Abstract
A brain tumor is a serious medical condition characterized by the abnormal growth of cells within the brain. It can cause a range of symptoms, including headaches, seizures, cognitive impairment, and changes in behavior. Brain tumors pose a significant health concern, imposing a substantial burden on patients. Timely diagnosis is crucial for effective treatment and patient health. Brain tumors can be either benign or malignant, and their symptoms often overlap with those of other neurological conditions, leading to delays in diagnosis. Early detection and diagnosis allow for timely intervention, potentially preventing the tumor from reaching an advanced stage. This reduces the risk of complications and increases the rate of recovery. Early detection is also significant in the selection of the most suitable treatment. In recent years, Smart IoT devices and deep learning techniques have brought remarkable success in various medical imaging applications. This study proposes a smart monitoring system for the early and timely detection, classification, and prediction of brain tumors. The proposed research employs a custom CNN model and two pre-trained models, specifically Inception-v4 and EfficientNet-B4, for classification of brain tumor cases into ten categories: Meningioma, Pituitary, No tumor, Astrocytoma, Ependymoma, Glioblastoma, Oligodendroglioma, Medulloblastoma, Germinoma, and Schwannoma. The custom CNN model is designed specifically to focus on computational efficiency and adaptability to address the unique challenges of brain tumor classification. Its adaptability to new challenges makes it a key component in the proposed smart monitoring system for brain tumor detection. Extensive experimentation is conducted to study a diverse set of brain MRI datasets and to evaluate the performance of the developed model. The model’s precision, sensitivity, accuracy, f1-score, error rate, specificity, Y-index, balanced accuracy, geometric mean, and ROC are considered as performance metrics. The average classification accuracy for CNN, Inception-v4, and EfficientNet-B4 is 97.58%, 99.56%, and 99.76%, respectively. The results demonstrate the excellent accuracy and performance of the previous proposed approaches. Furthermore, the trained models maintain accurate performance after deployment. The method predicts accuracy of 96.5% for CNN, 99.3% for Inception-v4, and 99.7% for EfficientNet-B4 on a test dataset of 1000 brain tumor images.
Similar content being viewed by others
Introduction
The human body’s most complex organ is the brain, which serves as the system of intelligence, interpreter of the senses, initiator of body movement, and behavior controller1. Tumors are one of the most harmful forms of cell growth, causing damage to body organs and resulting in patient’s death2. Cancer is one of the serious health problems and challenges that threatens the lives of humanity nowadays. After cardiovascular disorders, cancer is the second leading cause of death, and every sixth death is due to cancer. Among the different types of cancer, brain tumors are the most dangerous and deadly due to their heterogeneous characteristics, aggressive nature, and low survival rate3. A brain tumor, known as an intracranial tumor, is an abnormal tissue mass in which cells grow and multiply uncontrollably, and seems unchecked by the mechanisms that control normal cells4. Malignant (cancerous) or benign (non-cancerous) brain tumors are both possible, and while some tumors enlarge swiftly, others do so slowly. One-third of tumors are malignant. However, brain tumors can affect nervous system, blood vessels, tissue, health, and the normal functionality of the brain5.
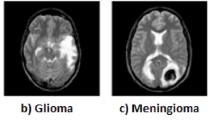
Although there are more than 150 known types of brain tumors, primary and metastatic are the two basic classifications. Primary brain tumors are divided into glioma and non-glioma tumor types. Gliomas are tumors that develop from glial cells. Gliomas come in variety of forms, including astrocytic tumors, such as astrocytoma, which begin in the cerebrum, oligodendr 3oglia tumors, which are found in the frontal and temporal lobes, glioblastomas, which originate in supporting brain tissue and are the most aggressive type6 and ependymomas, which develop from ependymal cells (called radial glial cells)5. On the other hand, non-glioma tumors are tumors that arise from cells in the brain that are not glial cells. These tumors are of several types: meningioma, usually starts in the meninges and is not malignant, schwannoma, a rare tumor that begins in the nerve sheath, or the lining of the nerves, medulloblastoma, the cerebellum is where medulloblastoma is hypothesized to originate from and is most common in children, pineal gland and pituitary gland tumors, tumors which start in the pineal gland and pituitary gland7.
The death rate of brain tumors is increasing every year. According to a report presented in8 from the years 1990-2019, the increasing death rates of brain cancer in different countries is presented in Fig. 1. Furthermore, based on the report of the estimated rate of cancer in 20239, it is reported that in 2023, an estimated 24,810 adults (14,280 men and 10,530 women) and 5,230 children under the age of 20 in the United States have been diagnosed with primary cancerous tumors of the brain and spinal cord.
Therefore, early and accurate brain tumor diagnoses are vital for effective treatment and prevention from disease3. Smart health care is an important aspect of connected living. It is one of the basic needs of human being. With the advent of new technologies and the fast pace of human life, patients today require a sophisticated and advanced smart healthcare framework that is tailored to suit their health requirements. The Internet of Things (IoT) is likely a potential solution for healthcare system challenges and has been extensively studied recently. Much of this research focuses on monitoring of patients with specific conditions. Research in related fields has demonstrated the feasibility of remote health monitoring, but its broader advantages are even more significant. Utilizing remote health monitoring could shift non-critical patient monitoring from hospitals to homes, easing the burden on hospital resources like doctors and beds. Moreover, it can enhance healthcare accessibility for rural residents and can promote independent living for the elderly. Ultimately, it enhances healthcare resource access, eases system strain, and empowers individuals to manage their health proactively10. The information of a patient is updated through IoT devices which help to administer medication in current situation in order to decrease the severity of disease11. Furthermore, deep learning (DL) is now considered as a key technology of the Fourth Industrial Revolution (also known as Industry 4.0) due to its ability to learn from data12. It outperforms conventional machine learning techniques by automating feature extraction, overcoming the drawbacks of manual feature engineering13. Through grouping and classification techniques which effectively handle the quality of information, deep learning is used to help people make better judgments.
Along with 5G and state-of-the-art smart Internet of Things (IoT) sensors, edge computing provides intelligent, real-time healthcare solutions that satisfy energy consumption and latency criteria14. IoT’s goal is the connection of humans with machines and smart technologies. The concept of smart Internet of Things (IoT) refers to the vast interconnected network of computer devices (e.g., sensors) that exchange large amounts of data at rapid speed15. Moreover, Edge AI is deploying AI applications on devices throughout the physical world. It is called “edge AI” because the AI computation is done near the user at the network’s edge, close to where the data is located, rather than centrally in a cloud computing facility or private data center16. In the field of healthcare, several sensors can be incorporated to collect data related to a patient’s health (including the patient’s images), and an emergent response can be triggered when required. Utilizing these devices makes it extremely simple to monitor a person’s health remotely and provides assistance. Smart systems which are built of AI and the IoT have proved to be immensely effective in healthcare. The healthcare sectors may be revolutionized by edge-enabled IoT, where many embedded sensors and IoT devices interconnect to provide different services to the community for the well-being of its citizens. With the current COVID-19 situation, chronic diseases, and an ageing population, interconnected IoT devices generate a huge amount of IoT data. The situation is changing so quickly that healthcare systems have not been able to keep up. It is essential to diagnose and detect diseases at an early stage to facilitate timely treatment. This also reduces the cost of healthcare. To this end, the convergence of edge AI and IoT has the potential to classify and cluster this massive amount of IoT data, make predictions, and deliver early insights17,18,19,20,21,22,23. This can address the global challenge of the pandemic and other related problems24,25.
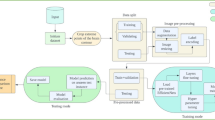
The proposed study is focused on combining IoT and deep learning approaches for the automatic detection and classification of brain tumor MRI images into ten classes. In the proposed framework, the smart health monitoring environment is used for the early prediction of a class of brain tumors for the life safety of patients. An overview of a smart brain tumor monitoring system based on smart IoTs and deep learning is presented in Fig. 2.
Motivation
The motivation behind this research lies in addressing the need for early and accurate detection of brain tumors. Brain tumors are concerned with serious health issues, and their timely diagnosis is important for effective treatment and improved patient outcomes. Conventional diagnostic methods may be limited in accuracy and efficiency. By harnessing the power of deep learning and IoT technologies, this research aims to create a smart monitoring system that can automatically classify and predict brain tumors from MRI images. This technology can significantly enhance medical imaging capabilities, enabling faster and more accurate diagnosis, ultimately leading to better patient care and survival rates.
Research problem
The research problem at the core of this study is the difficulty with accurately and promptly diagnosing brain tumors using conventional methods. Brain tumors can exhibit varied symptoms and share features with other neurological conditions, leading to delayed or incorrect diagnoses. This poses a substantial health risk and can impact treatment efficacy. The complexity of distinguishing between benign and malignant tumors further complicates the issue. Therefore, the research problem focuses on developing an intelligent system that can efficiently classify and predict brain tumor types from MRI images, improving diagnostic accuracy and ensuring timely intervention.
Research contributions
This research makes several significant contributions to the field of medical imaging and healthcare.
-
Firstly, it proposes a novel approach that combines IoT-based smart monitoring with deep learning techniques for brain tumor detection. By integrating IoT devices, the research envisions a more connected and accessible healthcare system.
-
Secondly, the study introduces a specialized custom convolutional neural network (CNN) model designed to accurately classify brain MRI images. This model is engineered to efficiently extract pertinent features from brain images, facilitating the precise classification of ten distinct tumor types, including Meningioma, Pituitary, Astrocytoma, Ependymoma, Glioblastoma, Oligodendroglioma, Medulloblastoma, Germinoma, No Tumor, and Schwannoma.
-
Additionally, incorporating two pretrained models, Inceptionv4 and EfficientNetB4, enhances the system’s classification accuracy, bringing a new dimension of accuracy and robustness to the research.
-
Overall, the research offers a comprehensive framework that contributes in advancing the early detection and classification of brain tumors, thus potentially revolutionizing the way healthcare professionals diagnose and treat this critical condition.
The rest of the paper is organized as follows: Section "Related work" presents the existing techniques, Section "Materials and methods" describes the proposed methodology with a description of the dataset and models. Results are described in Section "Results and discussion", and the conclusion is presented in Section "Conclusions and future work" with possible future research directions.
Related work
The literature review section aims to present a comprehensive overview of previous studies conducted in the field of brain tumor detection, focusing on various machine learning and deep learning models. Through a systematic analysis of these studies, this section will highlight the advancements, methodologies, and outcomes achieved in brain tumor detection, providing valuable insights for the current research endeavor.
Mohamed Amine Mahjoubi et al.26 propose a CNN-based model for brain tumor classification in medical image analysis. The model categorizes brain images into four classes: normal, glioma, meningioma, and pituitary, achieving an accuracy of 95.44%, a recall rate of 95%, and an F1-score of 95.36%, by utilizing the publicly available Kaggle database comprising 7022 MRI scans of the human brain. While the model demonstrates promising results, potential limitations such as the need for validation on diverse datasets, applicability to other tumor types, data requirements, and computational complexity should be considered for real-world practicality.
Abd El-Wahab et al.27 introduce the BTC-fCNN, a fast and efficient deep learning-based system for classifying different brain tumor types, including meningioma, glioma, and pituitary tumors. The proposed model utilizes 2D MRI images from a freely available dataset (Cheng J. Brain tumor dataset. Figshare), consisting of 3064 images, to evaluate the brain tumor classification networks. The proposed model consists of 13 layers: convolutional, 1x1 convolutional, average pooling, fully connected, and softmax layers. Through five iterations involving transfer learning and five-fold cross-validation, the model achieves an average accuracy of 98.63% and 98.86% using retrained five-fold cross-validation.
According to Emrah Irmak28, a convolutional neural network (CNN) is utilized for the multi-classification of brain tumors for early diagnosis purposes. Three CNN models are proposed for different classification tasks: tumor detection, tumor type classification (normal, glioma, meningioma, pituitary, and metastatic), and tumor grade classification (Grade II, Grade III, and Grade IV). The first CNN model achieves 99.33% accuracy for tumor detection, while the second model achieves 92.66% accuracy for tumor type classification, and the third model achieves 98.14% accuracy for tumor grade classification. Grid search optimization is employed to automatically tune the important hyper-parameters of the CNN models.
To identify and categorize brain tumors, Abdul Hannan Khan et al.29 presented the Hierarchical Deep Learning-Based Brain Tumor (HDL2BT) classifier. The model categorizes tumors into four types: glioma, meningioma, pituitary, and no-tumor. The system’s architecture comprises three essential layers: data acquisition, preprocessing, and prediction. The dataset used in the study was obtained from Kaggle. With a miss rate of 7.87%, the model achieves 92.13% accuracy.
An Intelligent Ultra-Light Deep Learning Model for Multi-Class Brain Tumor Detection is suggested by Shahzad Ahmad Qureshi et al.30. The proposed system is based on an Ultra-Light Deep Learning Architecture (UL-DLA) combined with Gray Level Co-occurrence Matrix (GLCM) textural features. It forms a Hybrid Feature Space (HFS) used for tumor detection with Support Vector Machine (SVM). The evaluation was conducted on a publicly available T1-weighted CE-MRI dataset, encompassing glioma, meningioma, and pituitary tumor cases. The system achieves accuracy, with an average detection rate of 99.23% (99.18%, 98.86%, and 99.67%) and an F-measure of 99% (99%, 98%, and 99%) for (glioma, meningioma, and pituitary tumors), respectively.
By combining deep-learning-based features with several classifiers, Ghazanfar Latif et al.31 provide a strategy for categorizing glioma tumors. In the study, classifiers including Random Forests (RF), Multi-Layer Perceptron (MLP), Support Vector Machines (SVM), and Naive Bayes (NB) were utilized. The aim is to classify glioma tumors into four classes: Edema, Necrosis, Enhancing, and Non-enhancing. The dataset used by the authors is the MICCAI BraTS 2018 dataset. The proposed method achieves a 96.19% accuracy for the HGG type using the Flair modality with the SVM classifier, and a 95.46% accuracy for the LGG type using the T2 modality with the same classifier.
Deep analysis of brain tumor identification using deep learning networks is presented by Md Ishtyaq Mahmud et al.32. For the accurate detection of brain tumors using MR images, the authors suggest a convolutional neural network (CNN) design. In addition, this study analyses several models, including ResNet-50, VGG16, and Inception V3, and compares them to the suggested design. The study uses a dataset of 3,264 MR images from Kaggle that includes cases without tumors as well as other forms of brain tumors, such as glioma tumors, meningioma tumors, and pituitary tumors. The accuracy of the CNN model in identifying brain tumors from MR images was 93.3%, while that of the pre-trained models ResNet-50, VGG16, and Inception V3 were 81.10%, 71.60%, and 80.00%, respectively.
Using 3D discrete wavelet transform (DWT) and the random forest classifier, Ghazanfar Latif et al.33 offer a method for classifying and segmenting low-grade and high-grade glioma tumors in multimodal MR images. MR images are preprocessed, normalized, and segmented into small blocks for feature extraction using 3D DWT. The random forest classifier is then utilized to classify the different types of glioma tumors, including necrosis, edema, non-enhancing tumors, and enhancing tumors. The MICCAI BraTS dataset, which includes cases of low-grade and high-grade gliomas, is used to assess the suggested method. The average accuracy of the random forest classifier is 89.75% for high-grade gliomas and 86.87% for low-grade gliomas.
Wadhah Ayadi et al.34 provides a novel method for classifying three different kinds of brain tumors (glioma, meningioma, and pituitary tumor) using multimodal MR imaging. The proposed technique combines image normalization, dense speeded-up robust features (DSURF), and a histogram of gradients (HoG) to improve picture quality and extract discriminative features. The support vector machine (SVM) is used as a classifier over a dataset of 233 patients with brain tumors. Furthermore, the system is tested, and the suggested technique obtains an overall accuracy of 90.27%. The accuracy of the suggested model for meningioma, glioma, and pituitary tumor, is 90.83%, 92.66%, and 97.06% respectively.
Heba Mohsen et al.35 focus on classifying four types of tumors: normal, glioblastoma, sarcoma, and metastatic bronchogenic carcinoma. The dataset consists of 66 brain MRIs, with 22 normal and 44 abnormal images representing the different tumor types. Image segmentation is performed using the fuzzy C-means clustering technique, and then the tumor features are extracted using DWT. The DNN classifier is trained on the extracted features and achieves an accuracy of 96.97%, recall of 0.97, precision of 0.97, F-measure of 0.97, and an AUC (ROC) of 0.984.
To distinguish between the three most prevalent forms of brain tumors, such as gliomas, meningiomas, and pituitary, a convolutional neural network (CNN) is used by the authors36. A publicly accessible dataset of 3064 T-1 weighted CE-MRI images of brain tumors from Figshare Cheng was used to train the CNN. The accuracy of brain tumor categorization was tested by the author using five different architectures. The validation and training accuracy of the suggested system are 84.19% and 98.51%, respectively. The highest validation accuracy is still provided by Architecture 2, which uses 32 filters in the convolution layers.
To determine brain tumor existence, based on MRI images, the authors in37 used two models: a customized CNN model created from scratch and a pre-trained VGG-16 model. The MRI scans in the training set fall into two categories: NO (tumor absent) and YES (tumor present). To increase the dataset, data augmentation techniques are used. On the test dataset, the system achieves an accuracy of about 90%, while on the validation set, it achieves an accuracy of 86%. The author concludes that the optimized VGG-16 model performs better than the modified CNN model.
Zar Nawab Khan Swati et al.38 suggests a new method for classifying images of brain tumors by combining transfer learning with fine-tuning methods. They use a deep CNN model that has already been trained and present a block-wise fine-tuning method based on transfer learning. Three different forms of brain tumors, including gliomas, meningiomas, and pituitary tumors, are included in the study’s CE-MRI dataset. Min-max normalization is used to preprocess the images. The VGG19 architecture, with 19 layers and 144 million trainable parameters, is used as the base model. Using five-fold cross-validation, the suggested method obtains an average accuracy of 94.82%.
The authors in39 present a study that utilizes four pre-trained models: DenseNet121, DenseNet201, VGG16, and VGG19 to classify brain tumor images. The dataset utilized in the study was obtained from Kaggle and consists of 257 MRI images, 157 of which have been labelled as brain tumors (BT) and 100 of which have not. Data augmentation increases the number of sample images to address the small dataset size. Evaluation of the models’ performance reveals that VGG16, VGG19, DenseNet121, and DenseNet201 achieve an accuracy of 94%, 98%, 96%, and 96%, respectively.
The MobileNetV2 architecture is used by Dheiver Santos et al.40 to classify brain tumors using the Android app. The dataset is comprised of 3,762 MRI images, of which 1,683 have tumors and 2,079 are without tumors. A test accuracy of 89% was achieved by the model used in this study.
The proposed study by Nilakshi Devi et al.41 suggests employing artificial neural networks to detect brain tumors and categorize the grades of astrocytomas. The system is divided into two stages: tumor detection using an ANN with features taken from a gray-level co-occurrence matrix (GLCM) and grade classification using a radial basis function neural network (RBFN) with energy, homogeneity, and contrast variables taken from MRI images. In identifying brain images with tumor and normal tissue, the proposed ANN had 99% accuracy. The RBFN, optimized using particle swarm optimization (PSO), classified astrocytoma grades using extracted features, with results compared to biopsy reports.
Using a radiomics analysis technique based on machine learning, Mengmeng Li et al.42 seek to distinguish between ependymoma (EP) and pilocytic astrocytoma (PA). A total of 135 MRI slices from the real preoperative dataset of 45 patients (age range: 0 - 14 years) was evaluated with 81 slices of EP and 54 slices of PA. 300 multimodal radiomics features, including texture, Gabor transforms, and wavelet transform-based features were extracted. For feature selection and tumor distinction, the support vector machines (SVM) and the Kruskal-Wallis test score (KWT) were used. The selected feature set achieved an accuracy of 87%, sensitivity of 92%, specificity of 80%, and an area under the receiver operating characteristic curve (AUC) of 86%. A technique for identifying glioblastoma tumor present in MRI images is created by Dr. P. Tamije Selvy et al.43. The MRI brain tumor dataset that was downloaded from GitHub has about 150 images. It follows a two-phase approach: feature extraction and classification. Gray Level Co-occurrence Matrix (GLCM) is employed for extracting texture features. The extracted features are then fed into a feed-forward Probabilistic Neural Network (PNN) classifier to predict the presence of a glioblastoma tumor. The system achieves an accuracy of approximately 90% in glioblastoma detection. The article44 provides a method for categorizing glioma brain tumors into different classifications using a combination of deep convolutional neural network (DCNN), k-means segmentation, and contrast enhancement. 350 MRI images, including those of oligodendroglioma, astrocytoma, ependymoma, and normal brain tissue are included in the dataset. Using a dataset of 270 images for training and 90 for testing, the DCNN model shows a testing accuracy of 95.5% and a training accuracy of 90%.
Table 1 displays a tabular comparison of the various machine learning and deep learning techniques used for brain tumor detection and classification.
Materials and methods
In this section, a proposed methodology is presented, providing a detailed description of the datasets utilized in this study and the preprocessing steps performed on the dataset. Additionally, the proposed models employed for detecting and classifying brain tumors are also introduced. Figure 3 illustrates the visual representation of the proposed methodology for automatic brain tumor detection and classification.
Dataset preparation phase
The first step in the model training process is the collection and processing of data. The details of the dataset and processing techniques are represented below.
Dataset description
In this study, we used a publicly available dataset obtained from various Kaggle sources. The dataset contains brain MRI images, both healthy and diseased, organized into ten different classes. One class is for normal brain MRI images, and the remaining nine classes represent various tumor types.
-
Tumor types: Astrocytoma, Ependymoma, Germinoma, Glioblastoma, Medulloblastoma, Meningioma, Oligodendroglioma, Pituitary, and Schwannoma are the tumor types that are included in the dataset.
-
Normal class: The normal class includes MRI images of individuals without brain tumors.
Data sources
The dataset was obtained from several Kaggle sources. Specifically, the Meningioma, Pituitary, and No tumor datasets were sourced from55 and56, The remaining 7 tumor types, namely Astrocytoma, Ependymoma, Glioblastoma, Oligodendroglioma, Medulloblastoma, Germinoma, and Schwannoma acquired from57. To ensure the model’s generalization ability, a separate test dataset was collected from58 and59.
Data preprocessing
The dataset undergoes two essential preprocessing techniques of data augmentation and cropping MRI images. The data augmentation technique is vital as it generates additional data from a limited dataset, effectively increasing the amount of data available for training. Secondly, it helps mitigate overfitting issues, enhancing the generalization capabilities of the model. Within our dataset, we encounter limitations in data availability for seven classes, namely Astrocytoma, Ependymoma, Germinoma, Glioblastoma, Medulloblastoma, Oligodendroglioma, and Schwannoma. Various augmentation techniques are applied including rotation, width, height shifting, zooming, shear transformation, horizontal and vertical flipping, and brightness adjustment. We expand the dataset by employing these augmentation methods, enabling our model to better learn from limited data and improve performance. Table 2 illustrates the number of images in the original dataset before and after augmentation for each brain tumor type. To extract only the brain portion from MRI images, we employ the OpenCV library to crop unwanted dark areas. This step is crucial, as it enables the model to effectively learn relevant information about the brain while avoiding irrelevant details.
Data splitting ratio
The dataset was split into training and validation sets, with 80% of the images used for training and 20% for validation. Additionally, we used a separate test set of 1000 images, not seen by the model during training, to rigorously assess the performance of models.
Training phase
The dataset is then passed to the models for training after being preprocessed. For brain tumor classification, we have selected two pre-trained models, Inception-v4 and EfficientNet-B4, based on transfer learning, a process of applying a previously learned model to a new situation with reduced training time and improved neural network performance60. In addition, we have developed a customized CNN model from scratch. The details of the models are presented below.
EfficientNet
EfficientNet is a convolutional neural network architecture and scaling method that uniformly scales all depth, width, and resolution dimensions using a compound coefficient. Unlike conventional practice that arbitrarily scales these factors, the EfficientNet scaling method uniformly scales network width, depth, and resolution with a set of fixed scaling coefficients. The key to the compound scaling method is based on EfficientNet to find a set of compound coefficients of depth, width, and image resolution to maximize the network’s performance. This optimization problem is mathematically formulated as in Eq. (1).
where, d, w, r are the scaling coefficients of network depth, width, and image resolution respectively, N(d, w, r) is the classification model, and \(max_{d,w,r}\) Accuracy is the maximum accuracy of the model.
To realize uniform scaling of d, w, r, this method introduces \(\varphi\), which is a user-specified coefficient that controls model scale, as shown in Eq. (2). The implementing steps of this scaling method are as follows: After determining the structure of the baseline network, this method first fixes the control coefficient \(\varphi\) as 1, then uses NAS technology to search coefficients d, w, r that maximize the classification accuracy, resulting in the final baseline model called EfficientNet-B0; Finally, this method specifies different \(\varphi\) from 2 to 7 and obtains corresponding models of different sizes, referred to as EfficientNet-B1, EfficientNet-B2,..., EfficientNet-B7, respectively61.
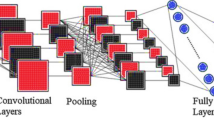
In this research, we choose the EfficientNet-B4 model, a specific variant of the EfficientNet model. The ”B4” in its name indicates its relative size within the EfficientNet series. It offers a powerful solution for various computer vision applications, balancing accuracy and computational resources. EfficientNet-B4 is well-suited for various computer vision tasks, including image classification, object detection, and segmentation62. Figure 4 illustrates the architecture of EfficientNet-B4 for detecting brain tumors and classifying them into ten classes, which consist of 19 layers, including a combination of convolutional, pooling, and fully connected layers, with various filter sizes and depth optimization.
Inception-v4
Inception-v4 is a convolutional neural network architecture that improves earlier versions of the Inception family by streamlining the architecture and utilizing more Inception modules than Inception-v363. Figure 5 depicts the overall architecture of Inception-V4 for brain tumor detection and classification into ten classes. It includes a pure Inception network with no residual connections, and all the key Inception V1 through V3 approaches are used. Regarding the number of layers, Inception-v4 comprises around 75 layers in its core architecture. This includes various convolutional, pooling, and fully connected layers and the unique ”Inception” modules that allow the network to efficiently learn features at different scales. The Inception-v4 architecture emphasizes the use of wider convolutional filters and factorizing convolutional layers to reduce computational complexity.
Custom CNN model
The proposed Custom Convolutional Neural Network (Custom CNN) model, illustrated in Fig. 6, is an intricately crafted architecture tailored to excel in the task of classifying brain tumors. It is designed with a specific focus on computational efficiency and adaptability to address the unique challenges of brain tumor classification. The architecture is particularly well-suited for IoT-based medical diagnostic systems where computational resources may be limited.
The significance of this model lies in its adaptability to new challenges, offering a fine-tuned structure and parameters designed to contribute to the overall success of the proposed smart monitoring system for early and accurate brain tumor detection. The design of the network focuses on achieving a balance between depth and computational efficiency, which is crucial for accurate and fast image classification.
Architecture overview
The architecture consists of five convolutional layers responsible for capturing intricate patterns and features from the input images. The convolutional layers progressively increase the number of filters from 32 to 512, allowing for the effective capture of complex spatial hierarchies within MRI images. This ensures that the model can learn a wide range of features, from simple edges to intricate patterns.
In this architecture, the input image is denoted as X. The convolutional operations are represented by \(\operatorname {Conv}()\), the max-pooling operations by MaxPool(), the fully connected layers by FC(), the Rectified Linear unit activation function by \(\operatorname {ReLU}()\) and the flattening operation by F(). The weights and baises for the i-th layer and \(\textrm{j}\)-th filter are denoted by \(W_{i j}\) and \(b_i\) respectively.
Convolution layers
The first convolutional layer, \(\operatorname {Conv1}\), applies a convolution operation to the input image X using 32 size 3 x 3 filters, which can be mathematically represented as:
The subsequent convolutional layers, Conv2 through Conv5, progressively increase the number of filters, starting from 32 and reaching 512. Following each convolutional operation, the Rectified Linear Unit (ReLU) activation function is applied to introduce non-linearity and capture essential image features:
The convolutional operations utilize a stride of 1, meaning the filter moves one pixel at a time across the input tensor. This allows the model to preserve the spatial resolution of the input images, which is critical for accurately capturing fine details, such as tumor boundaries, within MRI images. By retaining these details, the model can learn more precise features, leading to better classification performance.
Pooling layers
In tandem with the convolutional layers, max pooling operations are employed after each convolutional layer. Max pooling helps reduce the spatial dimensions of the feature maps, focusing on the most salient information:
Max pooling operations are typically performed with a 2x2 filter and a stride of 2, which halves the dimensions of the feature map, thereby further reducing the computational load while retaining the most critical features. This step is essential in reducing the size of the tensors passed through the network, allowing for more efficient processing without sacrificing the quality of feature extraction.
Fully connected layers
Transitioning to the fully connected layers, the flattened output of the last convolutional layer is fed into the first fully connected layer (FC1). FC1 transforms the flattened tensor into a 1024-dimensional vector and applies the ReLU activation function:
Subsequently, FC2 further compresses the vector into a 512-dimensional representation:
The final fully connected layer (FC3) takes the 512-dimensional vector and produces a 10-dimensional output, where each dimension corresponds to a distinct brain tumor class. This final transformation sets the stage for the ultimate predictions:
Softmax activation function
The softmax activation function is applied to the output of FC3, resulting in a probability distribution across the various classes:
The detailed overview of the Custom CNN architecture including layer specifications, output sizes, kernel sizes, strides, activation functions, and number of layers, is presented in Table 3.
The customized CNN architecture offers several distinct advantages over existing architectures, such as ResNet or VGG, particularly in the context of brain tumor classification using MRI images. The tailored design and careful selection of the number of filters and network depth resulted in a model that not only achieves high accuracy but also exhibits fast inference times and low memory usage.
In this study, we carefully considered the important details, such as input, tensor, kernel size, and the strides used. The convolutional layers use a stride of 1 when preserving spatial resolution is essential. We use the ReLU activation function, chosen for its simplicity and efficiency, throughout the network to introduce non-linearity, enabling the model to capture more complex patterns. These attributes are especially important for deployment in resource-constrained environments, such as IoT devices. The model’s performance, in terms of accuracy and computational efficiency, demonstrates significant advantages over existing architectures, making it a robust and efficient solution for brain tumor classification.
Smart IoT
Smart healthcare monitoring systems in hospitals and various health centers have witnessed significant growth, and the emergence of portable healthcare monitoring systems with advanced technologies has become a global concern. The advent of Internet of Things (IoT) technologies is facilitating a healthcare transformation from traditional face-to-face consultations to telemedicine.
Within the ___domain of brain tumor detection and prediction, this study explores the integration of IoT technologies in healthcare. In this innovative system, a dedicated IoT device, namely a computer, is the central component for brain tumor classification and prediction. This intelligent system harnesses deep learning techniques to automate the intricate process of categorizing brain tumors based on magnetic resonance imaging (MRI) scans.
Components of the IoT-based smart brain tumor classification system
-
1.
IoT Device - Computer:
The computer functions as the core IoT device and acts as an edge computing node in the IoT framework. It connects with hospital databases, cloud systems, or portable imaging devices through secure communication protocols (e.g., Wi-Fi, Bluetooth, or Ethernet). This ensures seamless integration with existing healthcare systems for data retrieval and processing.
-
2.
Data Input - MRI Image Acquisition
MRI images are input into the system via multiple channels:
-
Hospital Systems: MRI images are securely fetched from Picture Archiving and Communication Systems (PACS) or Radiology Information Systems (RIS).
-
Portable MRI Scanners: The system supports wireless data transmission from portable MRI scanners, enabling real-time processing in resource-constrained settings.
-
Direct Upload: Medical staff or users can upload images directly through a connected web interface or external devices like USB drives.
-
-
3.
Integrated Deep Learning Models:
The IoT device is equipped with optimized deep learning models, including a custom CNN and pre-trained models such as Inception-v4 and EfficientNet-B4. These models are converted to lightweight formats (e.g., TensorFlow Lite or PyTorch Mobile) to enable efficient operation on resource-constrained devices.
-
4.
Processing and Analysis:
The system processes input MRI images locally on the IoT device, utilizing the integrated models for feature extraction and classification. This localized processing minimizes latency and eliminates the need for constant connectivity to cloud servers, ensuring real-time decision-making.
-
5.
Diagnostic Results Display:
Classification results, including the predicted tumor type and associated probabilities, are displayed on the IoT device’s user interface (e.g., a web dashboard or connected mobile application). This allows healthcare professionals to access actionable insights promptly.
Prediction phase
The final phase of the proposed methodology involves deploying the trained model for practical use and evaluating its performance on previously unseen brain tumor images. This phase aims to assess how well the trained model can generalize to new, unknown images and accurately classify them into different brain tumor classes.
The flowchart for the prediction process is shown in Fig. 7. To carry out this evaluation, a dataset containing 1000 images representative of various brain tumor classes is employed. These images were not part of the training or validation data, ensuring that the model’s assessment remains unbiased and reflects its true generalization ability. Each image is fed into the deployed model, which processes the image through its learned feature extraction and classification mechanisms. As a result, the brain tumor class label with confidence score is shown with the test image.
Algorithm
The ”Brain Tumor Classification and Prediction Algorithm” as described in Algorithm 1, aims to automate the classification and prediction of brain tumors using a dataset. It takes as input the training dataset, the number of training epochs, model parameters, and a testing dataset. In Step 1, the algorithm begins by preparing itself and setting the initial variables necessary for execution. Step 2 iterates through each file in the dataset, counts images, and processes them through resizing. Moving to Step 3, the algorithm initializes Deep Neural Network (DNN) models such as Inception-v4, EfficientNet-b4, and a custom CNN, utilizing the provided model parameters. Proceeding to Step 4, the algorithm divides the training dataset into training and validation sets using k-fold splitting with an 8:2 ratio. Additionally, in the subsequent Step the algorithm extracts features from images for each epoch and saves them. If successful, it advances to the next step; otherwise, it repeats the preprocessing step. The process of classification and labeling is detailed in Step 6. Here, images are labelled based on the extracted features, determining tumor types. Corresponding labels are saved for future reference. In Step 7, the DNN models are trained using the training data, fitting the data to enhance the model’s performance. Finally, Step 8 covers Testing and Prediction. This step involves predicting tumor classes for images in the testing dataset, calculating confidence scores, and saving the obtained results.
Results and discussion
This section introduces the experimental setup employed for the research, the evaluation parameters used to assess the performance of the proposed models, and the results obtained from these models. Finally, the results are discussed in comparison with multiple techniques.
Implementation platform
The current study for brain tumor classification was implemented on Google Colab64, utilizing a system with an Intel(R) Core(TM) i5-2520M CPU @ 2.50 GHz and 4.00 GB of RAM, running on a 64-bit Windows 10 Pro environment. The model development and evaluation were conducted using the Fastai65 library and the Python programming language. The experiments were carried out on the Jupyter notebook platform within the Colab environment, ensuring a robust setup for the analysis.
Confusion matrix
In this study, the performance analysis was conducted using a confusion matrix66, which is a technique for summarizing the classification algorithm’s performance. True Positive (TP) represents cases where the model correctly predicts the positive class, indicating both the prediction and the actual class are positive. For example, this occurs when the model correctly identifies the presence of a tumor in an MRI image. True Negative (TN) indicates cases where the model correctly predicts the negative class, meaning both the prediction and the actual class are negative. An example of this is when the model correctly identifies the absence of a tumor in an MRI image. False Positive (FP) occurs when the model incorrectly predicts the positive class, meaning the model predicts a tumor is present when, in reality, it is absent. Whereas, the False Negative (FN) happens when the model wrongly predicts the negative class, with the predicted class being negative while the actual class is positive. A case where the model fails to identify the presence of a tumor in the MRI image67.
Evaluation parameters
The performance of the proposed models was evaluated using several commonly used evaluation parameters, including accuracy, precision, F1-score, specificity, error rate, sensitivity, Youden’s index, balanced accuracy, geometric mean, and ROC.
Accuracy (Acc): One of the most widely used metrics for classification performance is accuracy, which is calculated as the ratio of samples that were correctly classified to all samples as follows:
Error rate (Err): The error rate, often known as the misclassification rate, is a companion to the accuracy metric. This metric measures the number of samples from both positive and negative classes that were incorrectly categorized.
Sensitivity (Sens): Sensitivity, also known as the true positive rate or recall, of a classifier measures the proportion of positively classified samples to all positive samples, and it is calculated using Eq. (12).
Specificity (Spec): As shown in Eq. (13), specificity, true negative rate, or inverse recall is calculated as the proportion of correctly identified negative samples to all negative samples.
Precision (Prec): Positive prediction value, or precision, represents the proportion of positive samples that were correctly classified to the total number of positive predicted samples as indicated in Eq. (14)
F1-score (Fs): A balanced metric that incorporates both recall and precision is the F1-score. To assess the model’s overall effectiveness, it considers the harmonic mean of these two metrics. Equation (15) is used to get the f1-score.
Youden’s index (Y_index)): Youden’s Index, or Bookmaker Informedness Metric, is one of the well-known diagnostic tests. It evaluates the discriminative power of the test. The formula of Youden’s index combines the sensitivity and specificity as in the DOR metric, and it is defined as follows:
Balanced Accuracy (BAcc): This metric combines the sensitivity and specificity metrics, and it is calculated as follows:
Geometric Mean (GM): The geometric mean metric aggregates both sensitivity and specificity measures according to Eq. (18)68.
ROC: Combined metric based on the Receiver Operating Characteristic (ROC) space69.
Experimental results with Inception-V4
Table 4 presents the parameters utilized during the training of the Inception-v4 model for brain tumor detection and classification. These parameters were chosen to optimize the training process of the Inception-v4 model and achieve the best possible performance on the brain tumor dataset. The Inception-v4 model had a learning rate of 0.0001, resulting in rapid accuracy gains early in the training process. Around the 12th epoch, the model reached saturation, at which time additional training yielded minimal improvements in accuracy and validation loss, indicating that the model had fully matured.
The confusion matrix for the brain tumor classification using the Inception-v4 model is depicted in Fig. 8. In this confusion matrix, the diagonal values represent each class’s true positive (TP) counts, indicating the number of correctly classified instances. The values off the diagonal represent misclassifications or false positives for each class. The model achieved 368 true positives for astrocytoma, 351 for ependymoma, 362 for germinoma, 446 for glioblastoma, 371 for medulloblastoma, 527 for meningioma, 459 for no tumor, 398 for oligodendroglioma, 547 for pituitary, and 330 for schwannoma. The confusion matrix indicates that the model misclassified only nine images across all classes. This low number of misclassifications demonstrates the robustness and generalization ability of the Inception-v4 model for brain tumor classification. The model’s high true positive rates and minimal misclassification rate indicate its reliability in providing accurate diagnoses, which is critical in medicine.
Table 5 provides a comprehensive overview of the evaluation parameters for the Inception-v4 model’s performance in classifying various brain tumor types. Astrocytoma achieves remarkable results with 100% precision, 98.13% sensitivity, 99.83% accuracy, 99.04% F1-score, 100% specificity, a Youden’s Index of 98.13%, balanced accuracy and G-mean of 99.06%, an ROC score of 99.06%, and a minimal error rate of 0.17%. Ependymoma demonstrates 98.59% precision, perfect 100% sensitivity, 99.88% accuracy, 99.28% F1-score, 99.86% specificity, 99.86% Youden’s Index, 99.93% balanced accuracy, 99.92% G-mean, a 99.93% ROC score, and an error rate of 0.12%. Germinoma, Glioblastoma, Medulloblastoma, No Tumor, and Oligodendroglioma all excel with perfect scores across all metrics, indicating flawless classification. Meningioma achieves 100% precision, 99.62% sensitivity, 99.95% accuracy, 99.8% F1-score, 100% specificity, 99.62% Youden’s Index, 99.81% balanced accuracy, 99.8% G-mean, a 99.81% ROC score, and an error rate of 0.05%. Pituitary classification yields 99.63% precision, perfect 100% sensitivity, 99.95% accuracy, 99.8% F1-score, 99.94% specificity, 99.94% Youden’s Index, 99.97% balanced accuracy, 99.97% G-mean, a 99.97% ROC score, and an error rate of 0.05%. Lastly, Schwannoma achieves 99.39% precision, perfect 100% sensitivity, 99.95% accuracy, 99.68% F1-score, 99.94% specificity, 99.94% Youden’s Index, 99.97% balanced accuracy, 99.97% G-mean, a 99.97% ROC score, and an error rate of 0.05%. These comprehensive metrics affirm the model’s exceptional accuracy and its potential as a valuable tool in medical diagnostics.
Figure 9 shows the training and validation loss during the Inception-v4 model’s training for brain tumor classification. Both losses consistently decrease throughout training, indicating effective learning and improved prediction accuracy. As the number of epochs increases, the loss values steadily decrease. Importantly, the validation loss closely tracks the training loss, suggesting the model avoids overfitting and generalizes well to unseen data, making it reliable for real-world brain tumor image predictions.
Experimental results with efficientNet-b4 model
The parameters used to train the EfficientNet-B4 model for classifying brain tumors are listed in Table 6. With a learning rate of 0.001, the EfficientNet-B4 model was quickly optimized, showing significant improvements in the early epochs. The learning rate reached saturation around the 8th epoch, where accuracy and loss metrics plateaued, indicating that the model had effectively learned from the data and that further training would offer limited benefits.
Figure 10 displays the confusion matrix, presenting the EfficientNet-B4 model’s results for brain tumor classification. The model accurately predicted instances for various tumor types: 335 for astrocytoma, 377 for ependymoma, 364 for germinoma, 464 for glioblastoma, 347 for medulloblastoma, 495 for meningioma, 493 for no tumor, 420 for oligodendroglioma, 502 for pituitary, and 366 for schwannoma. With only five misclassifications, the model showcases high accuracy and efficiency in brain tumor classification.
The EfficientNet-B4 model achieved high precision values, ranging from 99.59% to 100%. Similarly, recall values consistently remained high, ranging from 99.4% to 100%. The model demonstrated an accuracy ranging from 99.92% to 100%, reflecting its exceptional predictive capability. The F1-Score, effectively balancing precision and recall, ranged from 99.54% to 100%. Additionally, the specificity values, ranging from 99.94% to 100%, indicated the model’s accurate identification of true negatives. Youden’s index values ranged from 99.37% to 100%. The model’s balanced accuracy ranged from 99.68% to 100%, while the geometric mean values remained within the range of 99.68% to 100%. ROC values spanned from 99.68% to 100%. The error rate values, ranging from 0.08% to 0.03%, highlighted the minimal misclassification achieved by the model. Table 7 presents the evaluation parameter results for the EfficientNet-B4 model.
In Fig. 11, the EfficientNet-b4 training and validation loss curve is displayed. The graph shows that the model performs well on both the training and validation datasets, as the losses are decreasing over the epochs. Additionally, the gap between the training and validation losses is very small, suggesting that the model is not overfitting severely.
Experimental results with custom CNN model
Table 8 provides a list of the input parameters used to train the Custom CNN model for classifying brain tumors. The optimal learning rate for the Custom CNN model was set at 0.001, allowing for quick convergence with steady improvements in accuracy and F1 score during the first 10 epochs. After this point, the learning rate saturated, and the model achieved stable performance with high accuracy and low error rates, indicating well-balanced training.
The confusion matrix for the Custom CNN for brain tumor classification is shown in Fig. 12. Looking at the confusion matrix, we can see that the model correctly classified 354 instances of astrocytoma, 349 instances of ependymoma, 369 instances of germinoma, 420 instances of glioblastoma, 348 instances of medulloblastoma, 523 instances of meningioma, 460 instances of no tumor, 419 instances of oligodendroglioma, 527 instances of pituitary, and 348 instances of schwannoma. However, there were instances of misclassification observed, signifying potential areas for improvement. Despite this, the model’s overall performance is promising, with high numbers of correctly classified instances for the majority of tumor types.
The evaluation results for the Custom CNN model in brain tumor classification are presented in Table 9, showcasing remarkable performance metrics. The model’s precision values range from 96.39% to 99.61%, while recall values range from 95.08% to 99.76%. With accuracy ranging between 99.25% and 99.90%, the model demonstrates its proficiency in accurate classification. Additionally, high F1 scores (95.72% to 99.60%), specificity values (99.65% to 99.94%), Youden’s Index values (94.73% to 99.65%), and balanced accuracy values (97.36% to 99.82%) further affirm its effectiveness. The geometric mean values fall within 97.33% to 99.82%, and the ROC values range from 97.39% to 99.82%. The model’s exceptional error rate values, ranging from 0.04% to 0.75%, highlight its minimal misclassification rate.
Figure 13 shows the training and validation loss graph for the Custom CNN model during the training process. As the model progresses, we observe a consistent decrease in both the training and validation losses. This loss reduction indicates that the model is learning from the data and improving its performance. A smaller gap between the training and validation losses indicates better generalization.
Hyperparameter table
The table 10 provides a summary of the key hyperparameters for the Inception-v4, EfficientNet-B4, and Custom CNN models. The Inception-v4 model achieved a high training accuracy of 99.56% with a training loss of 0.015, while its testing accuracy was 99.3% with a testing loss of 0.01. Similarly, the EfficientNet-B4 model showed excellent performance, with a training accuracy of 99.76% and a testing accuracy of 99.7%, along with a training loss of 0.004 and a testing loss of 0.0089. The Custom CNN, although slightly less accurate, still performed well with a training accuracy of 97.58% and a testing accuracy of 96.5%, with a training loss of 0.06 and a testing loss of 0.201.
Performance comparison
Figure 14 presents a comparison of the models, Inception-V4, Custom CNN, and EfficientNet-B4, in terms of accuracy for classifying brain tumor types.
Comparison of proposed methods with previous techniques
The results of proposed models for “Meningioma” and “Pituitary” detection are compared against techniques such as GN-SVM, GN-FT, RNGAP, 13-layer CNN, and VGG16. These techniques were applied in70,70,71,72, and73, respectively.
In the case of “No Tumor,” the outcomes of techniques employed by the authors in70,70,44, and74, specifically GN-SVM, GN-FT, DCNN, and NB Classifier, are contrasted with the proposed model results.
Similarly, for “Astrocytoma” and “Oligodendroglioma”, the proposed model results are compared with DCNN, NB Classifier, Ensemble Classifier, and Hybrid Feature Extraction techniques, which were respectively utilized in44,74,75, and76, respectively.
Likewise, the proposed model’s outcomes for “ependymoma” are compared with the outcomes of DCNN, NB Classifier, and Ensemble Classifier techniques utilized in44,74, and75, respectively.
Additionally, the “Glioblastoma” results are compared with the techniques NB Classifier, Ensemble Classifier, and Hybrid Feature Extraction utilized in74,75, and76, respectively.
Lastly, for “Medulloblastoma” the outcomes are compared with those from the ANN and PCA-ANN techniques, as detailed in77. These comparisons are visually depicted in Fig. 15.
As shown in Table 11, the proposed model’s performance is compared with previous studies using different techniques for brain tumor detection and classification.
Testing
The trained models are tested on 1000 test images for the prediction of brain tumor type. The predictions made by the proposed models on the test images show remarkable accuracy and robustness. The primary objective of this analysis was to evaluate the generalizability of our proposed approach across diverse datasets. Figure 16 depicts predictions made by the models for various tumor types.
Testing performance inceptionv-4
Figure 17 illustrates the testing confusion matrix of the Inception-v4 model using 1000 test images. Out of this set of 1000 images, the model accurately predicted the class of 993 images. However, it encountered challenges in precisely predicting the class of 7 images, resulting in minimal misclassifications.
By looking at the matrix, we see that the model’s performance was exceptional, with a 99.3% accuracy rate on the test images. The error rate is calculated at 0.007%. Both the precision and sensitivity rates are reported as 99.88% and 99.33%, respectively, as shown in Table 12. Overall, these findings validate the outstanding performance of the model.
Testing performance EfficientNet-b4
Figure 18 presents the testing confusion matrix of the EfficientNet-B4 model, depicting its performance on 1000 test images. Among these 1000 images, the model accurately predicted the class of 997 images, showcasing its robustness. However, the model faced challenges in precisely predicting the class of three images, resulting in minimal misclassifications. This exceptional performance highlights the effectiveness and reliability of the EfficientNet-B4 model in accurately classifying the test images.
As shown in the testing matrix, the model exhibits exceptional performance, achieving a 99.7% accuracy rate on the test images. The incredibly low error rate of 0.003% signifies the rarity of misclassifications, further emphasizing the model’s precision and sensitivity rates, both of which are 99.88% and 99.77%, respectively. These metrics are also presented in Table 13.
Testing performance custom CNN
Figure 19 displays the testing confusion matrix of a custom CNN model for 1000 test images. Among these images, the model accurately predicted the class of 965 images while encountering challenges in precisely classifying 35 images, resulting in misclassifications.
According to the testing matrix, the model attains an accuracy rate of 96.5% on the test images. Furthermore, the model exhibits a low error rate of 0.035%, indicating its high precision and sensitivity rates of 97.16% and 99.07%, respectively. These results are depicted in Table 14, demonstrating that the model has achieved exceptional performance.
Table 15 provides a comparison of the testing performance of the proposed models. The table contrasts how well the proposed models performed on the test dataset.
Statistical analysis
To provide a comprehensive evaluation of model performance, we calculated effect sizes, confidence intervals (CIs), and p-values. These statistical measures quantify the magnitude, precision, and statistical significance of the observed differences in performance.
Effect size
Effect size92 is a way of quantifying the difference between compared groups. It refers to the magnitude of a result and can be categorized into unstandardized effect sizes (e.g., the difference between group means, relative risk, or odds ratio) or standardized effect sizes (e.g., correlation coefficients or Cohen’s d).
Cohen’s \(d\) is used to compare the mean value of a numerical variable between two groups. It indicates the standardized difference between two means, expressed in standard deviation units. The formula for calculating Cohen’s \(d\) is shown in Eq. (20).
Where:
The interpretation of Cohen’s \(d\) depends on the context of the study, but general guidelines suggest:
-
\(d \le 0.2\): Small effect.
-
\(0.2 < d \le 0.5\): Medium effect.
-
\(d > 0.8\): Large effect.
Confidence interval (CI)
CIs93 provide a range of values within which the true metric is expected to lie, with a specified level of confidence. For accuracy, the CI is calculated using the formula:
Where:
-
\(\hat{p}\): Observed accuracy.
-
\(Z\): Z-score for the desired confidence level (e.g., 1.96 for 95% CI).
-
\(n\): Sample size.
P-value
P-value94 indicate whether observed differences are statistically significant. They are derived using the t-test. A p-value below 0.05 is generally considered statistically significant. The formula for the t-test is:
The results of comparisons among Inception-v4, EfficientNet-B4, and the Custom CNN are summarized in Table 16
The statistical analysis demonstrates that EfficientNet-B4 achieved the highest accuracy (99.36% to 100.04%), reflecting its stability and robustness. Similarly, Inception-v4 performed exceptionally well, with an accuracy range of 98.78% to 99.82% and a large Cohen’s d, signifying its reliability in classification tasks. In comparison, the Custom CNN achieved a reasonable accuracy range (95.36% to 97.64%), though its performance was notably lower than the pre-trained models.
The p-values for all comparisons were significantly below 0.05, indicating highly significant differences between the models. For instance, the p-value of \(7.87 \times 10^{-248}\) in the comparison between Inception-v4 and EfficientNet-B4 highlights the substantial distinction between these two high-performing models. These findings confirm the superiority of pre-trained models like EfficientNet-B4 and Inception-v4 for brain tumor classification tasks due to their higher accuracy and consistency. While the Custom CNN is less accurate, it offers a feasible alternative in computationally constrained environments and demonstrates potential for future optimization.
Comparison of model parameters for IoT suitability
Table 17 compares the number of parameters, model size, and inference time per image for the Inception-v4, EfficientNet-B4, and Custom CNN models, highlighting their suitability for deployment in IoT environments.
As shown in the table, the custom CNN model has 27.79 million parameters, a number strategically positioned between Inception-v4 and EfficientNet-B4. While it does not have the smallest parameter count, it strikes a balance that allows it to retain high performance while being less computationally expensive than Inception-v4. This makes it a strong candidate for IoT applications, where resources such as processing power and memory are often limited.
One of the standout features of the custom CNN is its inference time of 63.43 milliseconds per image. This is considerably faster than both Inception-v4 (1422.9 ms) and EfficientNet-B4 (180.6 ms). In IoT settings, where real-time data processing is often required, the custom CNN’s faster inference time is a significant advantage. It ensures that the model can quickly analyze and classify images, making it suitable for time-sensitive applications such as medical diagnostics.
The custom CNN has a model size of 318 MB, which, while larger than EfficientNet-B4, is still significantly smaller than Inception-v4. This moderate model size is beneficial for deployment on IoT devices, which may have constraints in terms of available storage. The custom CNN model size allows it to be deployed on a wide range of IoT devices without sacrificing performance.
Conclusions and future work
This study presents a novel approach for IoT-based smart automatic classification and prediction of brain tumor using deep learning techniques. The primary contribution of this research is the development of a custom CNN model and the utilization of a pre-trained model to achieve high accuracy in classifying various types of brain tumors from MRI images.
Accurate diagnosis and classification of brain tumors are important in medical practice, given the potential for variations in tumor types and imaging quality. Traditional methods can be labor-intensive and subject to human error. This research addresses these challenges by employing specialized models to enhance the accuracy and efficiency of tumor classification. Each model brings unique strengths to table, providing a comprehensive assessment of their performance in a variety of scenarios.
The experimental results highlight the effectiveness of each model individually. The Custom CNN model achieved a testing accuracy of 96.5%, demonstrating its capability to effectively distinguish between different tumor types and non-tumor cases. The Inception-v4 and EfficientNet-B4 models exhibited even higher accuracies of 99.3% and 99.7%, respectively, underscoring their superior performance in identifying brain tumors with minimal error rates.
These results are significant when compared to previous techniques. The Custom CNN model balances performance with computational efficiency, while pre-trained models such as Inception-v4 and EfficientNet-B4 offer exceptional accuracy. The findings affirm the potential of these models for integration into clinical workflows, aiding radiologists and medical professionals in making informed decisions.
The high accuracy and robustness of the models contribute significantly to medical diagnostics. By providing accurate and rapid classification of brain tumors, this approach supports early detection and diagnosis, which is crucial for improving patient outcomes. The efficiency of the models, especially in an IoT setting, enhances their applicability in clinical environments where quick and reliable analysis is essential.
Limitations
Despite the promising results, several limitations must be considered. The models were trained and tested on datasets from Kaggle, which, while comprehensive, may not fully capture the diversity of brain tumor cases encountered in real-world clinical settings. Variations in MRI imaging protocols and patient demographics could impact model performance. Additionally, while the models demonstrate high accuracy on the test datasets, their generalization to other medical imaging systems or different MRI scan types, such as those with varying resolutions or contrast settings, may be limited. Further validation on diverse datasets is needed to ensure the robustness of the models across various clinical scenarios.
Future work
There are several opportunities for further improvement in this research. Future work can explore integrating additional data sources, employing advanced preprocessing techniques, or developing more sophisticated model architectures to enhance performance. Expanding the scope to include a wider range of brain-related disorders such as Alzheimer’s disease, Parkinson’s disease, epilepsy, stroke, and brain infections will also be a key focus. This extended approach aims to create a comprehensive diagnostic tool that supports a broader array of neurological conditions, ultimately contributing to more effective management and treatment.
Data availability
The datasets used in this research are public datasets that are available on mentioned sources; https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri, https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset, https://www.kaggle.com/datasets/waseemnagahhenes/brain-tumor-for-14-classes, https://www.kaggle.com/datasets/fernando2rad/brain-tumor-mri-images-44c?sort=published, https://www.kaggle.com/datasets/adityakomaravolu/brain-tumor-mri-images.
Change history
26 May 2025
The original online version of this Article was revised: In the original version of this Article an incorrect email address for corresponding author Rana Othman Alnashwan was quoted. Correspondence and requests for materials should be addressed to [email protected]
References
National Institute of Neurological Disorders and Stroke (NINDS). Brain basics: Know your brain. https://www.ninds.nih.gov/health-information/public-education/brain-basics/brain-basics-know-your-brain, 2023. Accessed on 28 July 2023.
Latif, Ghazanfar, Brahim, Ghassen Ben, Awang Iskandar, D. N. F., Bashar, Abul & Alghazo, Jaafar. Glioma tumors’ classification using deep-neural-network-based features with svm classifier. Diagnostics 12(4), 1018 (2022).
Senan, Ebrahim Mohammed, Jadhav, Mukti E, Rassem, Taha H, Aljaloud, Abdulaziz Salamah, Mohammed, Badiea Abdulkarem, Al-Mekhlafi, Zeyad Ghaleb, et al. Early diagnosis of brain tumour mri images using hybrid techniques between deep and machine learning. Computational and Mathematical Methods in Medicine, 2022, (2022).
American Association of Neurological Surgeons (AANS). Brain tumors. https://www.aans.org/en/Patients/Neurosurgical-Conditions-and-Treatments/Brain-Tumors, n.d. Accessed on 28 July (2023).
Cleveland Clinic. Brain cancer and brain tumor. https://my.clevelandclinic.org/health/diseases/6149-brain-cancer-brain-tumor, 2022. Accessed on 28 July (2023).
Healthline. Understanding brain tumors. https://www.healthline.com/health/brain-tumor#benign-vs-malignant, 2022. Accessed on 30 July (2023).
American Society of Clinical Oncology (ASCO). Brain tumor: Introduction. https://www.cancer.net/cancer-types/brain-tumor/introduction, 2023. Accessed on 30 July (2023).
Max Roser. Death rate from cancer (2019).
American Society of Clinical Oncology (ASCO). Brain tumor statistics. https://www.cancer.net/cancer-types/brain-tumor/statistics, 2023. Accessed on 30 July (2023).
Ali, Abid et al. Multilevel central trust management approach for task scheduling on iot-based mobile cloud computing. Sensors 22(1), 108 (2021).
Srivastava, Jyoti, Routray, Sidheswar, Ahmad, Sultan & Waris, Mohammad. Internet of medical things (iomt)-based smart healthcare system: Trends and progress. Computational Intelligence and Neuroscience 1–17(07), 2022 (2022).
Sarker, Iqbal H. Deep learning: a comprehensive overview on techniques, taxonomy, applications and research directions. SN Computer Science 2(6), 420 (2021).
Allaoua Chelloug, Samia, Alkanhel, Reem, Saleh Ali Muthanna, Mohammed, Aziz, Ahmed & Muthanna, Ammar. Multinet: A multi-agent drl and efficientnet assisted framework for 3d plant leaf disease identification and severity quantification. volume 11, pages 86770–86789. IEEE, (2023).
Amin, Syed Umar & Shamim Hossain, M. Edge intelligence and internet of things in healthcare: A survey. Ieee Access 9, 45–59 (2020).
Gazis A. What is iot? the internet of things explained. Academia Letters, 2, (2021).
TIFFANY YEUNG. What is edge ai and how does it work? https://blogs.nvidia.com/blog/2022/02/17/what-is-edge-ai/, 2022. Accessed on 30 July (2023).
Jamil, Faisal & Hameed, Ibrahim A. Toward intelligent open-ended questions evaluation based on predictive optimization. Expert Systems with Applications, page 120640, (2023).
Jamil, Harun, Qayyum, Faiza, Iqbal, Naeem, Jamil, Faisal & Kim, Do Hyeun. Optimal ensemble scheme for human activity recognition and floor detection based on automl and weighted soft voting using smartphone sensors. IEEE Sensors Journal 23(3), 2878–2890 (2022).
Awais, Muhammad et al. Deep learning based enhanced secure emergency video streaming approach by leveraging blockchain technology for vehicular adhoc 5g networks. Journal of Cloud Computing 13(1), 130 (2024).
Jamil, Faisal, Ahmad, Shabir, Whangbo, Taeg Keun, Muthanna, Ammar & Kim, Do-Hyeun. Improving blockchain performance in clinical trials using intelligent optimal transaction traffic control mechanism in smart healthcare applications. Computers & Industrial Engineering 170, 108327 (2022).
Ahmad, Shabir et al. Design of a general complex problem-solving architecture based on task management and predictive optimization. International Journal of Distributed Sensor Networks 18(6), 15501329221107868 (2022).
Qayyum, Faiza, Jamil, Faisal, Ahmad, Shabir & Kim, Do-Hyeun. Hybrid renewable energy resources management for optimal energy operation in nano-grid. Comput. Mater. Contin 71, 2091–2105 (2022).
Jamil, Faisal, Qayyum, Faiza, Alhelaly, Soha, Javed, Farjeel & Muthanna, Ammar: Intelligent microservice based on blockchain for healthcare applications. Computers, Materials & Continua, 69(2), (2021).
Jamil, Faisal & Kim, DoHyeun. Enhanced kalman filter algorithm using fuzzy inference for improving position estimation in indoor navigation. Journal of Intelligent & Fuzzy Systems 40(5), 8991–9005 (2021).
Rathi, Vipin Kumar et al. An edge ai-enabled iot healthcare monitoring system for smart cities. Computers & Electrical Engineering 96, 107524 (2021).
Mahjoubi, Mohamed Amine, Hamida, Soufiane, El Gannour, Oussama, Cherradi, Bouchaib, El Abbassi, Ahmed & Raihani, Abdelhadi. Improved multiclass brain tumor detection using convolutional neural networks and magnetic resonance imaging. International Journal of Advanced Computer Science and Applications, 14(3), (2023).
Abd, Basant S., El-Wahab, Mohamed E., Nasr, Salah Khamis & Ashour, Amira S. Btc-fcnn: Fast convolution neural network for multi-class brain tumor classification. Health Information Science and Systems 11(1), 3 (2023).
Irmak, Emrah. Multi-classification of brain tumor mri images using deep convolutional neural network with fully optimized framework. Iranian Journal of Science and Technology, Transactions of Electrical Engineering 45(3), 1015–1036 (2021).
Khan, Abdul Hannan et al. Intelligent model for brain tumor identification using deep learning. Applied Computational Intelligence and Soft Computing 2022, 1–10 (2022).
Qureshi, Shahzad Ahmad et al. Intelligent ultra-light deep learning model for multi-class brain tumor detection. Applied Sciences 12(8), 3715 (2022).
Latif, Ghazanfar, Brahim, Ghassen Ben, Awang Iskandar, D. N. F., Bashar, Abul & Alghazo, Jaafar. Glioma tumors’ classification using deep-neural-network-based features with svm classifier. Diagnostics 12(4), 1018 (2022).
Mahmud, Md Ishtyaq, Mamun, Muntasir & Abdelgawad, Ahmed. A deep analysis of brain tumor detection from mr images using deep learning networks. Algorithms 16(4), 176 (2023).
Latif, Ghazanfar, Butt, M Mohsin, Khan, Adil H, Butt, Omair & Iskandar, DNF Awang. Multiclass brain glioma tumor classification using block-based 3d wavelet features of mr images. In 2017 4th International Conference on Electrical and Electronic Engineering (ICEEE), pages 333–337. IEEE, (2017).
Ayadi, Wadhah, Charfi, Imen, Elhamzi, Wajdi & Atri, Mohamed. Brain tumor classification based on hybrid approach. The Visual Computer 38(1), 107–117 (2022).
Mohsen, Heba, El-Dahshan, El-Sayed A., El-Horbaty, El-Sayed M. & Salem, Abdel-Badeeh M. Classification using deep learning neural networks for brain tumors. Future Computing and Informatics Journal 3(1), 68–71 (2018).
Abiwinanda, Nyoman, Hanif, Muhammad, Hesaputra, S Tafwida, Handayani, Astri, Mengko & Tati Rajab. Brain tumor classification using convolutional neural network. In World Congress on Medical Physics and Biomedical Engineering 2018: June 3-8, 2018, Prague, Czech Republic (Vol. 1), pages 183–189. Springer, (2019).
Singh, Vaibhav, Sharma, Sarthak, Goel, Shubham, Lamba, Shivay & Garg, Neetu. Brain tumor prediction by binary classification using vgg-16. Smart and Sustainable Intelligent Systems, pages 127–138, (2021).
Swati, Zar Nawab Khan. et al. Brain tumor classification for mr images using transfer learning and fine-tuning. Computerized Medical Imaging and Graphics 75, 34–46 (2019).
Sharma, Sarang, Gupta, Sheifali, Gupta, Deepali, Juneja, Abhinav, Khatter, Harsh, Malik, Sapna, Bitsue, Zelalem Kiros, et al. Deep learning model for automatic classification and prediction of brain tumor. Journal of Sensors, 2022, (2022).
Santos, Dheiver & Santos, Ewerton. Brain tumor detection using deep learning. medRxiv, pages 2022–01, (2022).
Devi, Nilakshi & Bhattacharyya, Kaustubh. Automatic brain tumor detection and classification of grades of astrocytoma. In Proceedings of the International Conference on Computing and Communication Systems: I3CS 2016, NEHU, Shillong, India, pages 125–135. Springer, (2018).
Li, Mengmeng et al. Ependymoma and pilocytic astrocytoma: Differentiation using radiomics approach based on machine learning. Journal of Clinical Neuroscience 78, 175–180 (2020).
Tamije Selvy, P., Dharani, V. P. & Indhuja, A. Brain tumour detection using deep learning techniques. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol 169, 175 (2019).
Rahmat, Romi Fadillah et al. Astrocytoma, ependymoma, and oligodendroglioma classification with deep convolutional neural network. IAES International Journal of Artificial Intelligence 11(4), 1306 (2022).
Garg, Ginni & Garg, Ritu. Brain tumor detection and classification based on hybrid ensemble classifier. arXiv preprint arXiv:2101.00216, (2021).
Thejaswini, P., Bhat, Ms Bhavya & Prakash, Mr Kushal. Detection and classification of tumour in brain mri. Int. J. Eng. Manufact. (IJEM) 9(1), 11–20 (2019).
Machiraju, Jaya Lakshmi, Rao, S Nagaraja. Brain tumor magnetic resonance image classification: A deep learning approach. (2021).
Tazin, Tahia, Sarker, Sraboni, Gupta, Punit, Ayaz, Fozayel Ibn, Islam, Sumaia, Monirujjaman Khan, Mohammad, Bourouis, Sami, Idris, Sahar Ahmed, Alshazly, Hammam, et al. A robust and novel approach for brain tumor classification using convolutional neural network. Computational Intelligence and Neuroscience, 2021, (2021).
Afshar, Parnian, Mohammadi, Arash, Plataniotis, Konstantinos N. Brain tumor type classification via capsule networks. In 2018 25th IEEE international conference on image processing (ICIP), pages 3129–3133. IEEE, (2018).
El-Feshawy, Somaya A., Saad, Waleed, Shokair, Mona & Dessouky, Moawad. Iot framework for brain tumor detection based on optimized modified resnet 18 (omres). The Journal of Supercomputing 79(1), 1081–1110 (2023).
Haq, Amin ul et al. Dacbt: Deep learning approach for classification of brain tumors using mri data in iot healthcare environment. Scientific Reports 12(1), 15331 (2022).
Deepak, S. & Ameer, P. M. Brain tumor classification using deep cnn features via transfer learning. Computers in biology and medicine 111, 103345 (2019).
Deeksha, KN, Deeksha, M, Girish, Anagha V, Bhat, Anusha S & Lakshmi, H. Classification of brain tumor and its types using convolutional neural network. In 2020 IEEE International Conference for Innovation in Technology (INOCON), pages 1–6. IEEE, (2020).
Ganesan, M., Sivakumar, N. & Thirumaran, M. Internet of medical things with cloud-based e-health services for brain tumour detection model using deep convolution neural network. Electronic Government, an International Journal 16(1–2), 69–83 (2020).
Bhuvaji, Sartaj. Brain tumor classification mri. https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri, 2020. 5 March (2023).
Nickparvar, Masoud. Brain tumor mri dataset. https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset, 2021. 5 March (2023).
WASEIM NAGAH HENNES. Brain tumor for 14 classes. https://www.kaggle.com/datasets/waseemnagahhenes/brain-tumor-for-14-classes, 2023. 1 July (2023).
FELTRIN, FERNANDO. Brain tumor mri images 44 classes. https://www.kaggle.com/datasets/fernando2rad/brain-tumor-mri-images-44c?sort=published, 2023. 1 July (2023).
KOMARAVOLU, ADITYA. Brain tumor mri images. https://www.kaggle.com/datasets/adityakomaravolu/brain-tumor-mri-images, 2022. 1 July (2023).
Analytics Vidhya. Understanding transfer learning for deep learning. https://www.analyticsvidhya.com/blog/2021/10/understanding-transfer-learning-for-deep-learning/, 2021. Accessed on 4 Aug (2023).
Huang, Zhihao, Lumei, Su., Jiajun, Wu. & Chen, Yuhan. Rock image classification based on efficientnet and triplet attention mechanism. Applied Sciences 13(5), 3180 (2023).
Tan, Mingxing & Le, Quoc. Efficientnet: Rethinking model scaling for convolutional neural networks. In International conference on machine learning, pages 6105–6114. PMLR, (2019).
Szegedy, Christian, Ioffe, Sergey, Vanhoucke, Vincent & Alemi, Alexander. Inception-v4, inception-resnet and the impact of residual connections on learning. In Proceedings of the AAAI conference on artificial intelligence, volume 31, (2017).
Akbar, Habib et al. Detecting rotational symmetry in polar ___domain based on sift. IEEE Access 11, 68643–68652 (2023).
Hassan, Mahmood Ul et al. Smart resource allocation in mobile cloud next-generation network (ngn) orchestration with context-aware data and machine learning for the cost optimization of microservice applications. Sensors 24(3), 865 (2024).
Brownlee, Jason. What is a confusion matrix in machine learning. https://machinelearningmastery.com/confusion-matrix-machine-learning/, 2020. Accessed on 5 Aug (2023).
Jayaswal, Vaibhav: Performance metrics: Confusion matrix, precision, recall, and f1 score. https://towardsdatascience.com/performance-metrics-confusion-matrix-precision-recall-and-f1-score-a8fe076a2262, 2020. Accessed on 5 Aug (2023).
Tharwat, Alaa. Classification assessment methods. Applied computing and informatics 17(1), 168–192 (2020).
Luque, Amalia, Gómez-Bellido, Jesús, Carrasco, Alejandro & Barbancho, Julio. Optimal representation of anuran call spectrum in environmental monitoring systems using wireless sensor networks. Sensors 18(6), 1803 (2018).
Rasool, Mohammed et al. A hybrid deep learning model for brain tumour classification. Entropy 24(6), 799 (2022).
Lokesh Kumar, R., Jagadeesh Kakarla, B., Isunuri, Venkateswarlu & Singh, Munesh. Multi-class brain tumor classification using residual network and global average pooling. Multimedia Tools and Applications 80, 13429–13438 (2021).
Kibriya, Hareem, Masood, Momina, Nawaz, Marriam & Nazir, Tahira. Multiclass classification of brain tumors using a novel cnn architecture. Multimedia Tools and Applications 81(21), 29847–29863 (2022).
Malla, Prince Priya, Sahu, Sudhakar & Alutaibi, Ahmed I. Classification of tumor in brain mr images using deep convolutional neural network and global average pooling. Processes 11(3), 679 (2023).
Gupta, Nidhi, Bhatele, Pushpraj & Khanna, Pritee. Identification of gliomas from brain mri through adaptive segmentation and run length of centralized patterns. Journal of Computational Science 25, 213–220 (2018).
Gupta, Nidhi, Bhatele, Pushpraj & Khanna, Pritee. Glioma detection on brain mris using texture and morphological features with ensemble learning. Biomedical Signal Processing and Control 47, 115–125 (2019).
Kirti Raj Bhatele and Sarita Singh Bhadauria. Multiclass classification of central nervous system brain tumor types based on proposed hybrid texture feature extraction methods and ensemble learning. Multimedia Tools and Applications 82(3), 3831–3858 (2023).
Sachdeva, Jainy, Kumar, Vinod, Gupta, Indra, Khandelwal, Niranjan & Ahuja, Chirag Kamal. Segmentation, feature extraction, and multiclass brain tumor classification. Journal of digital imaging 26, 1141–1150 (2013).
Ari, Ali & Hanbay, Davut. Deep learning based brain tumor classification and detection system. Turkish Journal of Electrical Engineering and Computer Sciences 26(5), 2275–2286 (2018).
Alsaif, Haitham et al. A novel data augmentation-based brain tumor detection using convolutional neural network. Applied Sciences 12(8), 3773 (2022).
Khan, Hassan Ali, Jue, Wu, Mushtaq, Muhammad & Mushtaq, Muhammad Umer. Brain tumor classification in mri image using convolutional neural network. Mathematical Biosciences and Engineering, (2021).
Seetha, J. & Selvakumar Raja, S. Brain tumor classification using convolutional neural networks. Biomedical & Pharmacology Journal 11(3), 1457 (2018).
Modiya, Priyanka & Vahora, Safvan. Brain tumor detection using transfer learning with dimensionality reduction method. International Journal of Intelligent Systems and Applications in Engineering 10(2), 201–206 (2022).
Ramtekkar, Praveen Kumar, Pandey, Anjana & Pawar, Mahesh Kumar. Accurate detection of brain tumor using optimized feature selection based on deep learning techniques. Multimedia Tools and Applications, pages 1–31, (2023).
Sajjad, Muhammad et al. Multi-grade brain tumor classification using deep cnn with extensive data augmentation. Journal of computational science 30, 174–182 (2019).
Nishat, Mirza Muntasir, Faisal, Fahim, Hasan, Tasnimul, Karim, Md Faiyed Bin, Islam, Zahidul & Shagor, Md Rafid Kaysar. An investigative approach to employ support vector classifier as a potential detector of brain cancer from mri dataset. In 2021 International Conference on Electronics, Communications and Information Technology (ICECIT), pages 1–4. IEEE, (2021).
Balasooriya, Narmada M & Nawarathna, Ruwan D. A sophisticated convolutional neural network model for brain tumor classification. In 2017 IEEE international conference on industrial and information systems (ICIIS), pages 1–5. IEEE, (2017).
Polly, FP, Shil, SK, Hossain, Mohammad Arif, Ayman, A & Jang, Yeong Min. Detection and classification of hgg and lgg brain tumor using machine learning. In 2018 International Conference on Information Networking (ICOIN), pages 813–817. IEEE, (2018).
Khan, Muneeb A. & Park, Heemin. A convolutional block base architecture for multiclass brain tumor detection using magnetic resonance imaging. Electronics 13(2), 364 (2024).
Mohammad Zafer Khaliki and Muhammet Sinan Başarslan. Brain tumor detection from images and comparison with transfer learning methods and 3-layer cnn. Scientific Reports 14(1), 2664 (2024).
Huda, Md Najmul, Olowolayemo, Akeem, Adeleye, Ayesha Dupe, Rashid, Amin Nur & Haque, Abrar Habib. Brain tumor classification using vanilla convolutional neural networks. International Journal on Perceptive and Cognitive Computing 10(2), 80–86 (2024).
Anantharajan, Shenbagarajan, Gunasekaran, Shenbagalakshmi, Subramanian, Thavasi & Venkatesh, R. Mri brain tumor detection using deep learning and machine learning approaches. Measurement: Sensors 31, 101026 (2024).
Aarts, Sil, Van Den Akker, Marjan & Winkens, Bjorn. The importance of effect sizes. The European Journal of General Practice 20(1), 61–64 (2014).
Foody, Giles M. Classification accuracy comparison: Hypothesis tests and the use of confidence intervals in evaluations of difference, equivalence and non-inferiority. Remote Sensing of Environment 113(8), 1658–1663 (2009).
Rochowicz Jr, John A. P-value approximations for t-tests of hypothesis. Spreadsheets Educ.(eJSiE), 5(5), (2012).
Acknowledgements
We are grateful to the Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2025R239), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Funding
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R239), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization, QUAI, RB, AA and YS; methodology, FJ, ROA, and MSAM; software, SAC; validation, ROA, FJ and AA; formal analysis, RB; investigation, YS; resources, RB; data curation, ROA; writing—original draft preparation, QUAI and AA; writing—review and editing, FJ, SAC; visualization, MSAM; supervision, RB, and QUA; project administration, YS; funding acquisition, RB, SAC, MSAM, and FJ.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ishfaq, Q.U.A., Bibi, R., Ali, A. et al. Automatic smart brain tumor classification and prediction system using deep learning. Sci Rep 15, 14876 (2025). https://doi.org/10.1038/s41598-025-95803-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-95803-3