, CT, USA). Meanwhile, 340 nm measurements were collected to get a NADH baseline for 1 min. Immediately following the centrifugation, 40 μL of clarified crude protein extract was added to the 1 mL cuvette and mixed with a stirring paddle. The NADH consumption was measured for 3 min with 0.1 s interval. The initial rates of the reaction were determined during first 20 s. The chemically activated assay followed the same steps as the initial, but the crude protein extract was initially activated with 15 mM NaHCO3 and 15 mM MgCl2 for 15 min before starting the reaction. The rate of the reaction was used to express the specific activity as μmoles CO2 m-2 s-1. The ratio of the initial and chemical activated rates were multiplied by 100 to resolve the activation state of rubisco on a percentage basis72. Activation state assays were performed with two technical replicates on five independent biological replicates using leaf tissue from different plants.
Catalase activity assay
The activity of CAT was determined in B. papyrifera by the production of O273,74 with the following modifications. The reaction mix containing 946 μL 50 mM K-phosphate buffer, pH 8.1, and 20 μL crude protein extract, was incubated for 1.5 min to determine the O2 baseline. The reaction was initiated with 5 mM H2O2 and the increase in O2 production (nmol/mL) was observed for 1.5 min with 1 s interval using a Oxygraph + Oxygen Monitoring System. The initial rate of reaction was determined during first 20 s and the specific activity was expressed as μmoles O2 produced m-2 s-1.
Protein quantification
Soluble protein content was determined in crude protein extract (Bio-Rad Protein Assay; BIO-RAD, USA) according to the manufacturer instructions using a SpectraMax M2 Microplate Reader (Molecular Devices, San Jose, CA, USA).
Total chlorophyll content
The supernatant from the clarified crude protein extract was used to quantify total chlorophyll content using a SpectraMax M2 Microplate Reader (Molecular Devices, San Jose, CA, USA). Chlorophyll content in the crude protein extract was determined by mixing 40 μL of the crude protein extract with 960 μL of 100% ethanol. After a few inversions and letting the crude protein extract and ethanol solution sit for 10 min, the solution was centrifuged to gather the precipitate at the bottom of the tube, and the clarified supernatant was measured at 649 nm and 665 nm. The absorption coefficient for 100% ethanol were taken from Table 1 of75. Chl a and Chl b equations used are below76,77,78:
Gas exchange
Temperature response data (10 °C, 20 °C, 30 °C, and 40 °C) of net carbon assimilation (A) under ambient oxygen conditions (21%) at high light (1800 μmol PAR m-2 s-1) intensity measured on the youngest, fully expanded leaves of B. papyrifera were provided by (Hammer et al., unpublished) using a LI-6800 or a LI-6400XT (LI-COR Biosciences, USA). Data of the rates of CO2 release from non-photorespiratory processes in the light (RL) at 25 °C measured using the common intersection method79,80 were taken from Schmiege et al.66. The temperature response of RL was estimated according to the Arrhenius equation,
81 (Supplemental Fig. 6). Where, c is the scaling constant (18.72), \({\Delta H}_{a}\) is the activation energy (46.39 J), R is the molar gas constant (8.314 J mol-1 K), and \({T}_{k}\) is the leaf temperature. The photorespiratory CO2 compensation point (Γ*) of B. papyrifera has not been measured, therefore was assumed to be similar to N. tabacum. The temperature response of Γ* was solved using an Arrhenius equation (above) with a c of 13.49 and a \({\Delta H}_{a}\) of 24.4682. CO2 response curves at 10 °C, 20 °C, 30 °C, and 40 °C were fit using an R-based ACi fitting tool to estimate mesophyll conductance (\({g}_{m}\)) while constraining previously resolved RL and Γ* parameters83 (see http://github.com/poales/msuRACiFit to access Rscript with user-friendly interface).
Estimating rates of v c and v o
Vc and vo for B. papyrifera were estimated according to
84. Where, the partial pressure of CO2 at the site of rubisco catalysis (Cc) was determined by
Modelling the temperature response of A, v c, and v o
The temperature response of A, vc, and vo were modelled to estimate key parameters (Vmax, Topt, Tmax) with the following equation
85. Where, \(Param\) is the parameter of interest (i.e., either A, vc, or vo), \({V}_{max}\) is an estimation of the maximum rate of the \(Param\) that occurs at the optimum temperature (\({T}_{opt}\)), \({T}_{max}\) is the temperature greater than optimum at which the biological rate drops to zero, and \({T}_{leaf}\) is the leaf temperature (i.e., 10 °C, 20 °C, 30 °C, or 40 °C). Practically, the estimate of Vmax indicates the peak performance of A, vc or vo at the most favorable thermal environment, which is estimated as Topt. Tmax, estimated from the same temperature response equation, indicates the thermal tolerance of A, vc or vo in the plant. Plants cannot sustain A, vc or vo above this temperature. These three output parameters (Vmax, Topt, Tmax) were compared between B. papyrifera across the six different growth environments (Supplemental Fig 6A–I).
Determining A, v c, and v o at leaf harvest
The B. papyrifera plants in this experiment took approximately 10 days to develop from young to fully expanded leaves. To establish the growth temperature of the leaves during their development, the temperature at solar noon (12:00 pm) across the 10-day range (i.e., September 27th to September 17th) was averaged. The average growth temperatures were 19.2 °C (T0AC), 19.4 °C (T0EC), 23.0 °C (T4AC), 23.1 °C (T4EC), 26.8 °C (T8AC), 26.4 °C (T8EC).
With this temperature information and the temperature response models, we resolved the photosynthetic parameters (i.e., A, vc, vo, and vo/vc) the leaves were reasonably experiencing at the time of harvest.
Determining safety factors of downstream photorespiratory enzymes
Safety factor was calculated using the equation below:
where, Capacity is enzyme activity and Load is vo37.
Data analysis
Gas exchange and biochemical data were analyzed and visualized using custom scripts in R86. We used lm() in the stats package to fit linear models, gnls() in the nlme package to fit non-linear models, and emmeans() in the emmeans package for mean and parameter comparison. Gas-exchange and biochemical data were checked for a normal distribution (assumption #1) using the shapiro.test() in stats() package and for homogeny of variance (assumption #2) using the leveneTest() in car() package before proceeding with parametric testing. Data that did not conform to normality were logarithmically transformed using log() or square root transformed using sqrt() to meet the assumption. Gas-exchange and biochemical data were analyzed using Two-way analysis of variance (ANOVA), accounting for growth CO2 concentration and temperature, to measure significance. Biochemical data was visualized using geom_boxplot() (i.e., box and whisker plot) to represent the distribution of enzyme activity. The box represents the first and third quartile, and the line inside of the box represents the median. The whiskers represent the minimum and maximum values, while the solid dots represent outliers (calculated as 1.5* interquartile range); however, these were not removed from the visualization or analysis. All ANOVA tests were followed with a Tukey’s post hoc test.
Data availability
The data that supports the findings of this study are available from the corresponding author upon request.
References
Wei, P. S. et al. Absorption coefficient of carbon dioxide across atmospheric troposphere layer. Heliyon 4, e00785. https://doi.org/10.1016/j.heliyon.2018.e00785 (2018).
Lee H. et al 2023 Climate change 2023: synthesis report In Kani B (eds) Contribution of working groups I, II and III to the sixth assessment report of the intergovernmental panel on climate change. The Australian National University, USA, 35–115
Bowes, G., Ogren, W. L. & Hageman, R. H. Phosphoglycolate production catalyzed by ribulose diphosphate carboxylase. Biochem. Biophys. Res. Commun. 45, 716–722. https://doi.org/10.1016/0006-291X(71)90475-X (1971).
Badger, M. R. & Andrews, T. J. Effects of CO2, O2 and temperature on a high-affinity form of ribulose diphosphate carboxylase-oxygenase from spinach. Biochem. Biophys. Res. Commun. 60, 204–210. https://doi.org/10.1016/0006-291X(74)90192-2 (1974).
Bowes, G. & Ogren, W. L. Oxygen inhibition and other properties of soybean ribulose 1,5-diphosphate carboxylase. J. Biol. Chem. 247, 2171–2176 (1972).
Laing, W. A. Regulation of soybean net photosynthetic CO(2) fixation by the interaction of CO(2), O(2), and ribulose 1,5-diphosphate carboxylase. Plant Physiol. 54, 678–685. https://doi.org/10.1104/pp.54.5.678 (1974).
Peisker, M. Model describing the influence of oxygen on photosynthetic carboxylation. Photosynthetica 8, 47–50 (1974).
Bassham, J. A., Benson, A. A. & Calvin, M. The path of carbon in photosynthesis. J. Biol. Chem. 185, 781–787 (1950).
Drake, B. G., Gonzàlez-Meler, M. A. & Long, S. P. More efficient plants: a consequence of rising atmospheric CO2?. Annu. Rev. Plant Biol. 48, 609–639 (1997).
Hermida-Carrera, C., Kapralov, M. V. & Galmés, J. Rubisco catalytic properties and temperature response in crops. Plant Physiol. 171, 2549–2561. https://doi.org/10.1104/pp.16.01846 (2016).
Hall, N. P. & Keys, A. J. Temperature dependence of the enzymic carboxylation and oxygenation of ribulose 1, 5-bisphosphate in relation to effects of temperature on photosynthesis. Plant Physiol. 72, 945–948 (1983).
Jordan, D. B. & Ogren, W. L. The CO2/O2 specificity of ribulose 1,5-bisphosphate carboxylase/oxygenase. Planta 161, 308–313. https://doi.org/10.1007/BF00398720 (1984).
Lorimer, G. H. & Miziorko, H. M. Carbamate formation on the epsilon-amino group of a lysyl residue as the basis for the activation of ribulosebisphosphate carboxylase by CO2 and Mg2+. Biochemistry 19, 5321–5328. https://doi.org/10.1021/bi00564a027 (1980).
Hammond, E. T., Andrews, T. J. & Woodrow, I. E. Regulation of ribulose-1,5-bisphosphate carboxylase/oxygenase by carbamylation and 2-carboxyarabinitol 1-phosphate in tobacco: insights from studies of antisense plants containing reduced amounts of rubisco activase. Plant Physiol. 118, 1463–1471. https://doi.org/10.1104/pp.118.4.1463 (1998).
Bhat, J. Y., Thieulin-Pardo, G., Hartl, F. U. & Hayer-Hartl, M. Rubisco activases: AAA+ chaperones adapted to enzyme repair. Front. Mol. Biosci. https://doi.org/10.3389/fmolb.2017.00020 (2017).
Spreitzer, R. J. & Salvucci, M. E. Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu. Rev. Plant Biol. 53, 449–475 (2002).
Portis, A. R. Rubisco activase—Rubisco’s catalytic chaperone. Photosynth. Res. 75, 11–27. https://doi.org/10.1023/A:1022458108678 (2003).
Carmo-Silva, A. E. & Salvucci, M. E. The regulatory properties of Rubisco activase differ among species and affect photosynthetic induction during light transitions. Plant Physiol. 161, 1645–1655. https://doi.org/10.1104/pp.112.213348 (2013).
Cen, Y.-P. & Sage, R. F. The regulation of Rubisco activity in response to variation in temperature and atmospheric CO2 partial pressure in sweet potato. Plant Physiol. 139, 979–990. https://doi.org/10.1104/pp.105.066233 (2005).
Crafts-Brandner, S. J. & Salvucci, M. E. Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Proc. Natl. Acad. Sci. 97, 13430–13435 (2000).
Salvucci, M. E. & Crafts-Brandner, S. J. Mechanism for deactivation of rubisco under moderate heat stress. Physiol. Plant. 122, 513–519. https://doi.org/10.1111/j.1399-3054.2004.00419.x (2004).
Kim, K. & Portis, A. R. Jr. Temperature dependence of photosynthesis in arabidopsis plants with modifications in rubisco activase and membrane fluidity. Plant Cell Physiol. 46, 522–530. https://doi.org/10.1093/pcp/pci052 (2005).
Galmés, J., Aranjuelo, I., Medrano, H. & Flexas, J. Variation in rubisco content and activity under variable climatic factors. Photosynth. Res. 117, 73–90. https://doi.org/10.1007/s11120-013-9861-y (2013).
Anderson, L. E. Chloroplast and cytoplasmic enzymes II. Pea leaf triose phosphate isomerases. Biochim. Biophys. Acta Enzymol. 235, 237–244. https://doi.org/10.1016/0005-2744(71)90051-9 (1971).
Flügel, F. et al. The photorespiratory metabolite 2-phosphoglycolate regulates photosynthesis and starch accumulation in Arabidopsis. Plant Cell 29, 2537–2551. https://doi.org/10.1105/tpc.17.00256 (2017).
Campbell, W. J. & Ogren, W. L. Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation in intact, lysed, and reconstituted chloroplasts. Photosynth. Res. 23, 257–268. https://doi.org/10.1007/bf00034856 (1990).
Cook, C. M., Mulligan, R. M. & Tolbert, N. E. Inhibition and stimulation of ribulose-1,5-bisphosphate carboxylase/oxygenase by glyoxylate. Arch. Biochem. Biophys. 240, 392–401. https://doi.org/10.1016/0003-9861(85)90044-x (1985).
Mulligan, R. M., Wilson, B. & Tolbert, N. E. Effects of glyoxylate on photosynthesis by intact chloroplasts. Plant Physiol. 72, 415–419. https://doi.org/10.1104/pp.72.2.415 (1983).
Chastain, C. J. & Ogren, W. L. Glyoxylate inhibition of ribulosebisphosphate carboxylase/oxygenase activation state in vivo. Plant Cell Physiol. 30, 937–944. https://doi.org/10.1093/oxfordjournals.pcp.a077837 (1989).
Dellero, Y., Jossier, M., Schmitz, J., Maurino, V. G. & Hodges, M. Photorespiratory glycolate–glyoxylate metabolism. J. Exp. Bot. 67, 3041–3052. https://doi.org/10.1093/jxb/erw090 (2016).
Dat, J. F. et al. Changes in hydrogen peroxide homeostasis trigger an active cell death process in tobacco. Plant J. 33, 621–632. https://doi.org/10.1046/j.1365-313X.2003.01655.x (2003).
Cavanagh, A. P. & Ort, D. R. Transgenic strategies to improve the thermotolerance of photosynthesis. Photosynth. Res. 158, 109–120. https://doi.org/10.1007/s11120-023-01024-y (2023).
Gregory, L. M., Roze, L. V. & Walker, B. J. Increased activity of core photorespiratory enzymes and CO2 transfer conductances are associated with higher and more optimal photosynthetic rates under elevated temperatures in the extremophile Rhazya stricta. Plant Cell Environ. 46, 3704–3720. https://doi.org/10.1111/pce.14711 (2023).
Taylor, C. R. & Weibel, E. R. Design of the mammalian respiratory system I. Problem and strategy. Respir. Physiol. 44, 1–10 (1981).
Suarez, R. K., Staples, J. F., Lighton, J. R. & West, T. G. Relationships between enzymatic flux capacities and metabolic flux rates: nonequilibrium reactions in muscle glycolysis. Proc. Natl. Acad. Sci. U. S. A. 94, 7065–7069. https://doi.org/10.1073/pnas.94.13.7065 (1997).
Weibel, E. R., Taylor, C. R. & Hoppeler, H. The concept of symmorphosis: a testable hypothesis of structure-function relationship. Proc. Natl. Acad. Sci. U. S. A. 88, 10357–10361. https://doi.org/10.1073/pnas.88.22.10357 (1991).
Diamond, J. Quantitative evolutionary design. J. Physiol. 542, 337–345. https://doi.org/10.1113/jphysiol.2002.018366 (2002).
Alexander, R. M. Factors of safety in the structure of animals. Sci. Prog. 67, 109–130 (1981).
Price, D. T. et al. Anticipating the consequences of climate change for Canada’s boreal forest ecosystems. Environ. Rev. 21, 322–365. https://doi.org/10.1139/er-2013-0042 (2013).
Collins M, et al. Long-term climate change: Projections, commitments and irreversibility. In Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change (eds. Stocker, T.F.) 1029–1136 (Cambridge University Press, 2013). https://doi.org/10.1017/CBO9781107415324.024.
Gauthier, S., Bernier, P., Kuuluvainen, T., Shvidenko, A. Z. & Schepaschenko, D. G. Boreal forest health and global change. Science 349, 819–822 (2015).
Dusenge, M. E., Madhavji, S. & Way, D. A. Contrasting acclimation responses to elevated CO(2) and warming between an evergreen and a deciduous boreal conifer. Glob. Change Biol. 26, 3639–3657. https://doi.org/10.1111/gcb.15084 (2020).
Salvador, A. & Savageau, M. A. Quantitative evolutionary design of glucose 6-phosphate dehydrogenase expression in human erythrocytes. Proc. Natl Acad. Sci. 100, 14463–14468 (2003).
Lawson, T. et al. C3 photosynthesis in the desert plant Rhazya stricta is fully functional at high temperatures and light intensities. New Phytol. 201, 862–873. https://doi.org/10.1111/nph.12559 (2014).
Timm, S., Woitschach, F., Heise, C., Hagemann, M. & Bauwe, H. Faster removal of 2-phosphoglycolate through photorespiration improves abiotic stress tolerance of arabidopsis. Plants https://doi.org/10.3390/plants8120563 (2019).
Timm, S. et al. Glycine decarboxylase controls photosynthesis and plant growth. FEBS Lett. 586, 3692–3697. https://doi.org/10.1016/j.febslet.2012.08.027 (2012).
López-Calcagno, P. E. et al. Overexpressing the H-protein of the glycine cleavage system increases biomass yield in glasshouse and field-grown transgenic tobacco plants. Plant Biotechnol. J. 17, 141–151. https://doi.org/10.1111/pbi.12953 (2019).
Somerville, C. R. & Ogren, W. L. Photorespiration mutants of Arabidopsis thaliana deficient in serine-glyoxylate aminotransferase activity. Proc. Natl. Acad. Sci. 77, 2684–2687 (1980).
Leegood, R. C., Lea, P. J., Adcock, M. D. & Häusler, R. E. The regulation and control of photorespiration. J. Exp. Bot. 46, 1397–1414. https://doi.org/10.1093/jxb/46.special_issue.1397 (1995).
Husic, D. W., Husic, H. D., Tolbert, N. E. & Black, C. C. Jr. The oxidative photosynthetic carbon cycle or C2 cycle. Crit. Rev. Plant Sci. 5, 45–100. https://doi.org/10.1080/07352688709382234 (1987).
Liepman, A. H. & Olsen, L. J. Alanine aminotransferase homologs catalyze the glutamate: glyoxylate aminotransferase reaction in peroxisomes of arabidopsis. Plant Physiol. 131, 215–227. https://doi.org/10.1104/pp.011460 (2003).
Betsche, T. Aminotransfer from alanine and glutamate to glycine and serine during photorespiration in oat leaves. Plant Physiol. 71, 961–965. https://doi.org/10.1104/pp.71.4.961 (1983).
Friso, G. & van Wijk, K. J. Posttranslational protein modifications in plant metabolism. Plant Physiol. 169, 1469–1487. https://doi.org/10.1104/pp.15.01378 (2015).
Keech, O., Gardeström, P., Kleczkowski, L. A. & Rouhier, N. The redox control of photorespiration: from biochemical and physiological aspects to biotechnological considerations. Plant Cell Environ. 40, 553–569. https://doi.org/10.1111/pce.12713 (2017).
Hodges, M. Photorespiration and improving photosynthesis. Prog. Bot. 84, 171–220. https://doi.org/10.1007/124_2022_64 (2022).
Timm, S. & Hagemann, M. Photorespiration-how is it regulated and how does it regulate overall plant metabolism?. J. Exp. Bot. 71, 3955–3965. https://doi.org/10.1093/jxb/eraa183 (2020).
Hodges, M., Jossier, M., Boex-Fontvieille, E. & Tcherkez, G. Protein phosphorylation and photorespiration. Plant Biol. 15, 694–706. https://doi.org/10.1111/j.1438-8677.2012.00719.x (2013).
Laxa, M. & Fromm, S. Co-expression and regulation of photorespiratory genes in Arabidopsis thaliana: A bioinformatic approach. Curr. Plant Biol. 14, 2–18. https://doi.org/10.1016/j.cpb.2018.09.001 (2018).
Salvucci, M. E., DeRidder, B. P. & Portis, A. R. Jr. Effect of activase level and isoform on the thermotolerance of photosynthesis in Arabidopsis. J. Exp. Bot. 57, 3793–3799. https://doi.org/10.1093/jxb/erl140 (2006).
Scafaro, A. P., Posch, B. C., Evans, J. R., Farquhar, G. D. & Atkin, O. K. Rubisco deactivation and chloroplast electron transport rates co-limit photosynthesis above optimal leaf temperature in terrestrial plants. Nat. Commun. 14, 2820. https://doi.org/10.1038/s41467-023-38496-4 (2023).
Sage, R. F., Sharkey, T. D. & Seemann, J. R. Regulation of ribulose-1,5-bisphosphate carboxylase activity in response to light intensity and CO(2) in the C(3) annuals Chenopodium album L. and Phaseolus vulgaris L.. Plant Physiol. 94, 1735–1742. https://doi.org/10.1104/pp.94.4.1735 (1990).
Ruuska, S. A., Andrews, T. J., Badger, M. R., Price, G. D. & Caemmerer, S. The role of chloroplast electron transport and metabolites in modulating rubisco activity in tobacco. Insights from transgenic plants with reduced amounts of cytochrome b/f complex or glyceraldehyde 3-phosphate dehydrogenase. Plant Physiol. 122, 491–504. https://doi.org/10.1104/pp.122.2.491 (2000).
Salvucci, M. E. & Crafts-Brandner, S. J. Relationship between the heat tolerance of photosynthesis and the thermal stability of rubisco activase in plants from contrasting thermal environments. Plant Physiol. 134, 1460–1470. https://doi.org/10.1104/pp.103.038323 (2004).
Salvucci, M. E. & Crafts-Brandner, S. J. Inhibition of photosynthesis by heat stress: the activation state of rubisco as a limiting factor in photosynthesis. Physiol. Plant. 120, 179–186. https://doi.org/10.1111/j.0031-9317.2004.0173.x (2004).
Farquhar, G. D., von Caemmerer, S. & Berry, J. A. A biochemical model of photosynthetic CO2 assimilation in leaves of C 3 species. Planta 149, 78–90. https://doi.org/10.1007/bf00386231 (1980).
Schmiege, S. C., Sharkey, T. D., Walker, B., Hammer, J. & Way, D. A. Laisk measurements in the nonsteady state: Tests in plants exposed to warming and variable CO2 concentrations. Plant Physiol. 193, 1045–1057. https://doi.org/10.1093/plphys/kiad305 (2023).
Shen, J. et al. Organelle pH in the Arabidopsis endomembrane system. Mol. Plant 6, 1419–1437. https://doi.org/10.1093/mp/sst079 (2013).
Heinze, M. & Gerhardt, B. In Plant Peroxisomes: Biochemistry, Cell Biology and Biotechnological Applications (eds Baker, A. & Graham, I. A.) 103–140 (Springer, 2002).
Liu, L. et al. Expression, purification, and initial characterization of human alanine aminotransferase (ALT) isoenzyme 1 and 2 in High-five insect cells. Protein Expr. Purif. 60, 225–231. https://doi.org/10.1016/j.pep.2008.04.006 (2008).
Kendziorek, M. & Paszkowski, A. Properties of serine:glyoxylate aminotransferase purified from Arabidopsis thaliana leaves. Acta Biochim. Biophys. Sin. 40, 102–110. https://doi.org/10.1111/j.1745-7270.2008.00383.x (2008).
Walker, B. J., South, P. F. & Ort, D. R. Physiological evidence for plasticity in glycolate/glycerate transport during photorespiration. Photosynth. Res. 129, 93–103. https://doi.org/10.1007/s11120-016-0277-3 (2016).
Sales, C. R. G., Degen, G. E., Silva, A. B. & Carmo-Silva, E. In Photosynthesis: Methods and Protocols (ed. Sarah, C.) 239–250 (Springer, 2018).
Zelitch, I. Selection and characterization of tobacco plants with novel O2-resistant photosynthesis. Plant Physiol. 90, 1457–1464. https://doi.org/10.1104/pp.90.4.1457 (1989).
Aebi, H. E. Catalase. In Methods of Enzymatic Analysis (ed. Bergmeyer, H. U.) 273–286 (Verlag Chemie, 1983).
Ritchie, R. J. Universal chlorophyll equations for estimating chlorophylls a, b, c, and d and total chlorophylls in natural assemblages of photosynthetic organisms using acetone, methanol, or ethanol solvents. Photosynthetica 46, 115–126. https://doi.org/10.1007/s11099-008-0019-7 (2008).
Ritchie, R. J. Consistent sets of spectrophotometric chlorophyll equations for acetone methanol and ethanol solvents. Photosynth. Res. 89, 27–41. https://doi.org/10.1007/s11120-006-9065-9 (2006).
Porra, R. J. The chequered history of the development and use of simultaneous equations for the accurate determination of chlorophylls a and b. Photosynth. Res. 73, 149–156. https://doi.org/10.1023/A:1020470224740 (2002).
Porra, R. J., Thompson, W. A. & Kriedemann, P. E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta Bioenerg. 975, 384–394. https://doi.org/10.1016/S0005-2728(89)80347-0 (1989).
Laisk, A. Kinetics of Photosynthesis and Photorespiration of C-3 Plants. Science, Moscow, 195 p. (In Russian) (1977)
Walker, B. J., Skabelund, D. C., Busch, F. A. & Ort, D. R. An improved approach for measuring the impact of multiple CO2 conductances on the apparent photorespiratory CO2 compensation point through slope–intercept regression. Plant Cell Environ. 39, 1198–1203. https://doi.org/10.1111/pce.12722 (2016).
Bernacchi, C. J., Singsaas, E. L., Pimentel, C., Portis, A. R. Jr. & Long, S. P. Improved temperature response functions for models of Rubisco-limited photosynthesis. Plant Cell Environ. 24, 253–259. https://doi.org/10.1111/j.1365-3040.2001.00668.x (2001).
Bernacchi, C. J., Portis, A. R., Nakano, H., Caemmerer, S. & Long, S. P. Temperature response of mesophyll conductance. Implications for the determination of rubisco enzyme kinetics and for limitations to photosynthesis in vivo. Plant Physiol. 130, 1992–1998. https://doi.org/10.1104/pp.008250 (2002).
Gregory, L. M. et al. The triose phosphate utilization limitation of photosynthetic rate: Out of global models but important for leaf models. Plant Cell Environ. 44, 3223–3226. https://doi.org/10.1111/pce.14153 (2021).
Walker, B. J., Kramer, D. M., Fisher, N. & Fu, X. Flexibility in the energy balancing network of photosynthesis enables safe operation under changing environmental conditions. Plants 9, 301 (2020).
Collier, C. J. et al. Optimum temperatures for net primary productivity of three tropical seagrass species. Front. Plant Sci. https://doi.org/10.3389/fpls.2017.01446 (2017).
R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Retrieved from https://www.R-project.org (2021)
Acknowledgements
We would like to thank Mauricio Tejera-Nieves for help with non-linear models for the temperature response curves.
Funding
L.M.G. and B.J.W were funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the United States Department of Energy (DE-FG02-91ER20021) and the National Science Foundation award from the Division of Integrated Organismal Systems (2030337). S.C.S received support from the Plant Resilience Institute of Michigan State University. D.A.W. acknowledges the support of an NSERC Discovery Grant RGPIN-2019–04677 and a Strategic Partnership Grant.
Author information
Authors and Affiliations
Contributions
BJW and DAW conceived the original research plans and supervised the research. JMH measured the temperature response of net carbon fixation of the Betula papyrifera in each growth condition and monitored the growth of the plants during the experiment. SCS collected the leaf material. LMG, and LAS performed the enzyme activity assays with input from LVR, while KFS performed the protein quantification and total chlorophyll content assays. LMG analyzed the results and wrote the paper with contributions from all authors. BJW serves as the author responsible for contact.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gregory, L.M., Scott, K.F., Sharpe, L.A. et al. Rubisco activity and activation state dictate photorespiratory plasticity in Betula papyrifera acclimated to future climate conditions. Sci Rep 14, 26340 (2024). https://doi.org/10.1038/s41598-024-77049-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-77049-7