Abstract
ETS variant transcription factor 5 (ETV5), a master transcription factor during development, exerts vital function on the occurrence and progression of various cancers. In order to systematically analyze and explore ETV5 potential specific regulatory mechanisms in pan-cancer, RNA sequencing data and clinicopathological features of patients with various tumors were obtained through the Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases, and an integrated data mining analysis was carried out, including the association of ETV5 expression with patient prognosis, drug sensitivity and epigenetic modification. The results revealed that abnormally highly expressed ETV5 resulted in unfavorable prognosis and differential drug sensitivity in multiple malignancies, and its expression was associated with epigenetic modification modulators including EZH2. ETV5 related genes were enriched in tumorigenesis biological processes and signaling pathways. In hepatocellular carcinoma, ETV5 expression was correlated with patients’ tumor pathological stage and resulted in adverse outcome of patients. Our further experiments evidences indicated that ETV5 facilitated cell proliferation and reduced sensitivity to GSK126 via regulating EZH2. Collectively, this study comprehensively elucidates the carcinogenic effects and molecular mechanisms of ETV5 in tumorigenesis and development, and provides theoretical basis and guidance for tumor diagnosis, targeted therapy for ETV5 and clinical epigenetic drug research.
Similar content being viewed by others
Introduction
As a worldwide disease problem, cancer poses a serious threat to human health due to its high incidence and mortality rate. It was estimated that the global cancer incidence will exceed 28.4 million annually, accounting for one in six deaths by 20401,2. The current treatment methods for tumors mainly include surgical resection, radiation therapy, chemotherapy3, targeted therapy4 and immunotherapy5. However, the heterogeneity of the tumors, individual patient variability6 and drug resistance during treatment7,8 lead to unsatisfactory therapeutic outcomes. With the development of epigenetics and epitranscriptomics, epigenetic therapy, a potential treatment for highly aggressive tumors, offers new hope for patients9. Single-cell sequencing and multi-omics analysis also revealed the importance of epigenetic modification in tumor heterogeneity. Currently, inhibitors targeting epigenetic modification modulators DNMTs, HDACs, and EZH2 have been approved by the FDA for various malignancies treatment, including 5-azacytidine, decitabine, romidepsin, vorinostat, belinostat, panobinostat and tazemetostat10. Besides, several inhibitors of LSD1, BET and DOT1L are also employed in experimental and preclinical researches to explore better anti-tumor effect10,11.
The tumorigenesis and tumor progression are accompanied by abnormal regulation of multiple genes, including transcription factors. ETS is one of the largest transcription factor families and involved in various developmental and physiological processes. It has been reported that ETS family members interact with chromatin remodeling complexes to regulate the chromatin accessibility of target genes12. Our previous studies identified ETV4, a member of ETS family, was activated by H3K27ac and facilitated angiogenesis, cell migration and invasion in hepatocellular carcinoma (HCC)13,14. ETV5 was a crucial driver gene in tumorigenesis15 and involved in varieties of cancer characteristic phenotypes, including cell proliferation16,17, epithelial-mesenchymal transition (EMT)18,19, migration and invasion20,21,22, angiogenesis23,24 and drug resistance25,26. Nevertheless, it remains unclear that ETV5 affects related biological processes and signaling pathways through regulating gene expression with epigenetic modification and contributes to the etiology of various cancers due to the complexity of tumorigenic mechanisms. Up to date, no pan-cancer analysis was reported to reveal the association between ETV5 and multiple cancers.
The present study indicated that ETV5 was a vital oncogenic transcription factor in tumorigenesis and development of various tumors, and ETV5 expression level may affect the epigenetic modification and tumor progression related genes expression, and was associated with drug sensitivity and patients’ unfavorable prognosis and tumor pathological stage, especially in hepatocellular carcinoma (HCC). In summary, this study offers a complete pan-cancer analysis for ETV5 gene and an important guidance for tumor diagnosis, anti-tumor targeted therapy and clinical drug research related to epigenetics.
Results
ETV5 expression profile and its association with prognosis evaluation in pan-cancer
To investigate ETV5 expression profile in pan-cancer, 26 types cancers from the Cancer Genome Atlas (TCGA) database were selected to analyze ETV5 differential expression pattern in tumor specimens compared with normal tissues. As shown in Fig. 1A, compared with adjacent normal tissues, ETV5 expression level was significantly higher in 14 types tumor tissues, including colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), stomach adenocarcinoma (STAD), liver hepatocellular carcinoma (LIHC), thyroid carcinoma (THCA) and rectum adenocarcinoma (READ). Only 6 types tumor tissues showed remarkably lower expression, containing lung adenocarcinoma (LUAD), breast invasive carcinoma (BRCA), and prostate adenocarcinoma (PRAD). Meanwhile, based on TCGA and Genotype-Tissue Expression (GTEx) databases (34 types cancers), it was identified that ETV5 expression level was obviously higher in 25 types tumor tissues compared with that in adjacent normal tissues (Supplementary Fig. S1A), which was consistent with the analysis from TCGA databases. Notably, only 2 tumor tissues, BRCA and PRAD, showed significantly lower expression level.
The different expression level of ETV5 in tumors and normal tissues among the 26 types cancers, and its association with prognosis. (A) The mRNA expression levels of ETV5 among various tumors and adjacent normal tissues according to The Cancer Genome Atlas (TCGA) database. (B) The forest map displayed the correlation between ETV5 expression level and overall survival (OS) in pan-cancer via univariate Cox regression analysis. (Hazard ratio > 1 represents unfavorable prognostic factors, while Hazard ratio < 1 represents favorable prognostic factors, CI: confidence interval.) (C) Subcellular localization of ETV5 in various cancer cells based on immunofluorescence staining images from The Human Protein Atlas (HPA) database. (ns: P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001)
To further assess the prognostic value of ETV5 in various tumors, univariate Cox regression was applied to analyze the effects of ETV5 expression level on overall survival (OS) and disease-specific survival (DSS) of patients. It was implied that LIHC, lower grade glioma and glioblastoma (GBMLGG), bladder urothelial carcinoma (BLCA), pan-kidney cohort (KIPAN), kidney chromophobe (KICH), and pancreatic adenocarcinoma (PAAD) patients with high expression level of ETV5 were associated with worse OS (Fig. 1B). In addition, DSS analysis suggested that highly expressed ETV5 was correlated with unfavorable prognosis in patients with GBMLGG, LIHC, BLCA, PAAD, kidney renal papillary cell carcinoma (KIRP), KIPAN, KICH, and colon/rectum adenocarcinoma (COADREAD) (Supplementary Fig. S1B). These results manifested that ETV5 expression level was abnormally high in various tumor tissues and associated with poor prognosis in some cancers.
In view of this, we preliminarily explored the subcellular localization of ETV5 to speculate its primary function. It was exhibited that ETV5 protein was mainly located in the nucleus in cancer cells from the Human Protein Atlas (HPA) database, which is consistent with its transcription factor identity (Fig. 1C). It suggested that it may function through transcriptional regulation in tumors.
Hypomethylation of ETV5 gene promoter in multiple malignancies
Considering ETV5 was abnormally high expressed in various tumors, we speculated the DNA methylation in its promoter region may be involved in it. As a common epigenetic modification, DNA methylation plays a critical role in regulating the expression of oncogenes. The differences of DNA methylation level in ETV5 promoter region were evaluated between adjacent normal tissues and tumor tissues in 22 types cancers based on the UALCAN database. Eight tumors with obvious differences were selected and it was identified that the overall gene promoter region was both hypomethylation in tumor and normal tissues, though DNA methylation level varied in malignancies (Supplementary Fig. S2A). It indicated that DNA methylation of promoter region may not be the major epigenetic mechanism affecting ETV5 expression.
DNA methylation profiles of ETV5 gene was further assessed in LIHC patients according to MethSurv database. The detection of 26 methylation probes indicated that there were 5 CpG sites with hypermethylation, including cg04451564, cg06390643, cg06518884, cg04400533 and cg07803284, which were mainly concentrated in the 3’ UTR and gene body regions (Supplementary Fig. S2B). Interestingly, TSS1500 and TS200 regions, which mainly affect gene transcription, exhibited consistent level of hypomethylation, containing 13 CpG sites. It implied that the transcription of ETV5 in LIHC patients was not affected by DNA methylation, which was also consistent with its high expression in tumor tissues. However, it needs further investigate whether the DNA methylation level of ETV5 gene locus can be serve as a potential biomarker for tumor prognosis.
Correlation between the expression of ETV5 and epigenetic modification-related modulators
Epigenetic modification exerts crucial effect on malignancies progression, which can affect gene expression through the regulation of gene transcription or post-translational modification and result in corresponding abnormal biological processes27,28,29,30,31. However, it remains unclear whether ETV5, as a transcription factor, regulates the epigenetic modification-related modulators, and further results in the abnormal expression of oncogenes and tumor suppressor genes. To explore the role of ETV5 in tumorigenesis, we analyzed the correlations between ETV5 expression and DNA methylation, RNA methylation and histone modification regulators associated with chromatin accessibility, respectively.
As shown in Fig. 2A, DNA methylation-related modulators were classified into 4 categories according to their functions, among which methyl-cytosine dioxygenases and glycosylases could both play a role in demethylation27. It was discovered that ETV5 expression level was highly correlated with these modulators in multiple cancers. For DNA methyltransferases, DNMT1 and DNMT3A, it was identified that ETV5 was positively associated with their expression in various malignant tumors, such as LIHC, KIRC, KICH, and BLCA (Fig. 2B, C). Notably, ETV5 expression level was an unfavorable prognostic factor in these tumors, suggesting that ETV5 may positively regulate DNA methyltransferase and inhibit tumor suppressor genes transcription, thereby promoting tumor progression and poor prognosis. For methyl CpG binding proteins, they bind with some co-repressors after binding the methylation site to further stabilize the methylation structure. For UHRF1 and MECP2, it was identified that ETV5 expression was similar correlated with them, implying that ETV5 may also be involved in succeeding DNA methylation stabilization (Fig. 2D, E). For demethylation-related regulators, TETs, TDG and UNG were also positively associated with ETV5 in most tumors, possibly due to the different target genes in the modification process (Fig. 2A).
Correlation between the expression level of ETV5 and DNA methylation modulators in pan-cancer. (A) The heatmap showed Pearson’s correlation coefficient between ETV5 and DNA methylation modulators expression levels. Modulators were classified based on their function. (B–E) The tumors with significant correlation were presented in volcano plots based on several representative DNA methylation modulators, including DNMT1 (B), DNMT3A (C), UHRF1 (D), MECP2 (E). (The absolute correlation value > 0.2 and P < 0.05)
As the most abundant RNA methylation modification, N6-methyladenosine (m6A) and 5-methylcytosine (m5C) play a crucial part in regulating the alternative splicing, output, stability, localization and translation of mRNA28. RNA methylation is regulated by “writers” (methyltransferases), “erasers” (demethylases), and “readers” proteins in general32. As demonstrated in Fig. 3A, ETV5 was associated with the expression of multiple m6A modulators, and there were opposite correlations in individual tumors, reflecting the heterogeneity among tumors. WTAP, the m6A writer, is responsible for recruiting METTL3 and METTL14, and was positively correlated with ETV5 expression in various tumors (Fig. 3B). It indicated that highly expressed ETV5 may up-regulate the expression of WTAP, and then affect the m6A modification. For methylated mRNAs, specific readers are required to perform subsequent biological processes. It was found that ETV5 was positively correlated with YTHDF2 and HNRNPC expression in a variety of tumors, such as LIHC, THCA, testicular cancer (TGCT), adrenocortical cancer (ACC) and KICH (Fig. 3C, D). It further implied that ETV5 may be involved in the m6A modification process of RNA. For FTO, the eraser, although it was also positively associated with ETV5 expression in multiple malignancies (Fig. 3E), it was necessary to further explore the relationship between its various functions and ETV5 regulatory functions due to it is also an obesity-related gene.
Correlation between the expression level of ETV5 and m6A modulators in pan-cancer. (A) The heatmap described Pearson’s correlation coefficient between ETV5 and m6A modulators expression levels. Modulators were classified based on their function. (B–E) Volcano plots showed the tumors with significant correlation in several representative m6A modulators, including WTAP (B), YTHDF2 (C), HNRNPC (D), FTO (E). (The absolute correlation value > 0.2 and P < 0.05)
It was similar with m6A that the correlations between ETV5 and multiple m5C modulators expression were also influenced by tumor heterogeneity (Fig. 4A). Notably, there were overlaps in m5C modulators and DNA methylation regulators, including DNMT and TET family members. However, ETV5 may indirectly regulate different target genes expression due to differences in the two modifications. For m5C writers, whether NSUN3 and NSUN4 from the NSUN family or TRDMT1 from the DNMT family, were both positively correlated with ETV5 expression in most tumors (Fig. 4B–D). It suggested that ETV5 may participate in the m5C modification process of RNA. In addition, it was also identified that the expression of ETV5 and ALYREF, the reader, was consistent in a variety of malignant tumors, such as LIHC, KIRC, ACC, KICH, HNSC (Fig. 4E). It further confirmed the possibility that ETV5 may be involved in m5C modification.
Correlation between the expression level of ETV5 and m5C modulators in pan-cancer. (A) The heatmap presented Pearson’s correlation coefficient between ETV5 and m5C modulators expression levels. Modulators were classified based on their function. (B–E) The tumors with remarkable correlation were exhibited in volcano plots based on several representative m5C modulators, including NSUN3 (B), NSUN4 (C), TRDMT1 (D), ALYREF (E). (The absolute correlation value > 0.2 and P < 0.05)
Chromatin needs to form open areas and expose DNA sequences during replication and transcription, and transcription factors or other regulatory elements will bind the sites, which is named as chromatin accessibility. Histone modifications can regulate chromatin accessibility, especially active marks, H3K4me3 and H3K27ac, and repressive marks H3K27me3 around the transcription start sites, and H3K4me1 and H3K27ac enriched around the active enhancer30. As exhibited in Fig. 5A, B, ETV5 was positively correlated with the expression of multiple H3K4me1 and H3K4me3 modulators. KMT2A and ASHIL are both involved in these two modifications, and their expression was consistent with ETV5 in various malignancies, such as KIRC, ACC, THCA, and thymoma (THYM) (Supplementary Fig. S3A, B). It suggested that ETV5 may regulate gene transcription in concert with H3K4me modification. In addition, KDM5B, the H3K4me3 demethylase, was also positively associated with ETV5 expression in multiple tumors (Supplementary Fig. S3C), indicating a possible diversity of transcription regulation. For H3K27ac, the expression of ETV5 and CREBBP and EP300, 2 common acetyltransferases, showed consistent positive associations across various malignancies (Fig. 5C and Supplementary S3D), implying that they may be involved in the transcription activation of related target genes. It was also verified in KAT2B, though the association was affected by tumor heterogeneity (Supplementary Fig. S3E). For HDAC2, although it was a deacetylase, its expression was also positively correlated with ETV5 in a variety of tumors (Supplementary Fig. S3F), possibly resulted from the different target genes in regulation. For the inactive marker H3K27me3, the correlation between ETV5 and modulators varied by tumor species, indicating the necessity to analyze the effects of ETV5 on epigenetic modification and target genes according to specific tumor (Fig. 5D). In summary, ETV5 was correlated with the expression of multiple epigenetic modification-related modulators in various malignant tumors, indicating that ETV5 may function as a transcription factor to co-regulate the target genes expression with multiple epigenetic modifications, thereby affecting the tumorigenesis and tumor progression.
Correlation analysis between the expression level of ETV5 and chromatin accessibility related modulators in pan-cancer. (A) Pearson’s correlation coefficient of ETV5 and H3K4me1 modulators was presented in heatmap. (B) The heatmap showed Pearson’s correlation coefficient between ETV5 and H3K4me3 modulators expression levels. (C) Pearson’s correlation coefficient of ETV5 and H3K27ac modulators was exhibited in heatmap. (D) The heatmap revealed Pearson’s correlation coefficient between ETV5 and H3K27me3 modulators expression levels. Modulators were classified based on their function.
Functional enrichment of ETV5 related genes in pan-cancer
In order to further explore the regulatory mechanism of ETV5 in tumorigenesis, we screened ETV5 related genes in pan-cancer and analyzed their biological functions. Based on the String database, we identified 50 proteins interacting with ETV5 (Fig. 6A). Subsequently, GEPIA2 database was applied to identify genes with similar expression trends in pan-cancer, and the top 200 genes were obtained. It was found that multiple genes involved in tumor progression were included, such as SOX10, CDK2, SOX5, CTNNB1, DVL2, and AKT3, and their expression was positively correlated with ETV5 (Fig. 6B). It indicated that ETV5 may participate in the occurrence and development of tumors together with these genes.
Biological function enrichment of ETV5 related genes in pan-cancer. (A) The network diagram of the top 50 proteins interacting with ETV5 through String database. (B) Pearson’s correlation between ETV5 and 6 representative genes from the top 200 co-expression genes. (C) Gene ontology (GO) analysis about biological process of ETV5 related genes. (D) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of ETV5 related genes.
ETV5 related genes were obtained after merging the interacting genes and co-expressing genes. Gene ontology for biological process analysis revealed that these genes were involved in the regulation of ERK and MAPK pathways, gene transcription, cell proliferation and apoptosis (Fig. 6C). In addition, Kyoto Encyclopedia of Genes and Genomes (KEGG)33 pathway analysis indicated that ETV5 related genes played a critical role in various tumor-related signaling pathways and transcriptional dysregulation (Fig. 6D). The genes involved in tumor-related pathways were exhibited in Supplementary Fig. S4. It was identified that they may play a crucial part in a variety of tumors, such as breast cancer, gastric cancer, prostate cancer, melanoma and some pan-cancer pathways. It further confirmed the speculation that abnormal ETV5 expression may influence tumor progression.
ETV5 expression level is associated with multiple drugs sensitivity
The relationship between ETV5 expression level and drug sensitivity in pan-cancer cells was evaluated through the CellMiner database. Eight drugs with strongest correlations were exhibited in Fig. 7A (P < 0.001), including RAF inhibitors TAK-632, MLN-2480, AZ-628, and MEK1/2 inhibitors Refametinib, ARRY-162, RO-4,987,655, and PD 184,352. Their sensitivity in tumor cells decreased with the increase of ETV5 expression, implying the inhibitory effect of ETV5 on drug therapeutic effect. The tumor cells were divided into ETV5-high and ETV5-low groups according to ETV5 expression level. It was worth noting that for the top 3 significant drugs in Fig. 7A, there was a remarkable difference in the drug sensitivity between 2 groups (Fig. 7B). The average z score of drugs was increased in ETV5-high group, which meant reduced sensitivity for drugs. In addition, 94 drugs were associated with ETV5 expression level when P < 0.05 was regarded as cut-off, such as inhibitors of RAF, MEK, ERK, PARP, GSK-3 and EGFR (Fig. 7C). The correlation between ETV5 expression and drug sensitivity may also change due to the differences in drug targets or functions. The impact of ETV5 on anticancer therapy needs to be evaluated based on specific drug.
Relationship between ETV5 expression level and drug sensitivity in cancer. (A) Correlation between 8 drugs sensitivity and the expression level of ETV5 in pan-cancer cells based on the CellMiner database (P < 0.001). (B) Comparison of the sensitivity for the top 3 significantly related drugs in panel A between high and low ETV5 expression groups. (C) The scatterplot showed the distribution of drugs whose sensitivity was significantly correlated with the expression level of ETV5 (P < 0.05). (D, E) Comparison of the IC50 for antitumor drugs between high and low ETV5 expression groups in HCC cells based on the GDSC and CCLE databases, including oxaliplatin (D), gefitinib (E). (*P < 0.05, ***P < 0.001)
The correlation between ETV5 expression level and the half maximal inhibitory concentration (IC50) of some commonly used chemotherapy and targeted drugs in HCC cells was further investigated based on GDSC and CCLE databases. HCC cells were divided into ETV5-high and ETV5-low groups according to ETV5 expression level. It was identified that ETV5 was obviously correlated with the IC50 of chemotherapy drug oxaliplatin and EGFR inhibitor gefitinib (Fig. 7D, E). The IC50 of drugs was significantly increased in ETV5-high group, suggesting that highly expressed ETV5 inhibited the sensitivity for drugs in HCC cells. Collectively, these findings indicated that ETV5 expression level could be applied for the anticancer drug selection reference, and its high expression inhibited the therapeutic effects of oxaliplatin and gefitinib in HCC cells.
Highly expressed ETV5 specifically facilitates hepatocellular carcinoma
It was revealed that ETV5 expression promoted various tumors malignant progression and adverse outcome in pan-cancer analysis. Additionally, based on the HPA database, Kaplan-Meier analysis of the optimal cutoff value identified ETV5 as a specific unfavorable prognostic factor in LIHC among 17 types tumors. It indicated that ETV5 may have a potential oncogenic impact on HCC carcinogenesis. Consequently, we evaluated the effect of ETV5 expression level on clinicopathological characteristics of HCC patients. Patients were categorized into 2 groups according to ETV5 expression level. Chi-square test analysis demonstrated that there were obvious differences in T status (the extent and diameter of primary tumor) and tumor stage between 2 groups (Fig. 8A). Patients with high ETV5 expression suffered from higher T status and tumor stage, implying larger tumor diameter and increased malignancy degree. Correspondingly, patients with high T status or tumor stage were accompanied with higher ETV5 expression level (Fig. 8B, C), indicating that ETV5 was involved in the malignant progression of HCC. Furthermore, it was identified that HCC patients with higher ETV5 expression were subjected to shorter OS durations in survival analysis, and their median survival was nearly 4 years shorter compared to patients with lower expression (Fig. 8D). In summary, these results suggested that ETV5 was a specific oncogenic factor in HCC, and its high expression may facilitate tumor proliferation and invasion, resulting in adverse outcome for HCC patients.
In HCC, the relationship between ETV5 expression level and patients’ clinicopathological features. (A) The heatmap presented the association between ETV5 expression level and clinicopathological features in HCC patients. (B) Differential expression level of ETV5 in HCC patients with different T status (the extent and diameter of primary tumor). (C) Differential expression level of ETV5 in HCC patients with different tumor stage. (D) The evaluation of ETV5 expression level on the prognosis of HCC patients through the Kaplan–Meier OS curve. The number of survival patients was shown in years. (*P < 0.05, **P < 0.01)
ETV5 co-expression genes are enriched in tumor-related pathways in hepatocellular carcinoma
In consideration of the oncogenic impact of ETV5 in HCC, we further screened the genes related to ETV5 in HCC and analyzed their biological functions to investigate the possible molecular mechanisms. Based on GEPIA2 database, the genes with similar expression trends in LIHC were identified and the top 200 genes were screened out. It was identified that the expression levels of tubulin alpha-related genes TUBA1A, TUBA1B, TUBA1C, matrix metalloproteinase gene MMP14, anti-inflammatory cytokine IL10 and SWI/SNF chromatin remodeling complex subunit SMARCB1 were obviously positively correlated with ETV5 (Fig. 9A). It suggested that ETV5 may be involved in HCC progression together with these genes. Gene ontology for biological process analysis revealed that co-expression genes participated in the regulation of mitosis related processes, cell migration and transcription factor activity (Fig. 9B). Moreover, Reactome pathway analysis showed that these genes played a crucial role in RHO GTPases related signaling pathways, the regulation of cell cycle and immune system (Fig. 9C). KEGG pathway analysis implied that co-expression genes were associated with cell apoptosis and immune-related phagocytosis pathways (Fig. 9D). Collectively, these results indicated that ETV5 may regulate the transcription of related genes and affect the cell cycle and immune regulation and other biological processes, thereby facilitating HCC development and progression.
Biological function enrichment of ETV5 co-expression genes in LIHC. (A) Pearson’s correlation between ETV5 and 6 representative genes from the top 200 co-expression genes. (B) GO analysis for biological process of ETV5 co-expression genes. (C) Reactome pathway analysis of ETV5 co-expression genes. (D) The genes in representative pathways from KEGG analysis of ETV5 co-expression genes.
ETV5 facilitates cell proliferation and reduces sensitivity to GSK126 by regulating EZH2 expression in HCC
Biological experiments were conducted to further reveal the oncogenic effect of ETV5 in HCC. The mRNA expression level of ETV5 was detected in 20 pairs of surgical specimens from HCC patients, and it was revealed that ETV5 was highly expressed in 75% tumor tissues compared with para-cancerous tissues (non-tumor) (Fig. 10A). Besides, ETV5 mRNA expression level in HCC cell lines was abnormally higher than that in L02 cells, the immortalized normal liver cells (Fig. 10B). Considering ETV5 co-expression genes were enriched in cell cycle related biological processes, we speculated that ETV5 may participate in the regulation of HCC cell proliferation. We screened 3 siRNA for comparison and identified si-1, which exhibited the highest efficiency, for the subsequent experiments (Supplementary Fig. S5A). Hep3B and HepG2.215 cells were transfected with ETV5 overexpression plasmid or ETV5 knockdown siRNA to investigate the impact of ETV5 on cell viability. The efficiency of ETV5 overexpression and knockdown at mRNA and protein expression level was verified by qPCR and Western Blot assay, respectively. (Fig. 10C and Supplementary Fig. S5B, C, D). It was identified that highly expressed ETV5 promoted cell proliferation while ETV5-knockdown inhibited cell proliferation through CCK-8 assay (Fig. 10D, E and Supplementary Fig. S5E, F). Correspondingly, colony formation assay revealed that ETV5-overexpression increased the number of cell clone foci, while ETV5-knockdown reduced it (Fig. 10F, G and Supplementary Fig. S5G, H). These results verified that ETV5 was highly expressed in tumor and facilitated cell proliferation in HCC, thereby driving the malignant progression of tumor.
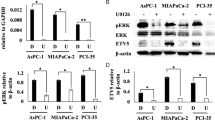
In HCC, function of ETV5 on cell proliferation and sensitivity to EZH2 inhibitor. (A) The mRNA expression level of ETV5 in 20 pairs HCC clinical tissue specimens. (T < N indicates the fold change (FC) of gene expression was less than two thirds in tumor compared with non-tumor tissues.) (B) The mRNA expression level of ETV5 in HCC cell lines compared with L02 cells. (C) The representative figure for the efficiency of ETV5-overexpression (up) and ETV5-knockdown (down) in Hep3B cells detected by Western Blot. (D, E) The efficiency of ETV5-overexpression (D) and ETV5-knockdown (E) in Hep3B cells detected by qPCR assay, and CCK-8 assay was applied to detecting the effect of ETV5 on cell viability. (F, G) Colony formation analyzed the impact of ETV5-overexpression (F) or ETV5-knockdown (G) on Hep3B cell proliferation. (H) Volcano plot presented the tumors with significant correlation between ETV5 and EZH2 (The absolute correlation value > 0.2 and P < 0.05). (I) The differential expression level of EZH2 in ETV5-high and ETV5-low expression HCC patients from TCGA database. (J) The assessment of EZH2 expression level on HCC patients’ prognosis via Kaplan–Meier OS curve. (K) The OS curve for HCC patients with ETV5/EZH2 co-expression. (L) Cell viability dose-response curve of HepG2.215 cells treated with GSK126 for 72 h. (M, N) The effect of ETV5-overexpression on EZH2 mRNA expression level (M) and cell viability treated with 5µM GSK126 (N) in HepG2.215 cells. The OD 450 value of day 4 was displayed separately in histogram. (O, P) The impact of ETV5-knockdown on EZH2 mRNA expression level (O) and cell viability treated with 5µM GSK126 (P) in HepG2.215 cells. The OD 450 value of day 4 was displayed separately in histogram. (N: non-tumor, T: tumor, NC: negative control, **P < 0.01, ***P < 0.001)
It was identified that ETV5 was highly associated with H3K27me3 methyltransferase EZH2 in LIHC (R = 0.407) in the analysis of correlation between ETV5 and chromatin accessibility related modulators (Fig. 10H). HCC patients from TCGA database were divided into 2 groups based on the median of ETV5 expression level, and it was discovered that the expression level of EZH2 in ETV5-high group was significantly higher than that in ETV5-low group (Fig. 10I). Survival analysis revealed that high expression of EZH2 was associated with unfavorable prognosis in HCC patients (Fig. 10J). Notably, HCC patients with high expression of both ETV5 and EZH2 suffered worse OS duration compared with high expression of single gene (Fig. 10K), implying that ETV5 and EZH2 may jointly contribute to poor prognosis for HCC patients. The Jaspar database (http://jaspar.genereg.net/) was applied to predict the transcription factor binding sites of EZH2 promoter region, and it was confirmed that ETV5 had multiple binding sites near the EZH2 transcription start site (Supplementary Fig. S6), indicating that ETV5 may regulate EZH2 at transcription level. Therefore, we speculated that ETV5 may affect the sensitivity to EZH2 inhibitor therapy in HCC cells. The IC50 value of HepG2.215 cells to GSK126, an EZH2 inhibitor34, was estimated according to the cell viability dose–response curve, and 5µM was selected as the drug treatment concentration for subsequent experiments (Fig. 10L). HepG2.215 cells were transfected with ETV5 overexpression plasmid or ETV5 knockdown siRNA to investigate the influence of ETV5 on EZH2 expression level and inhibitor sensitivity. Highly expressed ETV5 promoted EZH2 expression and reduced cell sensitivity to GSK126 (Fig. 10M, N). Correspondingly, ETV5 deficiency inhibited EZH2 expression and increased cell sensitivity to GSK126 (Fig. 10O, P). In particular, ETV5-knockdown combined with GSK126 treatment had a stronger inhibitory effect on cell viability. It indicated that targeting ETV5 may serve as a potential strategy for improving the therapeutic efficacy of EZH2 inhibitors.
Discussion
ETS is a transcription factor family with evolutionary conservation, and the proteins are characterized by an ETS DNA-binding ___domain which can interact with specific DNA sites to regulate gene expression35. ETV5, a member of the ETS family, is highly expressed in testis, lung and brain, indicating a critical role in development36,37,38. In addition, ETV5 also plays a crucial part in branching morphogenesis, energy metabolism and immune response39,40,41. It has been found that ETV5 affects the occurrence and prognosis of various diseases in recent studies, especially malignant tumors17,42,43,44. However, it remains unclear whether ETV5 influences varieties of tumor progression through common molecular mechanisms. The complete pan-cancer analysis for ETV5 gene is also lacking. In this study, we carried out a comprehensive systematic analysis of ETV5 based on the databases to explore its potential function in multiple cancers. The gene expression profile of tumor patients from TCGA and GTEx databases revealed that the expression level of ETV5 was significantly upregulated in most malignant tumor tissues compared to that in adjacent normal tissues. Moreover, for the analysis of patients’ OS and DSS, it was identified that highly expressed ETV5 was generally associated with unfavorable prognosis, especially in LIHC, GBMLGG, BLCA, PAAD, KICH and other malignancies. It indicated that ETV5 was a potential molecular marker for tumor diagnosis and prognosis evaluation.
It has been reported that ETV5 increased active histone marks H3K4me and H3K27ac and decreased repressive histone mark H3K27me through recruitment of histone modification modulators, thereby facilitating IL-17 production in TH17 cells45 and activating the repressed human telomerase (hTERT) gene promoter, leading to its abnormal activation during tumorigenesis46. Besides, it was identified that YTHDF2 recognized the m6A modification in the 5’UTR of ETV5 mRNA and recruited eIF3b to facilitate the translation47. It also has been reported that there are various regulatory mechanisms between ETS family members and epigenetic modification modulators, implying that ETV5 may be similarly involved in these regulatory mechanisms48,49,50. This study revealed that in various malignancies, including LIHC, ETV5 expression level was positively correlated with a variety of epigenetic modification modulators, with DNMT1 and DNMT3A prominent in DNA methylation, WTAP, YTHDF2, TRDMT1, ALYREF prominent in RNA methylation, KMT2A, EP300, EZH2 prominent in chromatin accessibility related modification. It suggested that ETV5 may function as a transcription factor to influence multiple epigenetic modifications and co-regulate the target genes expression with corresponding modulators. Subsequently, the functional enrichment analysis of ETV5 related genes demonstrated that they were involved in the development of various malignancies, including breast cancer, gastric cancer, prostate cancer and colorectal cancer. Moreover, except participating in regulating cell proliferation and apoptosis, the results also revealed that they exerted critical effect on the regulation of ERK and MAPK signaling pathways. Further, the drug sensitivity evaluation identified that ETV5 expression was associated with the sensitivity of multiple kinase inhibitors, especially RAF, MEK, ERK inhibitors. This was consistent with the result that ETV5 related genes participated in regulating ERK signaling pathway. Notably, it has been reported that ETV5 and JUN up-regulated CDK6 expression and promoted melanoma resistance to BRAF inhibitor PLX-4720 jointly51. In addition, ETV5 was recognized as a biomarker of tumor sensitivity to MEK inhibitors selumetinib52 and cobimetinib53. It indicated that ETV5 played a crucial role in malignancy progression, and its expression level could be applied to guide the selection of antitumor drugs.
HCC, accounting for approximately 90% of primary liver cancer, is one of the common malignancies posing a serious threat to human health54. Due to the heterogeneity and high metastasis of HCC, it is greatly significant for the guidance of HCC diagnosis and treatment strategies to explore potential biomarkers. It was identified that ETV5 promoted liver fatty acid metabolism through PPAR signaling pathway55 and facilitated the development of liver-resident memory CD8+ T-cell in liver through inducing liver injury41. These studies indicate that ETV5 exerts vital impact on the regulation of liver function, and its abnormal expression may affect the development of liver-related diseases. It has been reported that ETV5 was a downstream target gene of USF1 and involved in USF1-induced HCC progression56. However, the molecular mechanisms of ETV5 promoting the tumorigenesis and development of HCC remain indistinct. In this study, we discovered that ETV5 was highly expressed in tumor tissues from clinical samples and online databases, while the expression level was not associated with gene locus DNA methylation. HCC patients with highly expressed ETV5 were inclined to suffer from higher pathological stage and shorter median survival, implying that ETV5 was involved in the malignant progression and served as an unfavorable prognostic biomarker in HCC. Drug sensitivity analysis also identified that highly expressed ETV5 inhibited the sensitivity of HCC cells to oxaliplatin and gefitinib, suggesting its antagonistic effect on drug therapy. Further, ETV5 co-expression genes enriched in the regulation of cell cycle and immune function, thereby promoting the occurrence and development of HCC.
Subsequently, functional experiments were conducted to verify the results in vitro. It was identified that ETV5 overexpression facilitated cell proliferation in HCC. EZH2, the crucial H3K27me3 methyltransferase, may be regulated by ETV5 at the transcriptional level, jointly participating in the epigenetic modification of downstream target genes, resulting in worse prognosis for HCC patients. It has been reported that genetic or pharmacological inhibition of EZH2 increased the cell sensitivity to sorafenib in HCC57,58. Sorafenib was a first-line treatment in HCC patients with advanced stage. In addition, the combination treatments of EZH2 inhibitors (GSK-126) and DNMT inhibitors can synergistically activate therapeutic targets of HCC and facilitate the anti-tumor immune response59. It is suggested that EZH2 has an excellent therapeutic promise in HCC. In this study, knockdown of ETV5 increased cell sensitivity to GSK126, indicating that ETV5 may be employed as a target to improve epigenetic inhibitor therapeutic efficacy. Taken together, ETV5, as a critical oncogenic transcription factor in HCC, regulated related genes expression, impacted cell proliferation and drug sensitivity, facilitated tumor malignant progression and resulted in adverse outcome of patients.
Conclusions
In conclusion, this study demonstrated that abnormally upregulated ETV5 facilitated tumor progression and resulted in unfavorable prognosis, and ETV5 expression was associated with epigenetic modification, regulation of tumor-related gene expression and drug sensitivity in multiple malignancies. In HCC, ETV5 promoted tumor cell proliferation and resistance to drug therapy, affected patients’ pathological stage and facilitated the malignant progression of tumors. These findings contribute to elucidating the role of ETV5 in tumorigenesis more comprehensively, and provide theoretical basis and guidance for tumor diagnosis, targeted therapy and clinical drug research targeting ETV5 oncogenic molecular mechanisms.
Materials and methods
Gene expression profile of ETV5 in pan-cancer
The RNA sequencing data and related clinicopathological information of 34 types tumor patients (N = 22046) were obtained from TCGA (https://portal.gdc.cancer.gov/) and GTEx (https://gtexportal.org/home/) database and analyzed through the online tool Sangerbox (http://vip.sangerbox.com/). TCGA database included 26 types cancers (N = 11641). The expression value of ETV5 gene was transformed by Log2(x + 1). The differential expression of ETV5 between tumor tissues and adjacent normal tissues from various cancer patients was analyzed via parametric test. R language (version 4.3.0) was employed to present the results through “ggplot2” and “ggpubr” packages. The subcellular localization of ETV5 in human cancer cells was evaluated based on immunofluorescence staining images from HPA database (https://www.proteinatlas.org/).
Prognostic evaluation of ETV5 in pan-cancer
The prognostic value of ETV5 in various cancers was evaluated based on the RNA sequencing data and clinicopathological information obtained from TCGA database. ETV5 expression value was transformed through Log2(x + 1) and the cases whose follow-up time was less than 30 days were excluded. Univariate Cox regression analysis was applied to evaluate the hazard ratio of ETV5 expression level, and OS and DSS of patients were analyzed. Graphpad Prism (version 8) was employed in statistical analysis and forest map presentation.
DNA methylation status analysis of ETV5 gene
UALCAN (https://ualcan.path.uab.edu/) database was applied to explore the differential DNA methylation level in ETV5 gene promoter region between adjacent normal tissues and tumor tissues in various cancers. Beta value designates DNA methylation level ranging from 0 (unmethylated) to 1 (fully methylated). DNA methylation profiles of ETV5 gene in LIHC patients were further analyzed based on MethSurv (https://biit.cs.ut.ee/methsurv/) database60. The methylation distribution was assessed according to the methylation probes detection results.
Correlation between ETV5 expression and epigenetic modification modulators
TCGA Pan-Cancer (PANCAN) cohort was obtained via the UCSC Xena tool (https://xenabrowser.net/) and the TOIL RSEM FPKM in “gene expression RNAseq” was converted to Log2(FPKM + 1) format. Patients were classified according to phenotype and tumor type, excluding the duplicated data. DNA methylation, RNA methylation and chromatin accessibility related histone modification modulators27,28,29,30,31,61 were selected to analyze their Pearson’s correlation coefficients with ETV5 expression level in various tumor tissues, respectively. R packages “dplyr”, “psych”, “ggplot2”, “ggrepel” and “pheatmap” were applied for statistical analysis and heatmap and volcano plots presentation.
Functional enrichment analysis of ETV5 related genes
string (https://cn.string-db.org/) database was employed to screen the top 50 proteins interacting with ETV562. GEPIA2 (http://gepia2.cancer-pku.cn/) database was applied to obtain the top 200 genes with similar expression trends to ETV5 gene in TCGA tumor specimens. The interacting genes and co-expressing genes were summarized into ETV5 related genes. However, for HCC, ETV5 related genes only included co-expressing genes in LIHC tumor samples. The correlation between representative genes and ETV5 was analyzed through GEPIA2, and the expression values of genes were converted into Log2TPM format for scatter plot visualization. The functional enrichment analysis of ETV5 related genes was performed through the Database for Annotation, Visualization and Integrated Discovery (David, https://david.ncifcrf.gov/) to explore the biological processes and signaling pathways63. R package “ggplot2” and Origin (version 2021) were used for visualization of enrichment results and genes involved in representative pathways.
Correlation analysis between ETV5 expression and drug sensitivity
CellMiner (https://discover.nci.nih.gov/cellminer/) database was applied to obtain the gene expression profiles and drug activity data from 60 cancer cell lines listed by the National Cancer Institute (NCI)64. The drug sensitivity of cell lines was represented by the z-score, which is obtained from the experimental results after standardization. Drugs were only included for analysis if they had been approved by the FDA or were in clinical trials currently. R packages “magrittr”, “impute”, “limma”, “ggplot2”, “ggpubr” and Origin were employed to analyze the correlation between ETV5 expression level and drug sensitivity and visualize the results. Furthermore, the IC50 of common antitumor drugs in HCC cell lines were obtained from GDSC (https://www.cancerrxgene.org/) database65, and gene expression profiles of HCC cell lines were obtained through the CCLE (https://sites.broadinstitute.org/ccle/) database66. Subsequently, the correlation between ETV5 gene expression level and IC50 of antitumor drugs was analyzed to evaluate the effect of ETV5 on drug sensitivity in HCC. Graphpad Prism was used for statistical analysis and results visualization.
Analysis of ETV5 expression and HCC patients clinicopathological characteristics
For patients with primary HCC, gene expression profiles and clinicopathological information were obtained from TCGA database, and 371 tumor samples were included in subsequent analysis. Gene expression level was transformed through Log2(FPKM + 1). The samples with missing information were excluded. HCC patients were categorized into 2 groups according to expression level of ETV5. Chi-squared test was performed to assess the differences in clinicopathologic features between grouped cases. The differential expression level of ETV5 between different pathological grouped patients was analyzed through Student’s t-test. R package “pheatmap” and Graphpad Prism were used for visualizing results. Kaplan-Meier and log-rank tests were applied to assess the impact of ETV5 or EZH2 expression level on HCC patients’ OS probability via R package “survival” and “survminer”.
Clinical tissue specimens
The clinical tumor and para-carcinoma tissues were collected from HCC patients who underwent hepatic surgery at Zhongda Hospital affiliated with Southeast University, China. This research was authorized by the Ethics Committee of Zhongda Hospital13. Informed consent was obtained from all individual participants included in the study.
Cell culture and cell transfection
Human normal liver cell L02 and HCC cells HepG2, HepG2.215, Hep3B and PLC were acquired from Cell Bank, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gbico, 12100-061) with 10% fetal bovine serum (FBS, Wisent, 086–150) and penicillin-streptomycin solution (Biochannel, BC-CE-007) at cell incubator (Thermo Fisher, 3111) with 37 °C, 5% CO2. The recombinant plasmid containing human ETV5 gene coding sequences (ov-ETV5, Sangon Biotech Co., Ltd.) was constructed based on pcDNA3.1 vector plasmid. Small interfering RNA (siRNA, Sangon Biotech Co., Ltd.) was employed for ETV5 knockdown experiments. HCC cells were cultured to 60% confluence in 6-well plates, and plasmid or siRNA was mixed with transfection reagent (Yeasen, 40808ES02) to transfect cells. The relevant sequences were shown in Supplementary Table S1.
RNA extraction and reverse transcription quantitative PCR (qPCR)
Trizol reagent (Accurate, AG21102) was applied to extract total RNA from cells or tissue specimens, and tissues were ground with magnetic beads in advance. Reverse transcription kit (Accurate, AG11728) was utilized to obtain cDNA. Fluorescence quantitative experiment was carried out through SYBR Green chimeric fluorescence method (Accurate, AG11701). The expression level of the target genes was standardized by the internal parameter ACTB and calculated using 2−ΔΔCt method67. The primers sequences had been listed in Supplementary Table S2.
Western blot analysis
RIPA lysis buffer (APExBIO, K1020) was utilized to extract total protein from cells. The concentration of protein was quantified by bicinchoninic acid protein assay kit (Thermo Fisher, 23225). Protein samples were separated via sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto PVDF membrane (Millipore, ISEQ00010). After being blocked with 10% skim milk for 1.5 h at room temperature, the membranes were incubated with anti-ETV5 (Proteintech, 13011-1-AP) and anti-β-Actin (Sigma, A5441) overnight at 4℃, respectively. The HRP-conjugated secondary antibody (CST, 7074 or 7076) was applied for incubation at room temperature for 1.5 h. ECL luminescent solution (Vazyme, E423-01) was employed to expose membranes.
Drug sensitivity assay
EZH2 inhibitor GSK126 (APExBIO, A3446) was employed for drug treatment evaluation. HCC cells were cultured in 96-well plates at a density of 4 × 103 cells per well, and treated with multiple concentration gradients of GSK126 for 72 h. The absorbance value was detected at 450 nm wavelength after incubation with cell counting kit-8 (CCK-8) reagent (Fdbio, FD3788) for 2 h. Relative cell viability was calculated and the IC50 of GSK126 was assessed.
Cell proliferation assay
After transfected with plasmid or siRNA, HCC cells were seeded into 96-well plates at a density of 2 ~ 3 × 103 cells per well and treated with or without GSK126 at specific concentration. Cell viability was measured with CCK-8 reagent daily. Colony formation assay was performed for monitoring long-term cell proliferation. Transfected HCC cells were seeded into 6-well plates at a density of 1 × 103 cells per well for 12 ~ 14 days. The cells were fixed with 95% ethanol and stained with crystal violet solution (Beyotime, C0121).
Statistical analysis
For biological experiments, the results of each group were statistically analyzed through two-sided unpaired Student’s t-test or multiple comparisons of Two-way ANOVA and visualized utilizing Graphpad Prism. The error bars represented the mean ± standard deviation (SD) from three independent experiments at least. For this study, the statistically significant result was defined if P-value less than 0.05.
Data availability
The datasets used to investigate the research are available in online databases, and the specific sources were indicated in the Materials and Methods section. The materials and methods during the current study are available from the corresponding author on reasonable request.
References
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Deo, S. V. S., Sharma, J. & Kumar, S. GLOBOCAN 2020 report on global cancer burden: Challenges and opportunities for surgical oncologists. Ann. Surg. Oncol. 29, 6497–6500 (2022).
Bregni, G. et al. Adjuvant chemotherapy for rectal cancer: current evidence and recommendations for clinical practice. Cancer Treat. Rev. 83, 101948 (2020).
Huang, R. & Zhou, P. K. DNA damage repair: Historical perspectives, mechanistic pathways and clinical translation for targeted cancer therapy. Signal Transduct. Target. Ther. 6, 254 (2021).
Galon, J. & Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov 18, 197–218 (2019).
Zhou, C. et al. Disruption of SLFN11 Deficiency-Induced CCL2 signaling and macrophage M2 polarization potentiates Anti-PD-1 therapy efficacy in Hepatocellular Carcinoma. Gastroenterology 164, 1261–1278 (2023).
Song, H. et al. Epitranscriptomics and epiproteomics in cancer drug resistance: Therapeutic implications. Signal. Transduct. Target. Ther. 5, 193 (2020).
Ward, R. A. et al. Challenges and opportunities in cancer drug resistance. Chem. Rev. 121, 3297–3351 (2021).
Bezu, L., Chuang, A. W., Liu, P., Kroemer, G. & Kepp, O. Immunological effects of epigenetic modifiers. Cancers (Basel) 11, 1911 (2019).
Nepali, K. & Liou, J. P. Recent developments in epigenetic cancer therapeutics: Clinical advancement and emerging trends. J. Biomed. Sci. 28, 27 (2021).
Martinez-Useros, J., Martin-Galan, M., Florez-Cespedes, M. & Garcia-Foncillas, J. Epigenetics of most aggressive solid tumors: Pathways, targets and treatments. Cancers (Basel) 13, 3209 (2021).
Sandoval, G. J. et al. Binding of TMPRSS2-ERG to BAF chromatin remodeling complexes mediates prostate oncogenesis. Mol. Cell. 71, 554–566e557 (2018).
Zheng, C., Liu, M., Ge, Y., Qian, Y. & Fan, H. HBx increases chromatin accessibility and ETV4 expression to regulate dishevelled-2 and promote HCC progression. Cell Death Dis. 13, 116 (2022).
Su, H. et al. ETV4 facilitates angiogenesis in hepatocellular carcinoma by upregulating MMP14 expression. Biochem. Biophys. Res. Commun. 684, 149137 (2023).
Martinez-Jimenez, F. et al. A compendium of mutational cancer driver genes. Nat. Rev. Cancer 20, 555–572 (2020).
Peng, Y. et al. The role of E26 transformation-specific variant transcription factor 5 in colorectal cancer cell proliferation and cell cycle progression. Cell Death Dis. 12, 427 (2021).
DeSalvo, J. et al. ETV4 and ETV5 drive synovial sarcoma through cell cycle and DUX4 embryonic pathway control. J. Clin. Invest. 131, e141908 (2021).
Arase, M. et al. Dynamics of chromatin accessibility during TGF-beta-induced EMT of ras-transformed mammary gland epithelial cells. Sci. Rep. 7, 1166 (2017).
Puli, O. R. et al. The transcription factor ETV5 mediates BRAFV600E-Induced Proliferation and TWIST1 expression in papillary thyroid Cancer cells. Neoplasia 20, 1121–1134 (2018).
Monge, M. et al. ERM/ETV5 up-regulation plays a role during myometrial infiltration through matrix metalloproteinase-2 activation in endometrial cancer. Cancer Res. 67, 6753–6759 (2007).
Mus, L. M. et al. The ETS transcription factor ETV5 is a target of activated ALK in neuroblastoma contributing to increased tumour aggressiveness. Sci. Rep. 10, 218 (2020).
Li, D. et al. ETV5 transcriptionally activates TGFbeta1 and promotes cancer cell invasion and migration of NSCLC. J. Mol. Histol. 54, 419–426 (2023).
Cheng, X. et al. ETS variant 5 promotes colorectal cancer angiogenesis by targeting platelet-derived growth factor BB. Int. J. Cancer 145, 179–191 (2019).
Feng, H. et al. Targeting tumor cell-derived CCL2 as a strategy to overcome Bevacizumab resistance in ETV5(+) colorectal cancer. Cell. Death Dis. 11, 916 (2020).
Park, S. M. et al. Systems analysis identifies potential target genes to overcome cetuximab resistance in colorectal cancer cells. FEBS J. 286, 1305–1318 (2019).
Giri, A. K. Higher ETV5 expression associates with poor 5-Florouracil-based adjuvant therapy response in Colon cancer. Front. Pharmacol. 11, 620811 (2020).
Meng, H. et al. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 11, 604–617 (2015).
Wang, S., Chai, P., Jia, R. & Jia, R. Novel insights on m(6)a RNA methylation in tumorigenesis: A double-edged sword. Mol. Cancer 17, 101 (2018).
Guo, G. et al. Advances in mRNA 5-methylcytosine modifications: Detection, effectors, biological functions, and clinical relevance. Mol. Ther. Nucleic Acids. 26, 575–593 (2021).
Kimura, H. Histone modifications for human epigenome analysis. J. Hum. Genet. 58, 439–445 (2013).
Farria, A., Li, W. & Dent, S. Y. KATs in cancer: Functions and therapies. Oncogene 34, 4901–4913 (2015).
Zhao, Z. et al. Epitranscriptomics in liver disease: Basic concepts and therapeutic potential. J. Hepatol. 73, 664–679 (2020).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
McCabe, M. T. et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 492, 108–112 (2012).
Sizemore, G. M., Pitarresi, J. R., Balakrishnan, S. & Ostrowski, M. C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 17, 337–351 (2017).
Liu, D. et al. Brain-derived neurotrophic factor promotes vesicular glutamate transporter 3 expression and neurite outgrowth of dorsal root ganglion neurons through the activation of the transcription factors Etv4 and Etv5. Brain Res. Bull. 121, 215–226 (2016).
Herriges, J. C. et al. FGF-regulated ETV transcription factors control FGF-SHH feedback loop in lung branching. Dev. Cell 35, 322–332 (2015).
Wei, Y. et al. E26 transformation-specific transcription variant 5 in development and cancer: Modification, regulation and function. J. Biomed. Sci. 30, 17 (2023).
Zhang, Z. et al. Transcription factor Etv5 is essential for the maintenance of alveolar type II cells. Proc. Natl. Acad. Sci. U.S.A 114, 3903–3908 (2017).
Lin, H. et al. IP(6)-assisted CSN-COP1 competition regulates a CRL4-ETV5 proteolytic checkpoint to safeguard glucose-induced insulin secretion. Nat. Commun. 12, 2461 (2021).
Park, S., Park, J., Kim, E. & Lee, Y. The capicua/ETS translocation variant 5 axis regulates liver-resident memory CD8(+) T-cell development and the pathogenesis of liver Injury. Hepatology 70, 358–371 (2019).
Zhang, L. et al. Biological and prognostic value of ETV5 in high-grade serous ovarian cancer. J. Ovarian Res. 14, 149 (2021).
Chotteau-Lelievre, A. et al. Prognostic value of ERM gene expression in human primary breast cancers. Clin. Cancer Res. 10, 7297–7303 (2004).
Sun, M. C. et al. ETV5 overexpression promotes progression of esophageal squamous cell carcinoma by upregulating SKA1 and TRPV2. Int. J. Med. Sci. 19, 1072–1081 (2022).
Pham, D., Sehra, S., Sun, X. & Kaplan, M. H. The transcription factor Etv5 controls TH17 cell development and allergic airway inflammation. J. Allergy Clin. Immunol. 134, 204–214 (2014).
Zhang, F., Wang, S. & Zhu, J. ETS variant transcription factor 5 and c-Myc cooperate in derepressing the human telomerase gene promoter via composite ETS/E-box motifs. J. Biol. Chem. 295, 10062–10075 (2020).
Wen, J. et al. YTHDF2 is a therapeutic target for HCC by suppressing immune evasion and angiogenesis through ETV5/PD-L1/VEGFA axis. Adv. Sci. (Weinh) 11, e2307242 (2024).
Chen, Y. et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 18, 127 (2019).
Mao, X., Ji, T., Liu, A. & Weng, Y. ELK4-mediated lncRNA SNHG22 promotes gastric cancer progression through interacting with EZH2 and regulating miR-200c-3p/Notch1 axis. Cell Death Dis. 12, 957 (2021).
Taylor, M. A., Wappett, M., Delpuech, O., Brown, H. & Chresta, C. M. Enhanced MAPK signaling drives ETS1-mediated induction of miR-29b leading to downregulation of TET1 and changes in epigenetic modifications in a subset of lung SCC. Oncogene 35, 4345–4357 (2016).
Li, Z. et al. CRISPR screens identify essential cell growth mediators in BRAF inhibitor-resistant melanoma. Genom. Proteom. Bioinf. 18, 26–40 (2020).
Dry, J. R. et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244). Cancer Res. 70, 2264–2273 (2010).
Uitdehaag, J. C. M. et al. Combined Cellular and biochemical profiling to identify predictive drug response biomarkers for kinase inhibitors approved for clinical use between 2013 and 2017. Mol. Cancer Ther. 18, 470–481 (2019).
Llovet, J. M. et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 7, 6 (2021).
Mao, Z. et al. ETV5 regulates hepatic fatty acid metabolism through PPAR signaling pathway. Diabetes 70, 214–226 (2021).
Zeng, Y. L. et al. USF1 modulates transcription and cellular functions by regulating multiple transcription factors in Huh7 cells. Oncol. Lett. 26, 532 (2023).
Lai, Y. et al. EZH2 suppresses ferroptosis in hepatocellular carcinoma and reduces sorafenib sensitivity through epigenetic regulation of TFR2. Cancer Sci. 115, 2220–2234 (2024).
Wang, S. et al. Inhibition of EZH2 attenuates Sorafenib Resistance by Targeting NOTCH1 activation-dependent Liver Cancer Stem cells via NOTCH1-Related MicroRNAs in Hepatocellular Carcinoma. Transl Oncol. 13, 100741 (2020).
Zhang, L. et al. DNMT and EZH2 inhibitors synergize to activate therapeutic targets in hepatocellular carcinoma. Cancer Lett. 548, 215899 (2022).
Modhukur, V. et al. MethSurv: A web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics 10, 277–288 (2018).
Black, J. C., Van Rechem, C. & Whetstine, J. R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell. 48, 491–507 (2012).
Szklarczyk, D. et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Dennis, G. Jr. et al. Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 (2003).
Reinhold, W. C. et al. CellMiner: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 72, 3499–3511 (2012).
Yang, W. et al. Genomics of drug sensitivity in cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 41, D955–961 (2013).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001).
Acknowledgements
We acknowledge all the contributors of the public databases utilized in this study.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: HS and HF; Methodology, Data collection, analysis and interpretation: HS and LZ; Writing-original draft preparation: HS; Writing-review and editing: HS, TF and WH. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was authorized by the Ethics Committee of Zhongda Hospital affiliated with Southeast University, China. Informed consent was obtained from all individual participants included in the study. All research involving humans was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Su, H., Zhao, L., Fang, T. et al. Identification of ETV5 as a prognostic marker related to epigenetic modification in pan-cancer and facilitates tumor progression in hepatocellular carcinoma. Sci Rep 14, 29695 (2024). https://doi.org/10.1038/s41598-024-81642-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-81642-1