Abstract
To enhance the clinical utility of mass spectrometry (MS), lengthy dwell times on less informative regions of patient specimens (e.g., adipose tissue in breast) must be minimized. Additionally, a promising variant of MS known as picosecond infrared laser MS (PIRL-MS) faces further challenges, namely, lipid contamination when probing adipose tissue. Here we demonstrate on several thick non-sectioned resected human breast specimens (healthy and malignant) that reflection-mode polarimetric imaging can robustly guide PIRL-MS toward regions devoid of significant fat content to (1) avoid signal contamination and (2) shorten overall MS analysis times. Through polarimetric targeting of non-fat regions, PIRL-MS sampling revealed feature-rich spectral signatures including several known breast cancer markers. Polarimetric guidance mapping was enabled by circular degree-of-polarization (DOP) imaging via both Stokes and Mueller matrix polarimetry. These results suggest a potential synergistic hybrid approach employing polarimetry as a wide-field-imaging guidance tool to optimize efficient probing of tissue molecular content using MS.
Similar content being viewed by others
Introduction
Picosecond infrared laser mass spectrometry (PIRL-MS) profiles tissue molecular composition via mass-to-charge measurements of extracted water-soluble analytes (e.g., molecules in water-rich biological tissues). Advantageously, PIRL-MS can be performed under ambient conditions on specimens of various thicknesses and surface topologies without the need to prepare tissue homogenates or analyte extracts1,2,3,4. Notable demonstrations include classification of various unprocessed human brain and skin cancer specimens1,2,3,4. Successful clinical implementation of PIRL-MS (and potentially other ambient MS technologies) may enable rapid tumour margin delineation5,6, intraoperative cancer diagnostics7, and on-site biopsy adequacy assessment8.
A previous study has demonstrated the feasibility of PIRL-MS analysis of non-sectioned malignant breast tissue (in situ) using subcutaneous murine models9 to recover biomarker molecules unique to breast cancer similar to those reported in other MS studies10. Actual human breast tissue, on the other hand, presents a major challenge for PIRL-MS due to the often-significant presence of adipose tissue (i.e., fat) which absorbs the mid infrared laser light without efficient conversion of the thermal energy to ablation mode, resulting in an undesirable solid-to-liquid phase transition (melting) of the fat. This yields a thin lipid layer on the surrounding areas, jeopardizing the integrity of their PIRL-MS signal by contamination. Furthermore, lengthy data acquisition times exploring non-informative regions of a specimen (e.g., adipose tissue and other areas of ‘low-cellularity’) also limit the clinical appeal of on-site MS analysis (and any other point-sensing analytical sampling methods). Therefore, to optimize PIRL-MS for breast cancer diagnostic applications, (1) adipose regions must be avoided, and (2) analysis times must be shortened by guiding PIRL-MS probe to areas of high cellularity.
These objectives are achievable through the deployment of a guidance tool to avoid and target specific regions for MS probing. Polarimetry – an optical technique based on measured changes to polarized light after interacting with a sample11,12 – has shown promise as a guidance tool for oncological MS analysis in the past6,13,14. Indeed, polarimetry is emerging as a useful label-free cancer imaging approach through its sensitivity to cancer-associated structural changes to tissue (e.g., collagen remodelling and cellular proliferation), either as a stand-alone approach or in combination with other modalities11,12,15. Specifically to the latter, polarimetry and MS form a synergistic hybrid by complementing each other’s limitations: polarimetry is fast and wide-field, but not yet accurate enough to yield definitive diagnostics, whereas MS analysis may be slow, largely point-sensing (hence not wide-field) but is highly sensitive / specific to cancer-induced changes in tissue biochemistry and thus to molecular cancer signatures.
Our previous studies on polarimetric guidance of MS6,13,14 were demonstrated on thin tissue slices from humanized breast cancer mouse models to reveal tumourous regions for rapid MS analysis. Though interesting, those were proof-of-principle studies performed on ideal (clinically unrealistic) flat sectioned (< 50 μm thick) animal model tissues, benefitting from minimal depolarization noise, easy measurement geometry, and no variability in optical incidence angles. Importantly, clinical specimens are seldom flat nor thinner than 50 μm, and human breast tissue (as opposed to murine models) differs in adipose composition, thus the latter demonstrations remain distant from biomedical applicability. Due to the challenges associated with bulk tissue, polarimetric imaging of unprocessed bulk breast has seldom been performed; to the best of our knowledge in two studies16,17 and only one of which yielded spatial information16.
In this study, we take crucially important steps toward clinical feasibility through demonstration of polarimetric guidance of PIRL-MS on realistic, thick, non-sectioned (uneven) human breast tumour tissue in reflection-mode geometry. It is shown that circular degree of polarization (DOP) imaging – we posit via helicity flipping/preservation mechanisms – highlights adipose tissue regions (for PIRL-MS avoidance to minimize contamination). Additional polarimetric biomarkers beyond circular DOP are also explored. We note that this polarimetric imaging technique can directly optimize MS tissue sampling methods (and other sampling modalities), and may also inform on breast density which is an important breast cancer screening biomarker directly related to adipose composition18.
Materials and methods
Tissue specimens and histology
6 human breast specimens (2 healthy normals, 4 infiltrating ductal carcinomas) from surgical resections (3–4 mm thick) were obtained from the Princess Margaret Cancer Biobank (Toronto) under institutional authorization (UHN REB 18-5228). The specimens were frozen on dry ice and stored in − 80° C. The specimens were bisected while frozen to create two samples with matching surfaces. One was thawed at room temperature for ~ 10 min then polarimetrically imaged en face, and the other was kept frozen at − 80° C until subsequent polarimetrically-guided PIRL-MS probing was performed (on the matching surface side, ~ 30 s thaw). After thawing, the specimens were no longer flat, taking on clinically realistic topologies as seen in the image of specimen 1 in the side-view panel of Fig. 2.
Histological analysis was performed on the polarimetrically-imaged halves of each specimen utilizing a 4.5 μm slice taken from near the imaged surface (~ 10–20 μm below it to ensure a flat histology slice). We used two different stains: oil red O (ORO) for visualization of fat and cell nuclei; when ORO use was not successful in normal healthy tissues (stain pooling due to large fat aggregation), we used conventional hematoxylin and eosin (H&E) staining for visualization of cellular regions and normal non-adipose content. The H&E protocol prevented direct visualization of fat due to it being dissolved during the wash steps, but yielded lipid ‘footprint’ regions of empty spaces to identify its ___location (as is well known to those skilled in the art19). Thus, a combination of ORO and H&E staining yielded adequate identification of histopathologic regions of interest to contextualize the polarimetric and mass spectrometric analysis results.
Segmentation mask processing
The fat/non-fat segmentation masks (used to generate the histograms in Fig. 4, column v) were produced according to the workflow in Fig. 1 (examples shown for Healthy Specimen 2 and Cancer Specimen 1). The histology images were first annotated to contour the fatty regions (orange outline) and non-fat regions (purple outline) in consultation with a pathology expert (Fig. 1A). Only the most ‘homogenous’ regions were selected; ‘complex’ regions with intermixed fat and non-fat were disregarded. In order to achieve maximal co-registration between histology and polarimetry, the contoured histology images were superimposed onto the CC-DOP polarimetry images which enabled visualization of the tissue boundaries and their use as fiducial markers for alignment (Fig. 1B). Finally, a segmentation mask was produced by colour-filling the histology-based contours (orange = fat, purple = non-fat); the non-annotated regions (e.g., intermixed fat and non-fat) were marked as ‘undefined’ (grey) (Fig. 1C). These segmentation masks were then used as the ‘ground truth’ for fat and non-fat locations to quantify fat/non-fat classification performance of circular and linear DOP images.
Generation of the fat/non-fat segmentation mask for Healthy Specimen 2 and Cancer Specimen 1. (a) The histology image was manually annotated to contour the fat (orange) and non-fat (purple). (b) The contoured histology images were then superimposed onto the CC-DOP polarimetry image which visualize the tissue boundaries and enable their use as fiducial markers to achieve maximal co-registration. (c) The segmentation mask was produced by colour-filling the annotation outlines; the non-annotated regions (intermixed fat and non-fat) were marked as ‘undefined’ (grey).
Stokes and mueller matrix polarimetry calculus
Polarized light can be represented mathematically using Stokes vectors. A Mueller matrix (M) describes the polarimetric properties of an interacting material (e.g., tissue specimen), which relates incident (\(\:{\text{S}}_{\text{i}\text{n}}\)) and scattered (\(\:{\text{S}}_{\text{o}\text{u}\text{t}}\)) Stokes vectors according to
\(\:{\text{I}}_{\text{H}}\), \(\:{\text{I}}_{\text{V}}\), \(\:{\text{I}}_{+45}\), \(\:{\text{I}}_{-45}\), \(\:{\text{I}}_{\text{R}}\), and \(\:{\text{I}}_{\text{L}}\) are the light intensities of linearly polarized light at horizontal, vertical, \(\:+45\)°, and \(\:-45\)° orientations, and right-circular and left-circular polarizations. \(\:\text{I}\) is the total intensity of the light, typically measured by summing any two mutually orthogonal polarization states. \(\:\text{M}\)xx are the sixteen Mueller matrix elements.
Circular degree of polarization
The Stokes circular DOP describing the light beam ranges from \(\:-1\) to \(\:+1\) and was calculated as
where \(\:{\text{S}}_{\text{3,out}}/{\text{S}}_{\text{0,out}}\) is the standard calculation of circular DOP; however we multiply it by the term, \(\:{\text{S}}_{\text{3,in}}/\left|{\text{S}}_{\text{3,in}}\right|\), to assign a negative value in the case of helicity-flipping or a positive value in the case of helicity-preservation. Helicity-flipping occurs when \(\:{\text{S}}_{\text{3,out}}\) has an opposite sign to \(\:{\text{S}}_{\text{3,in}}\), indicating that the handedness of the scattered polarization state is opposite to the incident polarization state (i.e., more left-circular polarization intensity in the scattered state when the incident state was right-circular), whereas helicity-preservation occurs when \(\:{\text{S}}_{\text{3,out}}\) has the same sign as \(\:{\text{S}}_{\text{3,in}}\).
The Mueller DOP (DOCPMueller) is simply taken as the \(\:{\text{M}}_{44}\) element since it is closely related to circular polarization preservation20,21,22. This simplifies the analysis by avoiding the more complicated albeit more comprehensive calculation of ∆ (depolarization power) using Lu-Chipman polar decomposition23. When performing backscattering measurements, \(\:{\text{M}}_{44}\) becomes negative in the ‘helicity-flipping’ case24. Note that we use the term “degree of polarization” for the Mueller measurements to describe the sample’s polarization maintaining power (i.e., 1 minus depolarization power).
Since both of these DOPs showed good contrast between fat and surrounding tissues, we derived a ‘composite’ circular DOP (CC-DOP), calculated as the average of the two:
Linear degree of polarization
Stokes linear DOP is conventionally calculated as DOLPStokes\(\:=\surd\:\left({\text{S}}_{\text{1,out}}^{\text{2}}\text{+}{\text{S}}_{\text{2,out}}^{\text{2}}\right)/{\text{S}}_{0,\text{o}\text{u}\text{t}}\). Here in analogy with circular helicity preserving/flipping mechanisms described above, we calculate it such that it also ranges from \(\:-1\) to \(\:+1\):
where \(\:\left(-{\text{S}}_{1,\text{i}\text{n}}/\left|{\text{S}}_{1,\text{i}\text{n}}\right|\right)\cdot\:\left({\text{S}}_{1,\text{o}\text{u}\text{t}}/{\text{S}}_{0,\text{o}\text{u}\text{t}}\right)\) and \(\:\left({\text{S}}_{2,\text{i}\text{n}}/\left|{\text{S}}_{2,\text{i}\text{n}}\right|\right)\cdot\:\left({\text{S}}_{2,\text{o}\text{u}\text{t}}/{\text{S}}_{0,\text{o}\text{u}\text{t}}\right)\) become negative when there is higher co-linear polarization intensity than cross-linear polarization intensity in the scattered light (for example, when \(\:{\left\{{\text{I}}_{\text{V}}\right\}}_{\text{o}\text{u}\text{t}}\) is greater than \(\:{\left\{{\text{I}}_{\text{H}}\right\}}_{\text{o}\text{u}\text{t}}\) with incident linear vertically polarized light). This enables fair comparison between Stokes linear and circular DOP since both will now take on negative values upon direct backscatter events such as specular reflection and remain positive otherwise.
Mueller linear DOP is simply calculated as DOLPMueller\(\:={-1\cdot\:\text{M}}_{22}+{\text{M}}_{33}\) since these elements are strongly correlated to the linear polarization preservation20,21,22 (again, simplifying the analysis by avoiding polar decomposition). The negative sign is applied to the \(\:{\text{M}}_{22}\) element such that it becomes negative for direct backscatter / mirror-like reflections, akin to \(\:{\text{M}}_{33}\)24,25,26, thus enabling DOLPMueller to vary across the same dynamic range of \(\:-1\) to \(\:+1\) as DOCPMueller for fair comparison.
Analogously to Eq. (3), the composite linear DOP (LL-DOP) was calculated as the average between the Stokes and Mueller linear DOPs:
Retardance, linear diattenuation, and circular diattenuation
Retardance and linear and circular diattenuation were calculated using Lu-Chipman polar decomposition of the Mueller matrix as laid out in Ref23.
Experimental polarimetric imaging system
The polarimetric imaging system employed a helium-neon laser source (λ = 632.8 nm) and intensified-CCD (ICCD) camera detector (PI-MAX® 3, Princeton Instruments), configured into a 180° backscatter geometry detection scheme as shown in Fig. 2. Before tissue incidence, the light was (1) directed through a polarization state generator (PSG) consisting of a linear polarizer followed by a quarter wave retarder, then (2) reflected from a dielectric mirror oriented at 45° to steer the light towards the specimen for top-down illumination, and finally (3) passed through a glass beam splitter (BS) oriented at 45° (which transmitted ~ 90% of the light). The backscattered light from the specimen was reflected from the beam splitter and passed through a polarization state analyzer (PSA), consisting of a quarter wave retarder followed by a linear polarizer, then focused onto the camera via a lens to form an image.
The Stokes-Mueller equation to relate the incident Stokes vector (\(\:{\text{S}}_{\text{incident}}\)) to the detected Stokes vector (\(\:{\text{S}}_{\text{detected}}\)) in the polarimetric imaging system is:
\(\:{\text{M}}_{\text{P}\text{S}\text{A}}\) and \(\:{\text{M}}_{\text{P}\text{S}\text{G}}\) are the conventional Mueller matrices for the PSA and PSG depending on their configurations. \(\:{\text{M}}_{\text{B}\text{S},\text{t}\text{r}\text{a}\text{n}\text{s}}\), \(\:{\text{M}}_{\text{B}\text{S},\text{r}\text{e}\text{f}}\), and \(\:{\text{M}}_{\text{m}\text{i}\text{r}\text{r}\text{o}\text{r}}\) are the Mueller matrices for transmission through the BS, 45° reflection from the BS, and 45° reflection from the mirror, respectively. \(\:{\text{M}}_{\text{m}\text{i}\text{r}\text{r}\text{o}\text{r}}\) and \(\:{\text{M}}_{\text{B}\text{S},\text{r}\text{e}\text{f}}\) were measured to be roughly the same as the 90°-reflection Mueller matrix from a dielectric surface (this does not follow the theoretical calculation, as been observed previously26 and may be due to a thin-film coating27); \(\:{\text{M}}_{\text{B}\text{S},\text{t}\text{r}\text{a}\text{n}\text{s}}\) matched the theoretical calculation (see Ref26. for details). Thus we solved for Msample, the only unknown remaining in Eq. (6). To increase signal-to-noise, we used 36 measurements (6 \(\:{\text{S}}_{\text{i}\text{n}\text{c}\text{i}\text{d}\text{e}\text{n}\text{t}}\) and 6 \(\:{\text{S}}_{\text{detected}}\) polarization states versus the minimum 4 and 4\(\:)\) and the Mueller matrix calculation methodology outlined in Ref28.
Schematic of the 180°-reflection-mode polarimetric imaging system. The incident beam transmits through the PSG and is redirected by a mirror for top-down illumination of the specimen. Between the mirror and the specimen is a beam splitter oriented at 45°, which reflects the tissue-backscattered light through the PSA followed by the ICCD camera. The side view inset depicts the top-down illumination of the specimen and the beam splitter detection scheme to enable imaging in the exact backscattering geometry.
PIRL-MS system
PIRL-MS measurements were performed using a fiber-coupled laser unit (2,850 nm, 800 picoseconds, 1 kHz rep rate, fibre diameter = 0.425 mm) from Light Matter Interaction, Etobicoke, Canada, outputting ∼300 mW average power after fiber coupling. Mass spectra were recorded inside a biological safety cabinet on a Xevo-G2-XS time-of-flight mass spectrometer (Waters, Milford, MA, USA) using a dedicated tissue imaging station (Fig. 5, columnv, bottom graphic). The areas within the ellipses in Fig. 5, row B were sampled by continuously moving the PIRL-MS source within the area and averaging the mass spectral data. The signal was averaged over ~ 3–4 mm area (corresponding to 10–20 s of spectral collection). The ion block mass spectrometry analysis interface has been described previously in Ref2.
Results and discussion
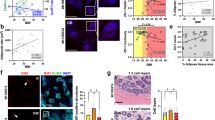
Porcine specimen: selecting optimal polarimetric signatures of adipose tissue
Porcine tissue was studied prior to the human breast specimens to explore polarization metrics that might yield useful contrast between fat and non-fat tissue components. Figure 3 shows the images, specifically its white-light photograph (Fig. 3A), retardance (B), linear diattenuation (C), circular diattenuation (D), Stokes linear DOP (E), Stokes circular DOP (F), Mueller linear DOP (G), and Mueller circular DOP (H).
Retardance, linear diattenuation, and circular diattenuation (Fig. 3B-D) obtained from polar decomposition analysis seem only modestly useful for fat discrimination. Retardance (Fig. 3B) is slightly higher in the muscle than in the fat, likely due to the birefringence of muscle fibres29. The fat induces more linear diattenuation than does the muscle (Fig. 3C) which is somewhat unexpected in light of previous reports29; however, this is not yet well-studied and perhaps diattenuation decreases in muscle as the porcine tissue ages29. Circular diattenuation appears to be slightly lower in fat compared to muscle (Fig. 3D). Overall, these three polar-decomposition-derived Mueller polarimetric biomarkers do not delineate the fat regions well, even in this simple biological system.
The most prominent fat/non-fat contrast is found in the linear and circular DOP images using both Stokes and Mueller approaches, as seen in Fig. 3E-H. The fat is seen to be far more depolarizing (positive values but lower magnitude). This has been previously observed30 and can be attributed to the higher scattering coefficient (µs) of fat compared to that of porcine muscle which typically results in higher depolarization31,32 (Ref33. reports fat µs ≈ 110 cm-1 vs. porcine muscle µs ≈ 74 cm-1). It is unclear why the linear and circular DOP effects are similar in the muscle area, as one expects the associated linear birefringence to induce more linear depolarization randomization34. This may be due to the relatively low difference in retardance between the muscle and fat (Fig. 3B).
Polarimetric imaging of the porcine tissue model, towards selecting the optimal fat-delineating polarimetric biomarkers in human pathologies. (A) White-light image, with the fat region on the left side and muscle on the right as labelled. (B) The retardance image, (C) linearly diattenuation image, (D) circular diattenuation image. (E-H) Stokes and Mueller DOP images using linear (E, G) and circular (F, H) polarization yields the most prominent fat discrimination.
Interestingly, the linear DOP images seem less uniform in an otherwise homogeneous region; for example, notice the higher values in the top left corners of each image (Fig. 3E and G). Additionally, the edges of the tissue, particularly on the right side, appear to increase sharply more so in the linear compared to circular DOP images. This may stem from the abrupt changes in curvature at the edges/corners of the specimen, resulting in varying angles of incidence which can induce strong linear diattenuation effects35. Indeed, this is corroborated by the linear diattenuation image Fig. 3C which also exhibits sharp value changes in the corners/edges of the tissue (compare to circular diattenuation of Fig. 3D). Linear polarization-based metrics, including linear DOP, are thus likely more sensitive to changes in curvature which become important when imaging the uneven human breast resections.
Overall, the porcine polarimetric imaging suggests that linear and circular DOP measurements offer the most robust fat/non-fat contrast, thus we now proceed to polarimetrically examine human breast normal and tumour resections through linear and circular DOP imaging.
Human breast specimens: polarimetric discrimination between fat and non-fat
Figure 4 shows the white-light images (row A) and stained histology slices (row B) of resected patient breast tissue specimens. Healthy specimens are H&E-stained and cancer specimens are ORO-stained (see Sect. 2.1 for details). Overall, it is clearly observed in the histology images that the cancer specimens are noticeably more heterogeneous than the healthy specimens, particularly exhibiting more intermixed fat regions. Also notice that the fatty areas are not visually identifiable in the white-light images of the cancer specimens compared to normal specimens, demonstrating the need for staining-free polarimetric identification.
The fat and non-fat region of each specimen are demarcated by the segmentation masks in row C with orange and purple contours, respectively (see Fig. 1 and related text for details on these masks). Since both Mueller- and Stokes-based DOP measurements yielded fat/non-fat contrast in porcine, we use composite metrics for linear and circular DOP which combine Mueller and Stokes quantities as defined in Eq. (5) and Eq. (3), respectively. Row D displays the composite linear degree of polarization (LL-DOP) and row F displays composite circular degree of polarization (CC-DOP) images of each specimen; all images are displayed within the same color bar range between − 0.7 and + 0.2.
The segmentation masks are laid over the LL-DOP and CC-DOP images to count the number of pixels that lie within the fat region (orange) and non-fat region (purple) of each specimen as shown in the corresponding histograms of row E and row G, respectively. We utilize the histograms, shown in row E and G, to gain a general indication of DOP-based differentiation between fat and non-fat by simply considering the differences (∆) between the mean values of fat (<F>) and non-fat (<NF>) pixels. To more rigorously evaluate fat discrimination performance, receiver operating characteristic (ROC) curves with accompanying area under the curve (AUC) values are shown for CC-DOP images in row H; we only show these curves for CC-DOP images since they yield larger ∆ values. The AUC values for the LL-DOP images were 0.42, 0.47, 0.84, and 0.83 for Healthy Specimens 1 and 2 and Cancer Specimens 1 and 2, respectively. Notice that LL-DOP only outperforms CC-DOP for one specimen, namely Cancer Specimen 2, while offering no fat discrimination for Healthy Specimens 1 and 2 (AUC < 0.5). These pixel-wise ROC curves were generated by plotting (x, y) = (1-specificity, sensitivity) for each given ‘classification threshold setting’ incremented from − 1 to + 1 (i.e., the full range of possible CC-DOP values) in steps of 0.05 (-0.95, -0.90, …, + 0.90, + 0.95). Thus, for each threshold setting, a sensitivity and specificity calculation is performed on a per-pixel basis using the numbers of true and false positives and true and false negatives as follows. For each iteration of a classification threshold setting, true positives are considered pixels which are located in the fat region (orange region of the segmentation masks; see row C) with values that are above the given threshold setting, while true negatives are considered pixels located in the non-fat region (purple region of the segmentation masks) with values below the given threshold setting. False positives are pixels in the non-fat region with values above the given threshold, and false negatives are pixels in the fat region with values below the given threshold. Note that pixels in the ‘undefined’ region (colour-coded gray in the segmentation masks) are not used in these calculations.
More quantitative comparisons and statistical tests of significance are possible, but will await more accurate contextualization with histology ‘ground truth’ results (issues of slight tissue deformation during slide preparation, usage of two stains, precise co-registration due to sub-surface ___location of the histology slice, etc.).
Investigation of fat discrimination via CC-DOP imaging of human breast tissues. White-light images (rows A) and stained histology slices (rows B) are shown for two healthy specimens (H&E-stained) and two cancer specimens (ORO-stained). Fat/non-fat segmentation masks (rows C) indicate the locations of the fat and non-fat regions of the specimens, which are superimposed on the LL-DOP images (row D) and on the CC-DOP images (row F) to enable the fat/non-fat histograms shown respectively in row E and row G. Row H shows ROC curves with corresponding AUC values to evaluate fat segmentation performance of CC-DOP images, indicating strong fat discrimination for each specimen with exception of Healthy Specimen 2 which shows some fat discrimination. Overall, CC-DOP values appear to correlate with fat content (higher values) and potentially dense-cellular content (high magnitude negative values).
Consistent with the porcine study summarized in Fig. 3, we conclude that human fat regions also tend to exhibit higher linear and circular DOP values. However, it is observed that CC-DOP offers greater and more consistent contrast between fat and non-fat, whereby the mean difference between fat and non-fat (denoted by ∆ on each histogram plot) is higher in CCDOP histograms. Again, it appears that linear DOP may be more sensitive to changes in curvature; for example, Healthy Specimen 1 is hemispherical in shape (e.g., see specimen image in Fig. 2) causing a ring-like band of specular reflection to appear (see arrows in Fig. 4, row D, col.i); although this effect is also somewhat seen in the CC-DOP image in Fig. 4, row F, col.i, but much less prominent. For circular DOP, interestingly, the highest values are seen in Healthy Specimen 1 which has the largest aggregation of fat, suggesting potential proportionality between CCDOP values and adipose content. Beyond fat discrimination, it is also noteworthy that dense cellular regions appear to exhibit high magnitude negative CC-DOP values. For example, notice that the ‘ductal cell cluster’ area of Healthy Specimen 2 (labelled in the histology image of Fig. 4, row B, col.ii) corresponds to highly negative values in the corresponding CC-DOP image, and similarly for Cancer Specimen 1’s densely packed cellular region in the upper left corner (see Fig. 4, row B, col.iii); also see Fig. 1 for more detailed views of the histology images. Therefore, higher CC-DOP values and high magnitude negative values may be useful in identifying fatty and dense cellular regions, respectively.
Circularly polarization’s sensitivity to scatterer size: the fat/non-fat contrast mechanism
The underlying mechanism responsible for the correlation between (1) fatty regions and higher CC-DOP values (mainly in the positive regime) and (2) cellular regions and lower CC-DOP values (in the negative regime) may be due to circular polarization’s sensitivity to ‘scatterer size’. Adipose tissue has been measured to have a far larger scatterer size than non-fat tissue in breast specimens, for example, by a factor of ~ 2.5 in Ref36. , and by larger factors in Ref37. (also, the size difference between fat cells and ductal cells can be somewhat seen in the histology image of Healthy Specimen 2 in Fig. 1). Fat cells (lipid globules) are the main scatterers in adipose breast tissue (diameter d ~ 20–120 μm38), whereas the scatterers in non-fat breast tissue are mainly cell nuclei (d ~ 5–15 μm) and smaller sub-cellular constituents (e.g., organelles with d < 2 μm39), and collagen fibres (d > 1 μm diameter40); see Refs36,37,38,41,42. for additional details.
Circular DOP takes on higher values through a higher preponderance of helicity-preserved light, whereas lower values arise from a higher fraction of helicity-flipped light (e.g., see Eq. (3)). Previous studies, including our own work31,43,44,45, have shown that helicity preservation occurs in media consisting of larger scattering particles, and helicity-flipped light dominates in media consisting of smaller particles (the actual sizes for this categorization depend on Mie scattering parameters, including laser wavelength, refractive index mismatch, and particle shape44). Thus the observed mostly positive values of the circular DOP corresponding to the adipose regions of each specimen suggest a helicity-preservation response of the larger fat cells. Conversely, since the lowest (negative) values generally corresponded to highly cellular regions, the observed helicity-flip responses may be responsible due to the smaller-sized nuclei along with sub-micron organelles which can act as Rayleigh scatterers46 (Rayleigh scatterers are known to induce strong helicity-flip responses via intense backward scattering (small g-factor)44). Also note that scattering from cylinders (i.e., collagen fibres) also results in negative circular DOP values, depending on their size and other parameters47, which may explain the somewhat negative CC-DOP values in collagenous regions.
We thus posit that circular polarization offers a unique advantage for MS guidance in differentiating fat from non-fat and in detecting highly cellular regions through its scatterer-size dependency.
Polarimetrically-guided PIRL-MS: fat avoidance
The finding that higher CC-DOP corresponds to fatty regions can be exploited to generate binarized polarimetry maps for PIRL-MS guidance. Such maps mark the presence of fat in each specimen by colour-coding each pixel of the CC-DOP images red if the pixel value is above the specified threshold of \(\:-\)0.05 and colour-coding pixels blue when below that threshold, as seen in Fig. 5, rows A and B. The threshold of \(\:-\)0.05 was chosen through examination of the CC-DOP images and their histograms in columnsiii and v of Fig. 4 which show that the histograms of fat pixels generally range from approximately \(\:-\)0.05 and greater for each specimen. Healthy Specimen 2 appears to contain ‘false positives’ where the non-fat regions exhibit CC-DOP values above \(\:-\)0.05; however, this is less problematic than ‘false negatives’ (i.e., fat not identified) which would result in a PIRL-MS contamination risk. The precise threshold choice is obviously somewhat arbitrary, and a more rigorous analysis of resultant sensitivity to chosen values is warranted; this will be pursued in future studies as the number of human samples is increased and histology correlation issues alluded to before are streamlined.
Polarimetric guidance maps enable efficient contamination-free PIRL-MS analysis of cancerous breast tissue by highlighting fat (to be avoided). Row A shows these guidance maps by discretizing and colour-coding the CC-DOP images through thresholding where red and blue indicate fat and non-fat respectively. Row B shows the colour-coded maps of Row A after automatic smoothing via maximum filtering. Rows C and D show PIRL-MS spectra from non-fat and fatty regions of Cancer Specimen 1, confirming that fatty tissue offers the weakest MS signal and non-fat regions offer the strongest, most feature-rich signal. Additionally, 6 known breast-cancer-correlated m/z peaks are detected in the non-fat region (orange font). Column v depicts the step-wise workflow of polarimetrically-guided PIRL-MS.
The resultant maps in Fig. 5, row A do not present uniformly colour-coded adipose regions due to inherent biological heterogeneity and analysis artefacts. Thus a maximum filter is applied which replaces each pixel with the brightest pixel that is found within a 2-pixel radius (provided by Python library SciPy48), yielding more continuous and uniform red-coloured fat regions, as seen in Fig. 5, row B. Note that a grey border was also placed along the outer edges of each specimen, labelled as ‘undefined’, due to the frequent appearance of edge artefacts and resultant uncertainty along the rim. These maps now provide polarimetric guidance away from fatty tissue (to avoid with PIRL-MS). The step-wise workflow of polarimetrically-guided PIRL-MS is depicted in column v.
As a representative assessment of these discretized maps, PIRL-MS analysis was performed on Cancer Specimen 1 according to the map in Fig. 5, row B, col. iii. This is achieved through user-mediated co-registering of the polarimetry map to the optical image of the specimen; in the future, this process can be automated using real time image co-registration and augmented reality as previously demonstrated with PIRL-MS9. As expected, the red-coloured fatty region yields the lowest MS signal (i.e., total ion count), whereas the blue-coloured non-fat region yields the highest and most feature-rich MS signal (Fig. 5, rows C and D). Additionally, PIRL sampling of the adipose region indeed resulted in lipid liquefaction and hence a contamination risk, demonstrating the need to avoid fatty regions.
Interestingly, PIRL-MS detected 6 known breast-cancer-correlated m/z peaks previously seen in desorption electrospray ionization MS studies10. These are labelled in orange (tentative assignments) and the rest of the abundant ions (to be characterized) in black. Importantly, notice that these peaks are least prominent or absent (e.g., m/z\(\:=\) 391.2 and 655.5) in the fat region’s spectral profile; the m/z\(\:=\) 572.5 peak corresponding to a chlorinated adduct of ceramide Cer(d34:1) seen previously in many PIRL-MS studies of other cancerous tissues1,2,3,9 is also absent from the adipose area.
These discretized maps therefore enable guidance away from adipose tissue to avoid lipid contamination whilst enabling rapid acquisition of pathological information to yield a diagnosis, saving significant MS analysis time. Both such feats significantly improve the clinical promise of PIRL-MS.
Conclusions
This study demonstrates that adipose and non-adipose tissue of clinically-realistic normal and cancerous human breast tissue (thick, uneven, non-sectioned, unstained) can be highlighted through label-free polarimetric imaging, towards time-efficient and contamination-free PIRL-MS oncological characterization. Polarimetric guidance was achieved through circular DOP imaging, which was advantageous relative to other polarization metrics potentially through its scatterer-size sensitivity mechanism, along with its apparent insensitivity to irregular surface topologies. A polarimetric guidance map was assessed with PIRL-MS, affirming the map’s tissue segmentation accuracy. The utility of the polarimetry-MS hybrid technology approach in unprocessed bulk human pathology tissues may prove advantageous in eventual clinical deployment.
Data availability
Data underlying the results presented in this paper are not publicly available at this time due to patient confidentiality but may be obtained from the corresponding author upon reasonable request.
Change history
05 December 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41598-024-81467-y
References
Woolman, M. et al. and A. Zarrine-Afsar, Picosecond infrared laser desorption mass spectrometry identifies medulloblastoma subgroups on intrasurgical timescales, Cancer Res 79(9), 2426–2434 (2019).
Woolman, M. et al. Lipidomic-Based Approach to 10 s classification of Major Pediatric Brain Cancer types with Picosecond Infrared Laser Mass Spectrometry. Anal. Chem. 96 (3), 1019–1028 (2024).
Katz, L. et al. Picosecond Infrared Laser Mass Spectrometry Identifies a Metabolite Array for 10 s Diagnosis of Select Skin Cancer Types: A Proof-of-Concept Feasibility Study. Anal. Chem. 94 (48), 16821–16830 (2022).
Woolman, M. et al. Rapid determination of medulloblastoma subgroup affiliation with mass spectrometry using a handheld picosecond infrared laser desorption probe. Chem. Sci. 8 (9), 6508–6519 (2017).
Ifa, D. R. & Eberlin, L. S. Ambient ionization mass spectrometry for cancer diagnosis and surgical margin evaluation. Clin. Chem. 62 (1), 111–123 (2016).
Woolman, M. et al. Optimized Mass Spectrometry Analysis Workflow with Polarimetric Guidance for ex vivo and in situ Sampling of Biological Tissues. Sci. Rep. 7 (1), 1–12 (2017).
Hänel, L., Kwiatkowski, M., Heikaus, L. & Schlüter, H. Mass spectrometry-based intraoperative tumor diagnostics. Future Sci. OA 5(3), (2019). https://doi.org/10.4155/fsoa-2018-0087
Woolman, M., Katz, L., Tata, A., Basu, S. S. & Zarrine-Afsar, A. Breaking Through the Barrier: Regulatory Considerations Relevant to Ambient Mass Spectrometry at the Bedside. Clin. Lab. Med. 41 (2), 221–246 (2021).
Woolman, M. et al. In situ tissue pathology from spatially encoded mass spectrometry classifiers visualized in real time through augmented reality. Chem. Sci. 11 (33), 8723–8735 (2020).
Calligaris, D. et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc. Natl. Acad. Sci. U S A. 111 (42), 15184–15189 (2014).
Singh, M. D., Ghosh, N. & Vitkin, I. A. Mueller Matrix Polarimetry in Biomedicine: Enabling Technology, Biomedical Applications, and Future Prospects, in Polarized Light in Biomedical Imaging and Sensing, (eds Ramella-Roman, J. C. & Novikova, T.) (Springer International Publishing, 61–103. (2023).
He, C. et al. Polarisation optics for biomedical and clinical applications: a review. Light Sci. Appl. 10 (1), 194 (2021).
Tata, A. et al. Wide-field tissue polarimetry allows efficient localized mass spectrometry imaging of biological tissues. Chem. Sci. 7 (3), 2162–2169 (2016).
Tata, A. et al. Rapid detection of necrosis in breast cancer with desorption electrospray ionization mass spectrometry. Sci. Rep. 6 (October), 1–10 (2016).
Qi, J. et al. Surgical polarimetric endoscopy for the detection of laryngeal cancer. Nat. Biomed. Eng. 7, 971–985 (2023). https://doi.org/10.1038/s41551-023-01018-0
Liu, T. et al. Comparative study of the imaging contrasts of Mueller matrix derived parameters between transmission and backscattering polarimetry. Biomed. Opt. Express. 9 (9), 4413 (2018).
Sharma, M. et al. Histopathological correlations of bulk tissue polarimetric images: Case study. J. Biophotonics. 14 (5), 1–12 (2021).
Nazari, S. S. & Mukherjee, P. An overview of mammographic density and its association with breast cancer. Breast Cancer. 25 (3), 259–267 (2018).
Berry, R. et al. Imaging of Adipose Tissue 1st edn 537 (Elsevier Inc., 2014).
Antonelli, M. R. et al. Mueller matrix imaging of human colon tissue for cancer diagnostics: how Monte Carlo modeling can help in the interpretation of experimental data. Opt. Express. 18 (10), 10200 (2010).
Novikova, T. et al. The origins of polarimetric image contrast between healthy and cancerous human colon tissue. Appl. Phys. Lett. 102 (24), 241103 (2013).
Sun, M. et al. Probing microstructural information of anisotropic scattering media using rotation-independent polarization parameters. Appl. Opt. 53 (14), 2949 (2014).
Lu, S. Y. & Chipman, R. A. Interpretation of Mueller matrices based on polar decomposition. J. Opt. Soc. Am. A. 13 (5), 1106 (1996).
Schwartz, C. & Dogariu, A. Backscattered polarization patterns determined by conservation of angular momentum. J. Opt. Soc. Am. A. 25 (2), 431 (2008).
Bohren, C. F. & Huffman, D. R. Absorption and Scattering by a Sphere, in Absorption and Scattering of Light by Small Particles (Wiley-VCH Verlag GmbH, 7(1), 82–129. (2007).
Chen, Z., Yao, Y., Zhu, Y. & Ma, H. Removing the dichroism and retardance artifacts in a collinear backscattering Mueller matrix imaging system. Opt. Express. 26 (22), 28288 (2018).
Gramatikov, B. I. A Mueller matrix approach to flat gold mirror analysis and polarization balancing for use in retinal birefringence scanning systems. Optik (Stuttg). 207 (February), 164474 (2020).
Baba, J. S., Chung, J. R., DeLaughter, A. H., Cameron, B. D. & Coté, G. L. Development and calibration of an automated Mueller matrix polarization imaging system. J. Biomed. Opt. 7 (3), 341 (2002).
Pardo, I. et al. Wide-field Mueller matrix polarimetry for spectral characterization of basic biological tissues: Muscle, fat, connective tissue, and skin. J. Biophotonics 17 (1), (2023).
Sankaran, V., Walsh, J. T. & Maitland, D. J. Comparative study of polarized light propagation in biologic tissues. J. Biomed. Opt. 7 (3), 300 (2002).
Singh, M. D. & Vitkin, I. A. Discriminating turbid media by scatterer size and scattering coefficient using backscattered linearly and circularly polarized light. Biomed. Opt. Express. 12 (11), 6831 (2021).
Nader, C. A. et al. Influence of size, proportion, and absorption coefficient of spherical scatterers on the degree of light polarization and the grain size of speckle pattern. Appl. Opt. 54 (35), 10369 (2015).
Sun, P. & Wang, Y. Measurements of optical parameters of phantom solution and bulk animal tissues in vitro at 650 nm. Opt. Laser Technol. 42 (1), 1–7 (2010).
Jacques, S. L., Roman, J. R. & Lee, K. Imaging superficial tissues with polarized light. Lasers Surg. Med. 26 (2), 119 (2000).
Pezzaniti, J. L. & Chipman, R. A. Angular dependence of polarizing beam-splitter cubes. Appl. Opt. 33 (10), 1916 (1994).
Wang, X. et al. Image reconstruction of effective Mie scattering parameters of breast tissue in vivo with near-infrared tomography. J. Biomed. Opt. 11 (4), 041106 (2006).
Wang, X. et al. Approximation of Mie scattering parameters in near-infrared tomography of normal breast tissue in vivo. J. Biomed. Opt. 10 (5), 051704 (2005).
Suárez–Nájera, L. E. et al. Castro– Reyes, Morphometric study of adipocytes on breast cancer by means of photonic microscopy and image analysis. Microsc Res. Tech. 81 (2), 240–249 (2018).
Bartek, M., Wang, X., Wells, W., Paulsen, K. D. & Pogue, B. W. Estimation of subcellular particle size histograms with electron microscopy for prediction of optical scattering in breast tissue. J. Biomed. Opt. 11 (6), 064007 (2006).
Bodelon, C. et al. Mammary collagen architecture and its association with mammographic density and lesion severity among women undergoing image-guided breast biopsy. Breast Cancer Res. 23 (1), 1–14 (2021).
Arifler, D., Pavlova, I., Gillenwater, A. & Richards-Kortum, R. Light scattering from collagen fiber networks: micro-optical properties of normal and neoplastic stroma. Biophys. J. 92 (9), 3260–3274 (2007).
Bevilacqua, F., Marquet, P., Coquoz, O. & Depeursinge, C. Role of tissue structure in photon migration through breast tissues. Appl. Opt. 36 (1), 44 (1997).
Singh, M. D. & Vitkin, I. A. Spatial helicity response metric to quantify particle size and turbidity of heterogeneous media through circular polarization imaging. Sci. Rep. 13 (1), 2231 (2023).
Macdonald, C. M., Jacques, S. L. & Meglinski, I. V. Circular polarization memory in polydisperse scattering media. Phys. Rev. E. 91 (3), 033204 (2015).
MacKintosh, F. C., Zhu, J. X., Pine, D. J. & Weitz, D. A. Polarization memory of multiply scattered light. Phys. Rev. B. 40 (13), 9342–9345 (1989).
Fang, H. et al. Noninvasive sizing of subcellular organelles with light scattering spectroscopy. IEEE J. Sel. Top. Quantum Electron. 9 (2), 267–276 (2003).
Mishchenko, M. I., Travis, L. D. & Macke, A. Scattering of light by polydisperse, randomly oriented, finite circular cylinders. Appl. Opt. 35 (24), 4927 (1996).
Virtanen, P. et al. T. Krauss, U. Upadhyay, Y. O. Halchenko, and Y. Vázquez-Baeza, SciPy 1.0: fundamental algorithms for scientific computing in Python, Nat Methods 17(3), 261–272 (2020).
Funding
Canadian Institutes of Health Research (CIHR, PJT-156110), the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-04930), and the New Frontiers in Research Fund (NFRFE-2019-01049).
Author information
Authors and Affiliations
Contributions
M.S. performed the polarimetric imaging experiments and subsequent data analysis, prepared all figures, and wrote the main manuscript. L.Y. procured and prepared the experimental specimens and oversaw histological processing, performed the PIRL-MS sampling experiments and subsequent data analysis, prepared parts of Fig. 5, and wrote parts of the Methods section. M.W. assisted in procuring the experimental specimens, helped/oversaw specimen preparation and histological processing, and aided in the PIRL-MS probing. M.W. also made foundational contributions to the development of the PIRL-MS system. F.T. assisted in the design of the PIRL MS set up used and Fig. 5 is displaying his design. A.V. and AZA were the supervising professors who oversaw the entire project, provided guidance, and revised major parts of the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
Arash Zarrine-Afsar and Michael Woolman are inventors of soft ionization used here for PIRL-MS analysis and are paid consultants with Point Surgical Inc. with financial interest. All the remaining authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained an error in the spelling of the author Arash Zarrine-Afsar which was incorrectly given as Arash Zarrine-Asfar.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Singh, M.D., Ye, L.A., Woolman, M. et al. Reflection mode polarimetry guides laser mass spectrometry to diagnostically important regions of human breast cancer tissue. Sci Rep 14, 26230 (2024). https://doi.org/10.1038/s41598-024-77963-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-77963-w