Abstract
Polyubiquitination of proteins serves distinct functions that are governed by the nature of polyubiquitin chains built on target proteins. Among the eight distinct type of ubiquitin chains, lysine 48 (K48)-linked chains are specifically associated with proteasomal degradation, while lysine 63 (K63)-linked chains are primarily involved in regulating signal transduction and protein trafficking. The ubiquitin-proteasome system (UPS) has recently been exploited in drug discovery and introduced PROTACs (Proteolysis Targeting Chimeras), or molecular glues (MGs), to hijack ubiquitin E3 ligases, to facilitate the targeted degradation of specific proteins. However, assessment of PROTAC or MG mediated endogenous target protein ubiquitination in a linkage-specific manner in high throughput format remains a challenge. In this study, we applied chain-specific TUBEs (Tandem Ubiquitin Binding Entities) with nanomolar affinities for polyubiquitin chains in HTS assays to investigate the ubiquitination dynamics of RIPK2, a key regulator of inflammatory signaling. Using L18-MDP to induce K63 ubiquitination of RIPK2 and RIPK degrader-2, a RIPK2 PROTAC to induce K48 ubiquitination, we demonstrate that chain-selective TUBEs can differentiate and unravel context dependent linkage specific ubiquitination of endogenous RIPK2. Potential application of this technology to other target proteins and cellular contexts will be discussed.
Similar content being viewed by others
Introduction
Ubiquitination is a versatile and highly regulated post-translational modification that involves the addition of ubiquitin (8 kDa) to target proteins, influencing various cellular functions such as proteolysis, cell cycle, DNA repair, apoptosis, and immune responses1,2. The process of ubiquitination involves a cascade of enzymes (E1, E2, and E3) wherein the E3 ubiquitin ligases select the target protein for ubiquitination and degradation. There are over 600 E3 ligases encoded in the human genome of which only a small fraction of E3s have been functionally characterized. Dysregulation of E3 ligases often leads to the development of diseases such as central nervous system disorders, inflammation, metabolic dysfunctions, and several cancers2,3,4,5. Ubiquitination can occur mainly through 8 different types of linkages (M1, K6, K11, K27, K29, K33,K48 and K63) among which the K48 and K63 linkages are most widely studied6,7. The initial evidence suggesting distinct functions of different ubiquitin chain linkages came from studies involving yeast mutants with mutations in either K48 or K63 of ubiquitin, where K63-linked polyubiquitin was found to be involved in DNA repair8 while K48-linked polyubiquitin was crucial for protein degradation and cell cycle progression9. These findings established the notion that specific chain linkages play unique roles in cellular processes. It is now well established that K48-linked chains target proteins for degradation, while K63-linked chains regulate protein function, signal transduction, subcellular localization, or protein-protein interactions10,11. K63 ubiquitination is involved in the formation of signalosomes that activate downstream signaling molecules, particularly in NF-κB and MAPK pathways12,13,14. NF-κB activation involves K63 ubiquitination of NF-κB essential modulator (NEMO), promoting IKK complex assembly and gene expression related to inflammation. K63 ubiquitination also contributes to the activation of the NLRP3 inflammasome, involved in pro-inflammatory cytokine production15.
Modulating the inflammatory response is important for therapeutic interventions due to the role of inflammation in various diseases16,17. Targeting K63 ubiquitination has emerged as a potential strategy to modulate inflammatory responses. Inhibiting enzymes involved in K63 ubiquitination, such as TRAF6, Ubc13, and Mms2, are potential targets for drug development18. Small molecule inhibitors of these enzymes have shown promise in reducing inflammation in preclinical models of diseases like rheumatoid arthritis and colitis19,20. Furthermore, the deubiquitinases (DUBs) that specifically cleave K63-linked ubiquitin chains have provided another avenue for therapeutic intervention. By removing K63 ubiquitin chains from target proteins, these DUBs can modulate inflammatory signaling and dampen the immune response21. Several DUB inhibitors and activators have been identified and tested in preclinical models, showing potential for regulating inflammation-associated diseases22.
PROTACs, are heterobifunctional small molecules, with remarkable efficacy in targeting proteasomal mediated degradation (through K48 ubiquitination) of a wide range of disease-related proteins including proteins that were considered undruggable23. Examples of such proteins include androgen receptor, estrogen receptor, BTK, BCL2, CDK8, RIPK and c-MET. Targeted degradation of these proteins has been achieved by leveraging a handful of E3 ligases such as cereblon (CRBN), Von Hippel-Lindau (VHL), IAP, and MDM2 in the PROTAC design. Additionally, PROTACs have been explored for Hepatitis C virus (HCV) protease, IRAK4, and Tau, targeting viral, immune, and neurodegenerative diseases, respectively24,25,26. However, the current focus on a limited number of E3 ligases hampers the broad application of PROTACs to the vast undruggable proteome. The lack of reliable tools for rapid evaluation of PROTACs with new ligases is a significant barrier to the progress. It is essential to develop screening platforms that accurately represent the physiological relevance of PROTACs, including their ability to induce target ubiquitination and degradation. Traditional cellular PROTAC screening methods often involve the use of cell lines containing reporter genes such as Nano-Luc (Promega, Nano Luciferase reporter) fusions of target proteins27,28 or rely on Western blotting. However, these approaches have limitations in terms of throughput, quantification, and sensitivity. Western blotting is low throughput, provides semiquantitative data, and lack sensitivity for detecting subtle changes. Reporter gene assays can overcome some of these challenges but can be affected by artifacts caused by internal lysines within the reporter tags.
The characterization of linkage specific ubiquitination on target proteins is challenging due to the complexity of the ubiquitin system and a lack of high affinity ubiquitin enrichment tools. Currently, the identification of specific polyubiquitin linkages relies mainly on affinity enrichment and mass spectrometry or the use of exogenously expressed mutant ubiquitins in which lysines are mutated to arginine29,30. However, these approaches have many limitations. Mass spectrometry is labor-intensive, requires sophisticated instrumentation, and has limited sensitivity for rapid changes in endogenous protein ubiquitination. Using mutant ubiquitins may not accurately represent modifications involving wild-type ubiquitin31. These challenges highlight the need for improved techniques to specifically capture, detect, and study linkage-specific ubiquitination of endogenous proteins, enabling better understanding of their functions and the development of targeted therapeutic strategies.
Recently, Gross, P et al. have shown the utility of Pan-selective TUBEs in developing high throughput assay for investigating PROTAC-mediated ubiquitination of endogenous target proteins32. Building upon this approach, in the present study we have developed high-throughput screening assays that leverage chain-specific TUBEs as ubiquitin-binding entities (Fig. 1).
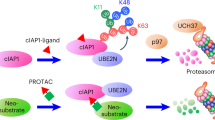
Graphical representation of ubiquitin chain-specific TUBE based assay platform. Cellular signaling or exogenous ligands such as PROTACs and MGs can induce ubiquitination of target proteins that are diverse in linkage configuration. Functionally, monoubiquitination and multimonoubiquitination are important for protein trafficking, K48 and K11 chains are associated with proteasomal degradation and K63-chains are involved in autophagy, inflammation and DNA repair and Linear chains mediate NF-κB activation. Polyubiquitin binding TUBEs facilitate selective capture and detection of context-dependent polyubiquitination of endogenous proteins in a high throughput manner.
These specialized affinity matrices facilitate the precise capture of chain-specific polyubiquitination events on native target proteins with high sensitivity. This method enables analysis of ubiquitin linkage diversity in response to inflammatory stimuli and/or PROTAC-mediated degradation. We applied this tool to investigate the ubiquitination dynamics of Receptor-Interacting Serine/Threonine-Protein Kinase 2 (RIPK2), a crucial regulator of inflammatory signaling pathways. Muramyldipeptide (MDP) is a peptidoglycan component of bacterial cell wall which binds to nucleotide-binding oligomerization ___domain (NOD)-like receptors. Upon MDP binding, NOD2 receptor oligomerizes, recruits RIPK2 and E3 ligases cIAP1, cIAP2, XIAP and TRAF2 inducing K63-Ubiquitination of RIPK233,34. K63 ubiquitin chains on RIPK2 serve as a signaling scaffold to recruit and activate the TAK1/TAB1/TAB2/IKK kinase complexes leading to NF-κB activation and production of proinflammatory cytokines. At the molecular level, XIAP binds RIPK2 via its baculovirus IAP repeat 2 (BIR2) ___domain and build K63-ubiquitination on multiple lysine residues of RIPK235. Our results demonstrate that inflammatory agent L18-MDP stimulated K63 ubiquitination of RIPK2 can be faithfully captured in a 96-well plate coated with K63-TUBEs or Pan-selective TUBEs but not using K48-TUBEs. On the other hand, RIPK2 PROTAC mediated ubiquitination was captured using K48-TUBEs and Pan-selective TUBEs while K63-TUBE did not capture appreciable PROTAC induced RIPK2 ubiquitination signals. Thus, chain-selective TUBEs can be applied to selectively capturing and quantifying distinct context-dependent ubiquitin linkages. Application of TUBE-based technologies not only contributes to a better understanding of the ubiquitin-proteasome system but also enhances the efficiency of PROTACs and MGs characterization. Overall, our novel approach offers a rapid, quantitative, and highly specific method for characterizing ubiquitin-mediated processes, paving the way for the development of the next generation of ubiquitin pathway drugs.
Results
Ponatinib inhibits RIPK2 polyubiquitination triggered by L18-MDP in THP1 cells
The RIPK2 expression and its ubiquitination was first assessed by immunoblotting using human monocytic THP1 cells treated with either water (vehicle control) or L18-MDP (Lysine 18-muramyldipeptide) at 200 ng/ml or 500 ng/ml for 30 min and 60 min. Cells were lysed in a lysis buffer optimized to preserve polyubiquitination (See materials and methods) and 50 µg of cell lysate was used in immunoblotting to assess RIPK2 using anti-RIPK2 antibody. Remarkably, RIPK2 ubiquitination was detectable only in L18-MDP stimulated cells. Moreover, RIPK2 ubiquitination was higher at 30 min of stimulation compared to the 60 min of stimulation (Fig. 2a). Overall, the data established that L18-MDP stimulation induces time-dependent ubiquitination of endogenous RIPK2.
L18-MDP induces RIPK2 polyubiquitination and Ponatinib inhibits RIPK2 Activation in THP1 Cells. (a) RIPK2 expression and ubiquitination were analyzed following L18-MDP (200 ng/ml and 500 ng/ml) treatment for 30 and 60 min. Cell lysates were probed with anti-RIPK2 antibody. GAPDH was used as a loading control. Original western blot images are presented in Supplementary Fig. 2. (b) Cells were pre-treated with 100 nM Ponatinib followed by stimulation with L18-MDP (200 ng/ml) for 30 min and 60 min. Cell lysates were subjected to Pan-selective TUBE pull down. Pull down samples were separated on SDS-PAGE and probed with anti-RIPK2 antibody.
Next, we evaluated the effect of RIPK2 inhibitor Ponatinib, on L18-MDP induced RIPK2 ubiquitination. THP-1 cells were pre-treated with either DMSO or Ponatinib (100 nM) for 30 min followed by the treatment with water or 200 ng/ml L18-MDP for 30 min and 60 min. Cells were lysed and polyubiquitinated proteins were enriched using TUBE1-conjugated magnetic beads (UM401M, LifeSensors Inc, )32,36 and probed with anti-RIPK2 antibody. As shown in Fig. 2b, polyubiquitinated RIPK2 was detectable only in cells treated with L18-MDP. Treatment with Ponatinib completely abrogated RIPK2 ubiquitination indicating that kinase inhibition has a profound negative effect on its ubiquitination. These results provide evidence that L18-MDP induces RIPK2 ubiquitination in THP1 cells and demonstrates the inhibitory effect of ponatinib on RIPK2 ubiquitination.
Delineation of RIPK2 polyubiquitin linkages using linkage-specific ubiquitin affinity matrices
Ubiquitin Binding Domains (UBDs) naturally occurring in cellular proteins such as Rad23A and Ubiquilin have been adapted to generate TUBEs that possess nanomolar binding affinity for polyubiquitin37. Pan-selective TUBEs that bind to.
High throughput ELISA assay to differentiate linkage-specific ubiquitination of RIPK2. THP1 cells were pre-treated with DMSO (vehicle control) or indicated doses of Ponatinib for 30 min followed by treatment with water (control) or 200 ng/ml L18-MDP for 30 min. Cell lysates were incubated on pan-selective TUBE (a and b), K63-selective TUBE (c and d) or K48-TUBE (e and f) coated plates. Ubiquitinated RIPK2 was detected using an anti-RIPK2 antibody and an HRP-conjugated secondary antibody followed by enhanced chemiluminescence (CL) signal generation. Relative increase in ubiquitination (top panel, relative CL intensity) and percent inhibition and IC50s of RIPK2 ubiquitination by Ponatinib (lower panel) were plotted using GraphPad Prism (n = 3). Error bars represent standard deviation. One-way ANOVA, ****p < 0.0001, ***p = 0.0004, ns = non-significant.
polyubiquitin chains without discriminating against linkage types have been used widely to characterize polyubiquitination of endogenous proteins. At least three linkage selective TUBEs (M1-TUBE, K48-TUBE and K63-TUBE) are available to study linkage-specific ubiquitination36,37,38,39. Here, we employed these TUBE matrices to develop a high throughput assay to characterize the linkage type of RIPK2 ubiquitination in response to L18-MDP stimulation as well as ponatinib treatment.
RIPK2 has been shown to undergo K63 ubiquitination upon L18-MDP stimulation40. We used previously reported Pan-selective TUBE, K63-specific TUBEs36,39 and K48-specific TUBEs, which were passively adsorbed onto 96-well plates, to capture and detect RIPK2 ubiquitination. As shown in Fig. 3a, L18-MDP stimulated RIPK2 ubiquitination in THP1 cells is readily detected when captured using Pan-selective TUBE (~ 12.0 fold, DMSO bar graph). Among the chain-selective TUBEs, K63-selective TUBE was able to capture L18-MDP induced RIPK2 ubiquitination similar to Pan-selective TUBE (~ 11.0 fold, Fig. 3c, DMSO bar graph). A modest (~ 2.0 fold, DMSO bar graph) level of RIPK2 ubiquitination signal was obtained using K48-selective TUBEs (Fig. 3e). This data suggests that L18-MDP treatment induces predominantly K63-ubiquitination of RIPK2, which agrees with the previous reports15.
Analysis of K63 polyubiquitination of endogenous RIPK2. THP1 cells were pre-treated with DMSO (vehicle control) or indicated doses Ponatinib for 30 min followed by treatment with water (control) or 200 ng/ml L18-MDP for 30 min. Cell lysates were subjected to pull downs using pan-selective TUBE (a), K63-selective TUBE (b) or K48-TUBE (c). 30 µg whole cell lysate (d) was used detect RIPK2 expression. Pull down samples and whole cell lysates were immunoblotted with anti-RIPK2 antibody. GAPDH was used as loading control for whole cell lysate. Original western blot images are presented in Supplementary Fig. 4.
Treatment with ponatinib prior to stimulation with L18-MDP (200 ng/ml, 30 min) showed strong dose-dependent inhibition of RIPK2 ubiquitination which was readily observed using Pan-selective and K63-selective TUBEs, (Fig. 3a, and c). We observed a comparable IC50 values of Ponatinib in both Pan and linkage-specific affinity matrices 4.3 nM and 4.5 nM respectively whereas the IC50 was > 1000 nM using K48-selective TUBEs (Fig. 3b, d and f). These results were further confirmed by immunoblotting of polyubiquitinated proteins pulled down using pan and linkage specific (K-63 and K-48) TUBEs (Fig. 4). Both Pan-selective and K63-selective TUBE pull downs of THP1 cells pre-treated with Ponatinib and stimulated with L18-MDP showed robust RIPK2 polyubiquitination (Fig. 4a and b, and c). Consistent with plate-based assay, the K48-selective TUBE pull down showed a modest increase in RIPK2 ubiquitination. Moreover, Ponatinib showed dose-dependent inhibition of RIPK2, which is also in agreement with the plate-based assays (Fig. 3). Treatment with L18-MDP or Ponatinib did not change the overall level of RIPK2 as shown in Fig. 4d. Thus, we have established a 96-well plate based method for efficient detection of stimulation dependent linkage-specific ubiquitination of RIPK2.
RIPK Degrader 2 PROTAC mediated K48-linked polyubiquitination facilitates RIPK2 degradation
Although PROTACs are known to promote K48-linked ubiquitination that leads to proteasomal degradation of target proteins, there is limited experimental evidence supporting the K48-ubiquitination of endogenous target proteins after PROTAC treatment. A high throughput method that facilitates rapid screening of PROTAC or molecular glue41 mediated K48-linked ubiquitination of endogenous proteins can be a valuable tool for the development of potent degrader molecules. Here we utilized linkage-specific TUBEs to study ubiquitination induced by RIPK degrader-2 (MedChem Express, HY-111866), a von Hippel-Lindau (VHL)-based PROTAC known to degrade RIPK2. Initially, we conducted a dose-response experiment in K562 cells and performed western blotting to establish the efficacy of the RIPK degrader-2. The K562 cells were treated with different doses (3.0 µM-3.0 nM ) of RIPK degrader-2 for 45 min (time point chosen based on previous report26) to assess RIPK2 degradation. As shown in Fig. 5a, RIPK2 degrader-2 induced dose-dependent degradation of RIPK2 in cells with a DC50 of 94 nM. The cell lysates obtained from RIPK degrader-2 treated K562 cells were then captured onto linkage-specific TUBE coated plates for overnight, washed with 1X PBS-T and ubiquitinated RIPK2 was detected using an anti-RIPK antibody. Relative intensities were calculated by dividing the raw signals from a given PROTAC dose with the signals from DMSO-treated control samples. As shown in Fig. 5b, dose dependent increase in RIPK2 ubiquitination was observed using Pan selective TUBE with peak ubiquitination (UbMax) at 100 nM of RIPK2 degrader-2. RIPK2 degrader-2 was previously shown to induce proteasomal degradation of RIPK2. In our experiments, co-treatment of RIPK2 degrader-2 with proteasomal inhibitor MG132 (1.0 µM) effectively blocked RIPK2 degradation (See Supplementary Fig. S1a) which further confirms that PROTAC mediated RIPK2 degradation requires proteasomal activity. Rescue of RIPK2 degradation by MG132 co-treatment also resulted in increased ubiquitination which was detected using Pan selective TUBE-based assay (See Supplementary Fig. S1b).
RIPK-Degrader 2 Mediates K48 Ubiquitination and Degradation of RIPK2. K562 cells (1.5 × 106 cells/ml) were treated with DMSO (vehicle control) or RIPK-degrader-2, PROTAC at indicated doses for 45 min. Cell lysates (30 µg) were separated on SDS-PAGE and probed with anti-RIPK2 antibody to detect RIPK2 degradation. Immunoblotting with anti-GAPDH antibody was used for loading control. Band intensities of RIPK2 normalized to GAPDH were used to quantitate RIPK2 degradation (%) and are shown below the western blot. Original western blot images are presented in Supplementary Fig. 5 (a) Lysates (15 µg/well) from RIPK degrader-2 treated cells were captured on Pan-selective TUBE. (b), K48-TUBE (c) and K63-TUBE (d) followed by detection with anti-RIPK2 antibody. Signal were developed using enhanced chemiluminescence (CL) substrate and luminescence was read using BMG LabTech Clariostar plate reader. Relative CL intensities were calculated by dividing raw CL signals from PROTAC treatment to the DMSO control and plotted using GraphPad Prism. Error bars represent standard deviation (n = 3).
Notably, RIPK2 in the K562 cell lysate appeared as a doublet in the western blots whereas THP1 cells showed a predominantly single band (Supplementary Fig. S2) likely due to differences in expression and/or post-translational modifications.
Most importantly, a similar increase in RIPK2 ubiquitination (UbMax at 100 nM) was observed in K48-Selective TUBE coated plates (Fig. 5c) whereas K63-selective TUBEs showed no appreciable RIPK2 ubiquitination signal (Fig. 5d). This result clearly demonstrates that the RIPK2 degrader-2 mediates K48-linked ubiquitination of RIPK2 which is then degraded by the proteasome. Interestingly, the ubiquitination signal increased with rising concentrations of the PROTACs until it reached a maximum value (referred to as UbMax), indicating the highest levels of ubiquitination. Subsequently, a decline in the ubiquitination signal was observed, indicating potential degradation of the target protein.
One of the challenges in drug discovery related to the ubiquitin-proteasome system (UPS) is quantitatively and reliably evaluating ubiquitin-driven degradation to understand the kinetics and stoichiometry of rate-limiting intermediates. UbMax or K48-UbMax as the peak ubiquitination level, enables the absolute quantification of the intermediate phase of PROTAC or MG-mediated target degradation. The UbMax values can be correlated with the DC50, which represents the condition at which the target protein is degraded by 50%, thus providing a means to evaluate the efficiency and potency of PROTACs32. Our result indicated an UbMax value for RIPK degrader-2 was 100 nM (Fig. 5b) which correlated well with the DC50 of 94 nM in our experiments. A fold increase greater than 2-fold can be considered reliable in assessing protein ubiquitination due to its dynamic and short-lived nature. Thus, UbMax values derived from Pan-selective or K48-TUBE-based assays represent as a surrogate measure for evaluating the efficiency and degradability of PROTACs, offering insights into their performance in cells and tissues.
UbiTest is another technique employed to determine chain-specific polyubiquitination of endogenous proteins42,43. In this method, polyubiquitinated proteins are enriched using affinity matrices and subsequently the ubiquitins are removed using chains-selective deubiquitinases (DUBs). An increase in the level of unmodified protein after being treated with a chain-selective DUB indicates the presence of specific polyubiquitin linkages on the protein. We performed the UbiTest analysis to further validate our plate-based observations of K48-linkage specific ubiquitination of RIPK2. K562 cells were pre-treated with MG132 (1.0 µM) for 60 min to block the proteasome. Cells were then treated with a dose range (0-1000 nM) of RIPK degrader-2 for 45 min. Cell lysates were subjected to pull down using pan-selective TUBEs and the eluted polyubiquitinated proteins were incubated in the presence or absence of a DUB cocktail, K48-selective DUB, or a K63-selective DUB. The samples were then analyzed by immunoblotting anti-RIPK2 antibody.
As shown in Fig. 6a, treatment with DUB cocktail which removes all ubiquitin modifications, completely abolished the ubiquitination signal confirming the presence of polyubiquitinated RIPK2 in PROTAC-treated samples. Notably, the disappearance of polyubiquitinated RIPK2 was accompanied by an increase in the level of unmodified RIPK2. On the other hand, with K48-linkage-specific DUB treatment, we observed a significant reduction in the ubiquitination smear, indicating that a substantial portion of the ubiquitin chains on RIPK2 were K48-linked. However, the loss of ubiquitination signal after K48-selective DUB treatment was not as complete as the DUB cocktail treatment suggesting the presence of other chain types that are not sensitive to K48-selective DUBs.
UbiTest analysis of RIPK Degrader-2 mediated ubiquitination of RIPK2. K562 cells were pre-treated with MG-132 (1.0 µM) for 60 min followed by treatment with DMSO (vehicle control) or indicated doses of RIPK degrader-2 for 45 min. Cell lysates (1.5 mg from each treatment condition) were incubated with pan-selective TUBE magnetic beads for overnight at 4 °C. Polyubiquitinated proteins were eluted from the beads, neutralized and incubated in the absence or presence of indicated DUBs (LifeSensors). (a) Treatment of samples with a cocktail of DUBs, (b) treatment with K48-selective DUB and (c) treatment with K63-selective DUB. After incubation with the DUBs, samples were separated on SDS-PAGE and probed with anti-RIPK2 antibody (n = 2). Original western blot images are presented in Supplementary Fig. 6.
In contrast, K63-linkage-specific DUB treatment had no effect on the ubiquitination pattern, confirming that RIPK degrader-2 does not induce K63-linked polyubiquitination of RIPK2. Collectively, this result indicates that RIPK degrader-2 induces predominantly K48-linked polyubiquitination of RIPK2 and promotes proteasomal degradation.
Discussion
Evolution of pan-selective TUBE-based technologies has facilitated characterization of protein ubiquitination with unprecedented precision leading to the identification of several novel proteins that are polyubiquitinated in various cell and tissue samples. While these efforts have enriched and expanded our knowledge of the ubiquitin proteome system, additional details such as ubiquitin linkage specificity and its functions remain less known. Development of ubiquitin chain-linkage specific high affinity TUBEs such as M1-TUBE, K63-TUBE and K48-TUBE-HF (K48-Tandem Ubiquitin Binding Entity-High Fidelity) aims to fill this gap in scientific knowledge.
In this study we demonstrated the utility of K63-TUBE and K48-TUBE HF to study context dependent K63-ubiquitination and K48-ubiquitination of RIPK2 in a reproducible and high throughput manner (Figs. 2 and 3). The linkage-specific TUBE platform clearly showed that L18-MDP induces predominantly K63-linked ubiquitination of RIPK2. Important, RIPK2 inhibitor ponatinib strongly inhibited RIPK2 K63-ubiquitination. Thus, the K63-TUBE coated 96-well plates can be employed to rapidly evaluate the cellular efficacy of RIPK2 inhibitors. Beyond its role in inflammation, RIPK2 shows promise as a drug target for cancer treatment35.
Previous studies demonstrated the utility of Pan-selective TUBEs for rapid development and characterization of PROTACs32 .Here, we show that chain-selective TUBEs can clearly differentiate the type of ubiquitin chain linkages generated on endogenous RIPK2 after treatment with the RIPK degrader-2 (a von Hippel-Lindau (VHL)-based PROTAC). Initially, we established that RIPK-degrader-2 induces a dose-dependent RIPK2 degradation as reported previously. Our 96-well plate based assay showed a robust ubiquitination peak (UbMax) at 100 nM of RIPK2 degrader-2. Remarkably, the UbMax was similar across the pan-selective TUBE and the K48-HF TUBEs. K63-selective TUBE did not show any appreciable PROTAC induced RIPK2 ubiquitination signal. These results strongly suggest that RIPK2 degrader-2 treatment results in K48-ubiquitination of RIPK2 which is consistent with proteasomal degradation of RIPK2.
Commonly used methods to assess polyubiquitin chain linkage in proteins involve immunoprecipitation of the protein from cells overexpressing epitope-tagged ubiquitin mutants or in vitro ubiquitination assays with the same mutants44,45. However, there are potential limitations with using ubiquitin mutants, including the possibility that E2 and/or E3 enzymes may be sensitive to lysine mutations on ubiquitin, alternative lysines may be utilized in the absence of the preferred lysine, and incomplete penetration of the mutant ubiquitin into the cellular ubiquitin pool may limit its ability to dominantly interfere with specific linkage-mediated events. Additionally, the impact of epitope tags on ubiquitin chain assembly remains unclear. The chain-selective “TUBEs” described in this study allows the researchers to investigate whether chain linkage plays a crucial role in determining the fate of endogenous polyubiquitinated proteins with natively expressed ubiquitin. This approach can be combined with other methods such as overexpression of various lysine mutants of ubiquitin to study linkage-specific ubiquitination of proteins.
One of the potential limitations of TUBE-based ELISA assay to determine context-dependent chain-selective target protein ubiquitination is the presence of mixed ubiquitin chains or branched ubiquitin chains on target proteins. In fact, CRBN E3 ligase has been shown to associate with a HECT type E3 ligase TRIP12 to promote K29/K48 branched ubiquitin chains on BRD4 in presence of ARV825, a CRBN-based BRD4 PROTAC46. Although TUBEs have remarkable specificity for binding to polyubiquitin chains, it is possible that they can interact non-specifically with some of the proteins in cell lysates. Additionally, it is critical that the primary antibody used in the TUBE-based assay is highly specific for the target and capable of detecting ubiquitinated target proteins. Ubiquitination, being a bulky post-translational modification, can potentially mask the epitopes and prevent antibodies from binding to the target protein efficiently. It is recommended to test multiple primary antibodies in the assay for optimal assay performance.
By leveraging ubiquitin chain-selective TUBE platform, researchers can rapidly gain crucial insights into the mechanisms of context dependent ubiquitination of endogenous proteins. The high-throughput screening method offers several notable advantages. Primarily, it exhibits high sensitivity, allowing for the detection and quantification of low levels of endogenous poly-ubiquitination. This enables the reliable assessment of PROTAC-mediated ubiquitination kinetics and provides valuable insights into the potency of different PROTACs with varying ligands and linkers opening up new avenues for the design and optimization of targeted therapies. We envision that K63-specific TUBE platform can be applied to rapidly evaluate non-proteasomal degrader technologies such as LYTACs. Development of chain selective polyubiquitin profiling is a simple and rapid alternative to identifying polyubiquitin signature by mass spec proteomics. Implementing ubiquitination assays in targeted protein degrader drug discovery will streamline the identification of effective molecules, deepen the understanding of their mechanisms of action, and ultimately facilitate the development of innovative therapeutic strategies.
Materials and methods
Chemicals and reagents
The THP1, and K562 cell lines obtained from ATCC were cultured in complete Iscove’s Modified Dulbecco’s medium (bioWorld, 30611046-1) and RPMI-1640 medium supplemented with heat-inactivated 10% fetal bovine serum, 500 U/mL penicillin, and 0.5 mg/mL streptomycin (Gibco). Cells were cultured at 37 °C in a humidified atmosphere flushed with 5.0% CO2. PBS and PBS-T refer to 1X Phosphate-buffered saline, with and without 0.1% Tween 20, respectively. Precast 4-20% Novex WedgeWell Tris-Glycine gels and iBlot 2 NC Mini Stocks were from Thermo. Anti-RICK antibody clone A-10 (sc-166765) to detect RIPK2 protein and HRP-Conjugated anti-GAPDH antibody (sc-47724 HRP) were purchased from Santa Cruz Biotechnology. HRP conjugated secondary antibodies (HAF007, R&D systems.) and Immobilon Western Chemiluminescent HRP substrate (WBKLS0500, EMD Millipore) were purchased. Odyssey XF Imager (LI-COR Biosciences) was used for western blot image acquisition. Streptavidin-Magnetic beads (UM401M), K48 Ubiquitin Linkage ELISA Kit (PA480), K63 Ubiquitin Linkage ELISA Kit (PA630), PROTAC Assay Plate Kit (PA950), K63 Biotin TUBE Elution Kit (UM411M) and UbiTest™ Magnetic TUBE Elution Kit (UM411M) were obtained from LifeSensors Inc. Ponatinib (HY-12047) and RIPK degrader-2 (HY-111866) was obtained from, MedChemExpress LLC). L18-MDP (tlrl-lmdp) was purchased from InvivoGen.
Cell treatments
THP1 suspension cells were maintained in complete Iscove’s medium containing 10% fetal bovine serum (FBS) together with 1X penicillin/streptomycin and 1X non-essential amino acids in a 37 °C/5.0% CO2 incubator. On the day of treatment, the maintenance medium was removed and replaced with fresh Iscove’s complete medium. Cells were then seeded in 100 mm2 dishes at a density of 10 × 106 cells per 10 ml (1.0 × 106 per ml) and placed in the incubator to equilibrate to 37 °C for at least 60 min. prior to treatment. Ponatinib was reconstituted to 10 mM in 100% anhydrous DMSO. Cells were pre-treated with either DMSO (vehicle) and/or Ponatinib at the indicated doses for 30 min. Following pre-treatment, cells were treated with sterile water (non-stimulated) or stimulated with L18-MDP at the indicated concentration for the indicated time point. The K562 cells (1.5 × 106 cells in a 1 mL) were maintained in RPMI complete medium and treated with various concentrations of RIPK degrader-2 a von Hippel-Lindau (VHL)-based PROTAC for 45 min. The cell lysates were prepared by adding 150 µL of RIPA lysis buffer to the treated cell pellets. After protein quantification using BCA method, the cell lysates were de-complexed by adding equal volume of 4 M urea (final concentration of 2.0 M urea) to dissociate protein complexes prior to capturing on TUBE coated plates.
Cell Lysis and immunoprecipitation
After treatment, cells were placed on ice. Cells were collected in 15 ml falcon tubes and centrifuged at ~ 120 x g for 5 min. Cell pellets were washed in ice-cold 1X PBS and either stored frozen at − 80 °C or used immediately for lysis. Cells were lysed using RIPA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1.0% NP40, 1.0% Sodium deoxycholate, 2 mM EDTA, 10% Glycerol, ) supplemented with UPS inhibitor cocktail containing 10 µM MG132, a proteasome inhibitor, 50 µM PR-619, a pan DUB inhibitor (SI9619, LifeSensors) and 5 mM 1,10-phenanthroline, a metalloprotease DUB inhibitor47 (SI9649,LifeSensors), 1 mM PMSF, 1:500 of Protease inhibitor cocktail (P8849, Sigma) and Phosphatase inhibitor cocktail (A32957, Thermo ). Lysis was carried for 15 min on ice with intermittent vortexing. Cell lysate was centrifuged at 13,000 g for 15 min at 4 °C, the supernatant was collected, and protein concentrations determined using BCA protein quantification method (232224, Thermo). Immunoprecipitation/pulldown was conducted using these lysates (1.0 mg each per pull down) to enrich for K63-ubiquitinated RIPK2 protein using Biotin- K63-TUBE beads (LifeSensors, UM-0304-0050) Inc). Briefly, 100 µl of K63-biotin-TUBE were added to 1.0 mg cell lysate and incubated overnight at 4 °C, followed by incubation with Streptavidin-conjugated Magnetic beads for 120 min at 4 °C. The enriched proteins on the beads were washed using the lysis buffer, eluted using elution buffer (LifeSensors) and neutralized by adding neutralization buffer. The samples were mixed with 6x SDS sample loading buffer, boiled for 10 min at 100 °C before performing SDS-PAGE.
UbiTest
UbiTest was conducted following manufacturer’s recommended protocol (UM411M, LifeSensors). Briefly, 100 µl of TUBE1-Magnetic beads from the kit were added to 1.0 mg of cell lysates and incubated overnight at 4 °C. The enriched proteins were eluted, neutralized, and divided equally into four fractions. Each fraction was treated separately with either a DUB cocktail containing USP2core, USP5 and USP20 (DB599, LifeSensors), and K48-linkage specific DUB, OTUB1 or K63-linkage specific DUB, AMSH at 37 °C as per the kit recommendations. One fraction was incubated without adding DUB. DUB digestions were stopped by adding 6X SDS sample loading buffer. Samples were boiled for 5–7 min and then subjected to immunoblot analysis anti-RIPK2 antibody.
Western blotting
Samples were resolved on Novex WedgeWell 10–20% gradient Tris-Glycine gels and transferred onto PVDF membranes using the iBlot2 transfer system. The blots were pre-incubated with blocking buffer containing 5% milk in 1X PBS containing 0.1% Tween20 (PBS-T) for 60 min. The anti-RICK2 antibody against RIPK2 was diluted in blocking buffer (1:250) and incubated on a shaker for overnight at 4 °C. Blots were washed 3 times with 1X PBS-T followed by incubation with HRP-conjugated secondary antibody (1:1000) for 60 min. After washing the blots 3 times with 1X PBS-T, the blots were developed with enhanced chemiluminescence substrates and images were acquired with Odyssey® Fc Imager (LI-COR) and processed using LI-COR Image Studio (version 5.2.5). Blots were stripped and incubated for 60 min with an HRP-conjugated anti-GAPDH antibody for obtaining protein loading control (Santa Cruz, sc47724 HRP, 1:1000).
Linkage specific TUBEs based assays
Pan-selective TUBE contains four tandem ubiquitin binding domains of human Rad23 and was prepared as described previously by Hjerpe et al.48. K63-TUBE was generated as described in Sims et al.39. Pan-selective and linkage specific TUBEs coated by passive adsorption onto the plates were purchased from LifeSensors and used according to the manufacturer’s instructions. Plates stored at − 80 °C were brought to room temperature prior to use. Cell lysates were diluted to 15.0 µg in 100 µL volume per well using 1X PBS-T and transferred to the assay plate. Lysates were incubated for 120 min at room temperature on an orbital shaker at 300 rpm. After incubation the plates were washed for 3 times with 200 µL 1X PBS-T. Antibody against RIPK2 (A-10, SC, 1:250), was diluted in 1X antibody dilution buffer (Superblock, Thermo) and 100 µL of diluted antibody were added to each well, and the samples were incubated at room temperature for 90 min on an orbital shaker at 300 rpm. After incubation, the plates were washed 3 times with 200 µL 1X PBS-T. Subsequently, 100 µL of 1:1000 anti-mouse IgG-HRP in 1X antibody dilution buffer were added to each well. Plates were incubated at room temperature for 45 min at 300 rpm on an orbital shaker. Plates were washed three times using 1X PBS-T. Excess wash buffer from wells were removed by gently tapping the plate on paper towels. To each well, 100 µL of Enhanced Chemiluminescence Substrate (Thermo) diluted in water was added and the plate was read on BMG LabTech Clariostar plate reader. The cell lysates obtained from PROTAC-treated cells were subjected to decomplexing with equal volume of 4.0 M urea (the final concentration is 2.0 M urea) for 15 min to disrupt any native complexes. Following urea pre-treatment, the samples were diluted to a concentration of 15.0 µg in a volume of 100 µL per well using 1X PBS-T. Subsequently, the diluted samples were transferred to the assay plate and incubated overnight at 4 °C. Plates were washed, incubated with anti-RIPK2 antibody, and detected using enhanced chemiluminescence substrates as described above.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 https://doi.org/10.1038/nm.3739 (2014).
Ali, M. S. et al. The giant HECT E3 ubiquitin ligase HERC1 is aberrantly expressed in myeloid related disorders and it is a novel BCR-ABL1 binding partner. Cancers (Basel). 13, 1–14 (2021).
Ali, M. S. et al. The downregulation of both giant hercs, HERC1 and HERC2, is an unambiguous feature of chronic myeloid leukemia, and HERC1 levels are associated with leukemic cell differentiation. J. Clin. Med. 11, 324 (2022).
Chaugule, V. K. & Walden, H. Specificity and disease in the ubiquitin system. Biochem. Soc. Trans. 44, 212–227 (2016).
Komander, D. & Rape, M. The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 (2012).
Mattern, M., Sutherland, J., Kadimisetty, K., Barrio, R. & Rodriguez, M. S. Using ubiquitin binders to decipher the ubiquitin code. Trends Biochem. Sci. 44, 599–615 (2019).
Spence, J., Sadis, S., Haas, A. L. & Finley, D. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15, 1265–1273 (1995).
Finley, D. et al. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 14, 5501–5509 (1994).
Alfano, C., Faggiano, S. & Pastore, A. The ball and chain of polyubiquitin structures. Trends Biochem. Sci. 41, 371–385 (2016).
Akutsu, M., Dikic, I. & Bremm, A. Ubiquitin chain diversity at a glance. J. Cell. Sci. 129, 875–880 (2016).
Hu, H. & Sun, S. C. Ubiquitin signaling in immune responses. Cell. Res. 26, 457–483 (2016).
Liao, Y., Sumara, I. & Pangou, E. Non-proteolytic ubiquitylation in cellular signaling and human disease. Commun. Biol. 5, 114 (2022).
Hrdinka, M. et al. Small molecule inhibitors reveal an indispensable scaffolding role of RIPK 2 in NOD 2 signaling. EMBO J. 37, 99372 (2018).
Chen, Z. J. & Sun, L. J. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 33, 275–286 (2009).
Turner, M. D., Nedjai, B., Hurst, T. & Pennington, D. J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Biophys. Acta (BBA)-Molecular Cell. Res. 1843, 2563–2582 (2014).
Libby, P. Inflammatory mechanisms: The molecular basis of inflammation and disease. Nutr. Rev. 65, S140–S146 (2007).
Yin, X. et al. Emerging roles of non-proteolytic ubiquitination in tumorigenesis. Front Cell. Dev. Biol 10, 944460 (2022).
Brenke, J. K. et al. Targeting TRAF6 E3 ligase activity with a small-molecule inhibitor combats autoimmunity. J. Biol. Chem. 293, 13191–13203 (2018).
Cockram, P. E. et al. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell. Death Differ. 28, 591–605 (2021).
Panda, S. & Gekara, N. O. The deubiquitinase MYSM1 dampens NOD2-mediated inflammation and tissue damage by inactivating the RIP2 complex. Nat. Commun. 9, 4654 (2018).
Lopez-Castejon, G. & Edelmann, M. J. Deubiquitinases: Novel therapeutic targets in immune surveillance? Mediators Inflamm. 2016, 3481371 (2016).
Lu, D. et al. Applications of covalent chemistry in targeted protein degradation. Chem. Soc. Rev. 51, 9243–9261 (2022).
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Yu, X. et al. Development of a RIPK1 degrader to enhance antitumor immunity. Nat. Commun. 15, 1–14 (2024).
Bondeson, D. P. et al. Catalytic in vivo protein knockdown by small-molecule protacs. Nat. Chem. Biol. 11, 611–617 (2015).
Ji, X. et al. A rapid and accurate method for evaluating the degradation of pan-Akt in cells by protacs using nanoluc luciferase. Biol. Methods Protoc. 9, bpae014 (2024).
Grohmann, C. et al. Development of NanoLuc-targeting protein degraders and a universal reporter system to benchmark tag-targeted degradation platforms. Nat. Commun. 13, 1–13 (2022).
Kim, D. Y., Scalf, M., Smith, L. M. & Vierstra, R. D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant. Cell. 25, 1523–1540 (2013).
Romero-Barrios, N. et al. Advanced cataloging of lysine-63 Polyubiquitin networks by genomic, interactome, and sensor-based proteomic analyses. Plant. Cell. 32, 123–138 (2020).
Kliza, K. & Husnjak, K. Resolving the complexity of ubiquitin networks. Front. Mol. Biosci. 7, 21 (2020).
Gross, P. H. et al. Accelerating PROTAC drug discovery: Establishing a relationship between ubiquitination and target protein degradation. Biochem. Biophys. Res. Commun. 628, 68–75 (2022).
Hasegawa, M. et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-κB activation. EMBO J. 27, 373–383 (2008).
Damgaard, R. B. et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol. Cell. 46, 746–758 (2012).
You, J., Wang, Y., Chen, H. & Jin, F. RIPK2: A promising target for cancer treatment. Front. Pharmacol. 14, 1192970 (2023).
LeNoue-Newton, M. et al. The E3 ubiquitin ligase-and protein phosphatase 2A (PP2A)-binding domains of the Alpha4 protein are both required for Alpha4 to inhibit PP2A degradation. J. Biol. Chem. 286, 17665–17671 (2011).
Chen, L., Shinde, U., Ortolan, T. G. & Madura, K. Ubiquitin-associated (UBA) domains in Rad23 bind ubiquitin and promote Inhibition of multi-ubiquitin chain assembly. EMBO Rep. 2, 933–938 (2001).
Mori, Y. et al. Intrinsic signaling pathways modulate targeted protein degradation. Nat. Commun. 15, 5379 (2024).
Sims, J. J. et al. Polyubiquitin-sensor proteins reveal localization and linkage-type dependence of cellular ubiquitin signaling. Nat. Methods 2012. 93 (9), 303–309 (2012).
Yang, Y. et al. NOD2 pathway activation by MDP or Mycobacterium tuberculosis infection involves the stable polyubiquitination of Rip2. J. Biol. Chem. 282, 36223–36229 (2007).
Chandler, F. et al. Molecular glues that inhibit deubiquitylase activity and inflammatory signaling. Nat. Struct. Mol. Biol. 2025, 1–13. https://doi.org/10.1038/s41594-025-01517-5 (2025).
Mevissen, T. E. T. & Komander, D. Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 (2017).
van Wijk, S. J. L., Fulda, S., Dikic, I. & Heilemann, M. Visualizing ubiquitination in mammalian cells. EMBO Rep. 20, e46520 (2019).
Wang, C. et al. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 (2001).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 (2004).
Kaiho-Soma, A. et al. TRIP12 promotes small-molecule-induced degradation through K29/K48-branched ubiquitin chains. Mol. Cell. 81, 1411–1424e7 (2021).
Cooper, E. M. et al. K63-specific deubiquitination by two JAMM/MPN + complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 28, 621–631 (2009).
Hjerpe, R. et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 (2009).
Author information
Authors and Affiliations
Contributions
M.S.A. performed the experiments with assistance from C.R., prepared the figures and wrote the main manuscript draft. J.P. performed some of the plate-based experiments with assistance from M.S.A. D.E.S. and H.W contributed to the making of vectors, and reagents. T.R.B. and K.S. conceived the study, contributed to the experimental design, interpretation of the data, generation of figures and final manuscript editing. All the authors contributed to the study, reviewed the manuscript and approved the submission.
Corresponding author
Ethics declarations
Competing interests
M.S.A.,C.R., D.E.S., H.W., K.S., and T.B. are employees of Progenra Inc. These authors declare no competing interests. J.P. was an employee of LifeSensors Inc. which is the commercial owner of TUBE technology and their applications along with chain-selective TUBE-based assay plates.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ali, M.S., Rainville, C., Pedroza, J. et al. Unraveling chain specific ubiquitination in cells using tandem ubiquitin binding entities. Sci Rep 15, 22961 (2025). https://doi.org/10.1038/s41598-025-07242-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-07242-9