Abstract
We aimed to investigate the genomic and tumor microenvironmental (TME) profiles in non-muscle invasive bladder cancer (NMIBC) and explore potential predictive markers for Bacillus Calmette–Guérin (BCG) treatment response in high-risk NMIBC patients (according to European Association of Urology (EAU) risk stratification). 40 patients with high-risk NMIBC (cTis-T1N0M0) who underwent en bloc resection followed by BCG instillation were retrospectively enrolled. Surgical samples were subjected to Next Generation Sequencing (NGS) and multiplex immunofluorescence (mIF) assay. Genomic profiling revealed high prevalences of alterations in TERT (55%), KDM6A (32.5%), FGFR3(30%), PIK3CA (30%), TP53(27.5%) and ARID1A (20%). TME analysis showed different proportions of macrophages, NK cells, T cells subsets in tumoral and stromal compartment. Multivariate analysis identified TERT C228T and alteration in KDM6A as two independent factors associated with inferior RFS. The study comprehensively depicted the genomic and TME profiles in NMIBC and identified potential predictive biomarkers for BCG treatment.
Similar content being viewed by others
Introduction
Bladder cancer (BCa) is the seventh most common malignant tumor in men worldwide, and it ranks tenth when both genders are considered1. In 2012, there were an estimated 430,000 newly diagnosed cases and 165,000 bladder cancer deaths2, and these numbers changed to 573,278 and 212,536 in 2020, which indicated a growing disease burden3.
Non-muscle invasive bladder cancer (NMIBC) accounts for 75% of bladder cancer cases. Tumor resection followed by subsequent adjuvant intravesical treatment is the standard treatment for intermediate- and high-risk NMIBC1. The complete resection can be performed by either traditional transurethral resection of bladder tumor (TURBT) or en-bloc resection of bladder tumor (ERBT). Compared to TURBT, ERBT can achieve complete tumor excision while minimizing the risk of leaving behind residual cancer cells or disrupting the tumor boundaries, thus reducing the risk of tumor implantation, bleeding, obturator nerve reflex, and bladder perforation. Besides, ERBT provides high-quality resected specimens and can preserve the original structure and local microenvironment of BCa1,4.
Intravesical bacillus Calmette-Guérin (BCG) immunotherapy is the most common adjuvant intravesical treatment after tumor resection. BCG is a live attenuated form of Mycobacterium bovis, which can induce innate and adaptive immune responses to prevent BCa recurrence and progression after resection5. A meta-analysis has reported that compared with intravesical chemotherapy, BCG could improve the recurrence-free rate from 26% to 47% (odds ratio, 0.41 [95% CI, 0.30–0.56]) and the progression-free rate from 20% to 15% (OR, 0.74 [95% CI, 0.45–1.22]) in patients with carcinoma in situ (CIS)6. Another meta-analysis also reported BCG maintenance treatment could lead to a 32% reduction in the risk of BCa recurrence compared to mitomycin C (MMC)7. However, approximately 42% of the NMIBC patients will still experience recurrence, and 14% will experience progression within five years after BCG treatment8,9. Therefore, it is essential to find out predictive factors associated with recurrence after BCG treatment.
Many biomarkers have been proven to be associated with recurrence after BCG treatment. Telomerase reverse transcriptase gene promoter (TERTp) mutations are considered the most common mutations in BCa. And Batista et al. found c.1-146 G > A TERTp mutation was an independent predictor of nonrecurrence after BCG treatment (HR, 0.382 [95% CI, 0.150–0.971])10. Fibroblast growth factor receptor alterations (FGFRa) also occur frequently in patients with NMIBC11,12. However, studies have found FGFRa was not significantly associated with RFS and PFS in high-grade NMIBC patients with or without BCG treatment13. Since BCG acts mainly by activating immunity, the heterogeneity of immune cells in the tumor microenvironment (TME) is tightly associated with the efficacy of BCG treatment. Strandgaard et al. found T cell exhaustion, especially CD8 + T cell exhaustion, significantly correlated with a high risk of post-BCG high-grade recurrence9. Tumor-associated macrophages (TAMs) might also play a vital role in the response to BCG. Suriano et al. found M2 macrophages infiltration could be a prognostic predictor in patients with NMIBC after BCG treatment14.
Next generation sequencing (NGS) is very popular nowadays and has been applied in NMIBC to identify the latent biomarkers in European cohorts15. However, no previous studies have comprehensively investigated the genomic profile of NMIBC patients in the Chinese cohort to predict the efficacy of BCG treatment, and most studies used samples acquired by fractioned resection for NGS, which could possibly influence the intrinsic character and TME of the original tumors. Therefore, in this study, using NGS and multiplex immunofluorescence (mIF) assays, we sought to investigate genomic and TME profiles in NMIBC and explore potential predictive markers for BCG treatment following en-block resection.
Methods
Patients and samples
The research was performed with the approval of the Ethics Committee of the Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology (Grant number: TJ-IRB2021S055), and was conducted with the approval of China’s human genetic resources in international cooperation scientific research (Reception number: 2021SLGH2769). This is a retrospective study waiving informed consent and was conducted in accordance with the Declaration of Helsinki.
A total of 40 patients who were diagnosed with high-risk NMIBC and underwent en-bloc resection followed by BCG instillation were retrospectively enrolled. The inclusion criteria were: (1) Diagnostic confirmation of high-risk NMIBC (cTis-T1N0M0) by a professional genitourinary pathologist according to the EAU risk stratification. (2) Received ERBT. (3) Received a scheduled BCG intravesical instillation. (4) With good organ function. (5) With an ECOG score of 0–2. Patients accepting adjuvant intravesical chemotherapy instillations such as mitomycin or receiving other adjuvant perioperative therapies were excluded. The bladder tumor tissues were fixed by 4% paraformaldehyde and embedded by paraffin immediately after resection. The patients received regular follow-up based on NCCN guidelines (bladder cancer 2020.v6) and were divided into two groups according to bladder cancer recurrence.
DNA extraction and DNA library construction
Tissue DNA was isolated utilizing the QIAamp DNA formalin-fixed paraffin-embedded (FFPE) tissue kit from Qiagen (Valencia, CA) according to the manufacturer’s protocol. The concentration of DNA derived from the tumor tissue was determined using the Qubit 2.0 Fluorometer and the Qubit double-stranded DNA assay kit from Life Technologies (Carlsbad, CA). A minimum of 50 ng of DNA was employed for the preparation of the NGS library. DNA fragmentation was accomplished using the Covaris M220 focused ultrasonicator (Covaris, Inc., Woburn, MA), followed by procedures such as end repair, phosphorylation, dA addition, and adaptor ligation to construct the library. Subsequently, the DNA library was purified using Agencourt AMPure beads from Beckman Coulter (Fullerton, CA).
Capture-based targeted sequencing
Capture-based targeted sequencing was conducted on samples at a Clinical Laboratory Improvement Amendments-certified, College of American Pathology (CAP)-accredited laboratory. A panel including 520 genes spanning 1.67 M of human genomic regions (OncoScreenPlus, Burning Rock Biotech, Guangzhou, China) was employed for the analysis16. Indexed samples were subjected to sequencing on a Nextseq500 platform (Illumina, Inc., San Diego, CA) with paired-end reads, achieving an average sequencing depth of 958.26X. The sequencing data underwent analysis using a custom bioinformatics pipeline designed to detect various types of genomic alterations, including single-nucleotide variants, small insertions and deletions, large genomic rearrangements (LGRs), copy number variants (CNVs), and genomic fusions. Raw sequencing data were initially preprocessed using bcl2fastq. The preprocessed data were then aligned to the human genome (hg19) using Burrows-Wheeler Aligner 0.7.10, resulting in the generation of BAM files. Variant calling was performed using Vardict to identify single-nucleotide variants, insertions, and deletions. ANNOVAR and SnpEff v3.6 were utilized for variant annotation. The maximum allele frequencies of the genomic alterations were calculated. Genomic fusions were identified and their abundance determined using Factera. CNVs and LGRs were estimated based on an in-house algorithm previously described17,18 that takes into account the sequencing depth. We calculated the tumor mutation burden (TMB) using the total number of mutations, except for synonymous mutations, which was also performed by Burning Rock.
Multiplex immunofluorescence (mIF) assays
Serial FFPE slides were subjected to multiplex immunofluorescence (mIF) assays to visualize tumor-infiltrating lymphocytes (TILs) using the PANO 7-plex IHC kit (Panovue, Beijing, China). The mIF protocol involved sequential application of primary antibodies against cell differentiation markers, including CD3, CD8, CD56, CD68, CD163, PD-1, and PD-L1. After primary antibody incubation, horseradish peroxidase-conjugated secondary antibodies and tyramide signal amplification were employed. Nuclei were stained with 4′-6′-diamidino-2-phenylindole (DAPI) following the labeling of all antigens.Two consecutive FFPE sections derived from the same tissue sample were used for mIF analysis. One section was stained with CD3 (indigo), CD8 (red), PD-1 (green), PD-L1 (yellow), panCK (purple), and DAPI (blue), while the other section was stained with CD56 (yellow), CD163 (red), CD68 (green), panCK (purple), and DAPI (blue). The stained slides were scanned using the Mantra System (PerkinElmer, Waltham, Massachusetts), which captures fluorescent spectra at 20-nm wavelength intervals ranging from 420 to 720 nm. Subsequently, image analysis was conducted using inform image analysis software (PerkinElmer, Waltham, Massachusetts)19. TILs present within tumor cell nests and in the tumor stroma were quantified separately.
Statistical analysis
The study employed various statistical methods to analyze and compare data between two groups. Categorical variables were assessed using either the Chi-square test or Fisher’s exact test. For continuous variables, the Wilcoxon rank-sum test or Mann–Whitney U-test was utilized. The Spearman correlation coefficient was employed to examine the association between continuous variables. The hazard ratios (HR) with corresponding 95% confidence intervals (CI) were calculated using Cox’s proportional hazards model. Statistical significance was determined by a p value threshold of less than 0.05.
Furthermore, the relationship between TILs and survival outcomes was investigated using Kaplan–Meier curves and the Log-rank test. All statistical analyses were performed using R 4.0.5 (https://cran.r-project.org/src/base/R-4/), a widely-used free software environment for statistical computing and graphical analysis provided by The R Project for Statistical Computing (https://www.r-project.org/).
Results
The clinical and molecular characteristics of the patients included in this study
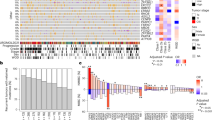
As shown in Fig. 1A, between June 2016 and September 2020, a total of 40 patients with high-risk NMIBC were retrospectively enrolled. The patients received regular BCG instillation treatment about one month after ERBT. The basic characteristics of these patients were shown in Table 1 and Supplementary Data 1. The cohort had a median age of 63 years, and 80% were male. After a median follow-up of 21.8 months, 19 patients recurred, which did not reach the median recurrence-free survival (RFS) (Fig. 1B). The 3-months, 6-months, 9-months and one-year RFS were 92.5%, 75%, 62.5%, and 57.5%, respectively, which means that with longer follow-up, the recurrence rate essentially ceases to change, and short-term recurrence-free patients have a relatively long RFS rate. All tumors were microsatellite stable and showed a median TMB of 7.98 muts/Mb, which was situated in the middle of the pan-cancer spectrum.
Genomic landscape of NMIBC in small Chinese cohort
Genomic profiling revealed a high prevalence of alterations in TERT (55%), KDM6A (32.5%), KMT2D (32.5%), FGFR3(30%), PIK3CA (30%), TP53(27.5%), KMT2C (25%), and ARID1A (20%) (Fig. 2A, Supplementary Fig. 1A). The most frequently altered genes in small Chinese cohort were consistent with those in European cohorts15. There existed no significant differential genes between recurrence and non-recurrence patients. Two FGFR3 fusions were identified in this cohort, which were both FGFR3-TACC3 fusions. And one intergenic (C8orf86, RNF5P1)-FGFR1 was also TERT. As for KDM6A, the loss of function (LoF) variant was most commonly identified. Then we analyzed the associated molecular pathways of these altered genes, and we found these mutated genes primarily located in five signaling pathways: TP53-RB1-pathway, RTK-RAS-PI3K-pathway, Epigenetic Modifiers pathway, SWI/SNF complex pathway and DNA damage repair (DDR) pathway. Alterations in TP53-RB1-pathway and RTK-RAS-PI3K-pathway were present in 70% (28/40) and 72.5% (29/40) of NMIBC tumors, which were associated with cell cycle regulation and tumor proliferation, respectively. Alterations in the Epigenetic Modifiers pathway, SWI/SNF complex pathway were present in 67.5% (27/40) and 27.5% (11/40) of NMIBC tumors, which were chromosome remodeling-related pathways. The DDR pathway happened in 60% of these patients, which was connected with DNA damage repair.
A An overview of the genomic landscape of NMIBC in small Chinese cohort. B Correlations of genomic alterations between these genes. C The percentage of patients with EP300 mutations and wildtype in different genders (female and male). D The percentage of patients with FGFR3 mutations and wildtype in different genders (female and male). E The percentage of patients with PIK3CA mutations and wildtype in different tumor grades (low, intermediate and high grade).
Next, we aimed to explore the co-occurrence and mutually exclusive patterns of genomic alterations in NMIBC (Fig. 2B). We found that TP53 mutation was mutually exclusive with FGFR3/TERT alteration and co-mutated with RB1. Alterations in 11q13.3 region (CCND1, FGF3, FGF4) significantly co-occurred with ERCC2 mutations. Co-occurrence patterns were also identified in ARID1A and ERCC2 mutations, KDM6A and APC mutations, and CDKN2A and PTPRD, CDKN2B, and MTAP mutations.
Then we investigated the association between genetic alterations and clinical characteristics. We discovered that EP300 (Fig. 2C) and FGFR3 (Fig. 2D) alterations were relatively enriched in female patients, which indicated that targeted therapies focused on these targets might be more effective in female patients. We also found that PIK3CA was relatively enriched in patients with low-grade disease, which might indicate a better prognosis (Fig. 2E). However, the number of patients in this subgroup was relatively limited, and large cohort validation was necessary.
The exploration of multidimensional biomarkers of the efficacy of postoperative BCG treatment
In our cohort, we didn’t find a statistically significant difference in TMB between recurrence and nonrecurrence subgroups (Fig. 3A). Then, we would like to explore whether TMB was connected with recurrence after BCG treatment. When setting different cut-off values, we divided the patients into high-TMB and low-TMB two subgroups (Supplementary Data 2). However, there still lacked of statistical significance when we tried different cut-off value, which was consistent with the previous results20, and the p value was minimum when the cut off value was 2. Although lacking statistical significance, patients with low TMB seemed to have better relapse-free survival than those with high TMB (HR = 2.78, 95% CI: 0.91–8.46, p = 0.0602; Fig. 3B). But, the number of patients in the low TMB subgroups was limited, so the results needed to be interpreted with caution. Previous studies also found ARID1A mutations could significantly increase the risk of recurrence after BCG treatment in European cohorts20,21. However, in our study, we found that ARID1A mutations were not associated with relapse-free survival after BCG treatment (HR = 0.68, 95% CI: 0.2–2.32, p = 0.5331; Fig. 3C). Besides, ARID1A protein truncation also had no interaction with the relapse-free survival after BCG treatment (HR = 1.66, 95% CI: 0.38–7.2, p = 0.494; Fig. 3D). Although there existed a trend in the interaction between RFS after BCG treatment and ARID1B mutation (HR = 2.64, 95% CI: 0.87–8, p = 0.077; Fig. 3E), the sample size was too small to reach statistical significance.
A Comparison of TMB between patients in the recurrence and non-recurrence groups. B Survival analyses for patients in high and low TMB groups using Kaplan–Meier curves. C Survival analyses for patients in ARID1A mutations and wildtype groups using Kaplan–Meier curves. D Survival analyses for patients with ARID1A truncation and without ARID1A truncation using Kaplan–Meier curves. E Survival analyses for patients in ARID1B mutations and wildtype groups using Kaplan–Meier curves. F Survival analyses for patients in TERT mutations and wildtype groups using Kaplan–Meier curves. G Survival analyses for patients in TERT promoter mutations and wildtype groups using Kaplan–Meier curves. H Survival analyses for patients with TERTp C250T and without TERTp C250T using Kaplan–Meier curves. I Survival analyses for patients with TERTp C228T and without TERTp C228T using Kaplan–Meier curves. J Survival analyses for patients in KDM6A mutations and wildtype groups using Kaplan–Meier curves. K Survival analyses for patients in low and high HRD score groups using Kaplan–Meier curves.
TERT promoter (TERTp) mutation has been considered one of the most common mutations in bladder cancer. Previous studies have found that TERTp mutation was an independent predictor of BCG therapy success and significantly associated with recurrence after BCG therapy10,22. In our cohort, we found both TERT mutation (HR = 2.27, 95% CI: 0.86–5.99, p = 0.0874; Fig. 3F) and TERTp mutation (HR = 2.21, 95% CI: 0.87–5.64, p = 0.0864; Fig. 3G) were not significantly associated with recurrence after BCG therapy, but it seemed patients without TERT or TERTp mutation had relatively better RFS. Then, we explored the relationship between recurrence and two common mutation types in the upstream of TERTp, TERT C250T and TERT C228T. Although TERT C250T was not significantly connected with recurrence after BCG therapy (HR = 0.49, 95% CI: 0.07–3.69, p = 0.4817; Fig. 3H), TERT C228T was significantly associated with worse RFS (HR = 2.72, 95% CI: 1.05–7.04, p = 0.0242; Fig. 3I), which was consistent with previous reports23.
Mutations in lysine-specific demethylase 6 A (KDM6A) were very common in several cancer types, including bladder cancer24. Previous studies have found KDM6A was tightly connected with poor prognosis of bladder cancer patients24, but was not an independent predictor for recurrence after BCG therapy15. In our study, we found KDM6A mutation was significantly associated with worse RFS after BCG therapy (HR = 2.51, 95% CI: 1.01–6.21, p = 0.0395; Fig. 3J).
Homologous recombination deficiency score (HRD score) was proven to have the ability to predict the response of platinum-containing neoadjuvant chemotherapy in patients with BRCA1/2 nonmutated triple-negative breast cancers25. Therefore, we would like to investigate whether the HRD score could predict the efficacy of BCG immunotherapy in bladder cancer. Interestingly, in our cohort, we found that a high HDR score was significantly associated with worse RFS after BCG therapy (HR = 2.55, 95% CI: 1.01–6.41, p = 0.0394; Fig. 3K).
Then, we used univariate Cox regression to evaluate the relationship between genomic alterations and recurrence after BCG therapy, and the results with statistical significance were shown in Table 2. NF2 was mutated in only 3 patients, all in the KDM6A mutation group, and PTPN11 was also mutated in only 3 patients, and all in the TERT C228T mutation group. Considering the correlation of these mutations, these two genomic alterations were excluded in the subsequent multivariate Cox regression analysis. We also evaluated the relationship between several clinical factors and recurrence after BCG therapy, such as tumor stage, gender, age, tumor percentage, and TMB. We found these factors were all not significantly associated with RFS after BCG therapy (Table 3). Then, we performed multivariate Cox regression analyses and included factors with statistical significance in univariate Cox regression analysis, including HRD score, TERT C228T mutation, and KDM6A mutation (Fig. 4A). And we found TERT C228T mutation and KDM6A mutation were independent predictors for recurrence after BCG therapy, respectively. When we divided these patients into four subgroups according to the mutation status of TERT C228T and KDM6A, we found the survival curves of TERT C228T and KDM6A single mutation subgroups almost overlapped. Therefore, we divided all patients into three subgroups: co-mutation, single mutation, and wild type. The clinical characteristics of patients in different subgroups were shown in Table 4. We found patients in the co-mutation subgroup had the worst RFS, and patients in the wild-type subgroup had the best RFS (Fig. 4B).
A Hazard ratio of HRD score, TERTp C228T and KDM6A mutation after multivariate Cox regression analysis (*p < 0.05). B Survival analyses for comparison between wild type and mutation groups (left) and comparison between co-mutation and single mutation groups (right) using Kaplan–Meier curves. The mutation groups showed significantly worse relapse free survival than wild type groups (p = 0.0027). C The percentage of patients with PIK3CA mutations and wildtype in different groups (wild type, single mutation and co-mutation). D Comparison of tumor purity between patients in different groups (wild type, single mutation and co-mutation). E Comparison of ploidy between patients in different groups (wild type, single mutation and co-mutation). F Comparison of HRD score between patients in different groups (wild type, single mutation and co-mutation).
Following this, we would like to explore the differences in molecular characteristics in different subgroups. We found patients with PIK3CA mutation were more centralized in a single mutation subgroup, which might be associated with co-mutation with KDM6A (Fig. 4C). Patients in the co-mutation subgroup had the highest tumor purity, whilst those in the wild-type subgroup had the least, although lacking statistical significance, the trend was significant (Fig. 4D). The ploidy and HRD scores in the co-mutation subgroup were significantly higher than the other two subgroups, which indicated a higher level of chromosome instability (Fig. 4E, F).
The landscape of tumor microenvironment in bladder cancer
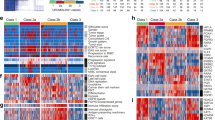
Next, using multiplex immunofluorescence (mIF) assays, we would like to investigate the landscape of TME of patients in our cohort. Except for tumor cells, there existed many immune cells in the TME, such as macrophages, T cells, and NK cells. NK cells were defined as a cluster of CD3-CD56+ cells and could be divided into CD56dim and CD56bright two subgroups according to the CD56 density on the cell surface26. According to the expression of CD68 and CD163, TAMs could be divided into M1 (CD68 + CD163-) and M2 (C68 + CD163 + ) subtypes27. TILs were a group of intratumor lymphocytes and were marked as CD3+ cells28. According to the results of mIF assays, we found the immune cells infiltrating the parenchymal area of the tumor were predominantly M1 macrophages, followed by TILs and CD56dim NK cells (Fig. 5A, B). The immune infiltrating cells in the stroma area of the tumor were predominantly TILs cells, followed by M1 macrophages and CD56dim NK cells (Fig. 5C, D). In terms of spatial distribution, TILs, CD8 + T cells, CD56bright NK cells, M2 macrophages, and exhaustion T cells (Tex) infiltrated significantly more in the stroma area. The number of M1 macrophages and CD56dim NK cells were comparable in the stroma and parenchymal areas of the tumor, suggesting that the distribution of immune cells was spatially heterogeneous in bladder cancer (Fig. 5E). Next, we investigated the correlation between different immune cells in the parenchymal and stroma areas of the tumor (Fig. 5F). We found the correlations of M2 macrophages, CD56dim NK cells, and Tex cells in the parenchymal and stroma areas of the tumor were relatively high (r = 0.759, 0.786, 0.778, respectively), but the correlation coefficients of other immune cells were low, which indicated that the ability to penetrate from the stroma to the parenchymal area of the tumor for different immune cells was different. Further analyses revealed that TILs in the parenchymal region of the tumor significantly correlated with RFS (Fig. 5G), and CD56bright NK cells in the parenchymal region of the tumor as well as macrophages in the stroma region of the tumor also correlated with RFS although lacking of statistical significance (Fig. 5H, I). In conclusion, these results illustrated the correlation between tumor immune infiltration and RFS in NMIBC patients treated with BCG instillation.
A Relative abundance of different immune cell types in the parenchymal of tumor in different patients. B The percentage of different immune cell types in the parenchymal of tumor. C Relative abundance of different immune cell types in the stroma of tumor in different patients. D The percentage of different immune cell types in the stroma of tumor. E Comparison of the percentage of different markers in the stroma and tumor area. F Correlations of different immune cells between stroma and tumor area. G Survival analyses for high and low CD3+ cells infiltration in the tumor area using Kaplan–Meier curves. H Survival analyses for high and low CD56bright NK cells infiltration in the tumor area using Kaplan–Meier curves. I Survival analyses for high and low CD68+ cells infiltration in the stroma area using Kaplan–Meier curves.
Furthermore, we would like to investigate the relationship between immune cells in the TME and two mutations that were proved to affect RFS after BCG therapy previously. We found the TME of patients in the KDM6A WT subgroup was characterized by two features and can be categorized into two groups. In the parenchymal area of the tumor, one group was characterized by relatively high infiltration of CD8 + T cells, and the other one was characterized by relatively high infiltration of M1 macrophages and NK cells (Fig. 6A, B). But, in KDM6A mutation subgroup, the M1 macrophages and NK cells rarely infiltrated in the parenchymal area of the tumor. In the stroma area of the tumor, NK cells were more enriched in the KDM6A WT group compared to the mutation group. All these results could partially explain the reason why patients in the KDM6A WT group had better RFS after BCG therapy. Similarly, in the stroma area of the tumor, we also found NK cells, M1 macrophages, and TILs were more enriched in the TERT C228T WT group compared to the mutation group. Meanwhile, the density of PD1 + CD3+ as well as PD1 + CD8 + T cells was lower in the TERT C228T WT group than that in the mutation group (Fig. 6A, B).
A Heatmap of different immune cells infiltration in patients grouped by KDM6A mutations and TERTp C228T in the tumor area. B Heatmap of different immune cells infiltration in patients grouped by KDM6A mutations and TERTp C228T in the stroma area. C Multiplex immunofluorescence images of immune cell infiltration in patient numbered RS21083210FFP.
Finally, we would like to report a patient with TERT C228T and KDM6A co-mutation. The patient was 67 years old when he was diagnosed with bladder cancer and received an ERBT. Postoperative pathology showed the tumor was T1 stage and high grade, and the tumor percentage was 90%. The patient’s risk classification is high, and he started to receive BCG instillation one month after the surgery. He received a total of 7 times BCG treatment, and the BCG treatment lasted for 84 days until he was confirmed recurrence by cystoscopy and ultrasonography. He was microsatellite stable and was grouped into a high TMB and high HRD score group. Except for TERT C228T and KDM6A co-mutation, he also had TP53 and RB1 mutations. From his stained slide, we could find the TILs primarily concentrated in the stroma area of the tumor, especially CD8 + T cells (Fig. 6C). And the proportion of CD3 + PD1+ and CD8 + PD1+ cells was relatively high in this patient. Besides, the NK cells and M1 macrophages were rare in the parenchymal area of the tumor (Fig. 6C). The multiplex immunofluorescence images and clinical characteristics of the other two patients were showed in Supplementary Fig. 2 and Supplementary Fig. 3, respectively.
Discussion
BCG immunotherapy has become a standard treatment for NMIBC for over 40 years. However, there still exists no specific biomarkers to predict its efficacy. Although many previous studies have used the NGS method to explore the molecular landscape in NMIBC and the association between molecular heterogeneity and different clinical outcomes, to the best of our knowledge, this is the first study to investigate genomic biomarkers associated with the efficacy of BCG treatment in the Chinese cohort. Here in our study, we included 40 patients with high-risk NMIBC who received regular BCG instillation treatment after ERBT. Using the NGS technique, we figured out the genomic profile of these patients with NMIBC in a small Chinese cohort and then correlated it with several clinical characteristics of them. Next, we investigated biomarkers associated with the RFS after BCG treatment from the genome level. Finally, we also explored the landscape of immune cell infiltration in TME in these patients using mIF assay, and tried to find out the correlation between TME and therapeutic efficacy of BCG.
Specifically, we found high-frequency mutated genes in this small cohort of Chinese patients with NMIBC were similar to those of the European population. Nowadays, the gold standard for BCa diagnosis and surveillance is cystoscopy, which is invasive and painful. Therefore, non-invasive screening methods such as liquid biopsy based on urine have attracted more and more attention. So, it is more and more important to discover appropriate and novel biomarkers for the detection of BCa. It has been demonstrated that the telomerase activity was enhanced in most cancer types, and the mutations in the promoter of TERT were most common29. Previous studies have reported TERT promoter mutations were very frequent in BCa, occurring in 60-80% cases of all stages and grades of BCa30, and two single nucleotide substitutions C228T and C250T, were the most common TERT promoter mutations types in BCa, accounting for nearly 99% of all TERT promoter mutations cases in BCa. In our study, we found the most frequent mutation in NMIBC was TERT, and TERT promoter mutations were the most common mutation type. Because of its high frequency and cancer specificity, TERT promoter mutations (C228T and C250T) have been previously used for detection in the DNA of urinary exfoliated tumor cells to monitor the recurrence and predict the prognosis. Allory et al. found the sensitivity and specificity of using TERT mutations in urine samples to detect NMIBC recurrence or progression was 42% and 73%, respectively31. Some studies also investigated the relationship between TERTp mutations and the efficacy of BCG treatment. Batista et al. found TERTp mutation c.1-146 G > A was an independent factor in predicting the efficacy of BCG treatment, and patients with TERTp mutation c.1-146 G > A had significantly longer RFS than noncarriers10. Besides, studies have found TERT mutations were very early events in BCa tumorigenesis and highly conserved during BCG treatment21. Here in our study, we found TERTp C228T was significantly associated with worse RFS after BCG treatment in the Chinese cohort. To sum up, although having not been recommended both by the European Association of Urology (EAU) and American Urological Association (AUA) guidelines considering the cost and performance, TERTp mutations, especially TERTp C228T, would be a potential biomarker for diagnosis and prognosis prediction in BCa in Chinese population in the future. We will continue our research to validate the feasibility and effectiveness of TERTp C228T as a biomarker for predicting BCG treatment response.
FGFR3 genomic alterations were potent oncogenic drivers in BCa, and over 50% of BCa cases have genomic alterations in the FGFR3 coding sequence. FGFR3-target therapies have begun to conduct early clinical trials. FGFR3-target therapies are comprised mainly of tyrosine kinase inhibitors (TKIs), which could be divided into two classes according to their mechanism of action: multi-targeting TKIs and selective TKIs. The former could target many other receptors except for FGFR3, including dovitinib and derazantinib, and the latter had a specifical affinity for FGFRs kinase ___domain, including erdafitinib and rogaratinib. In 2019, erdafitinib was granted approval by the FDA and used for treating patients with cisplatin-refractory FGFR2/FGFR3-altered bladder cancer. Although the future looks bright, there still exist many challenges, including drug resistance, target selectivity, and treatment-related adverse events32. Drug combination and patient selection might be practical solutions for these concerns. In our study, we found a rare FGFR1 fusion in a patient with a high-grade tumor, which could be applied in the development of TKIs in the future. We also found FGFR3 mutations were more frequent in female patients, which might provide some evidence to support the use of TKI therapy in female bladder cancer patients.
ARID1A alterations was common in BCa and often occurred in FGFR3 wildtype, poor-prognosis BCa33. Previous studies have found that ARID1A mutations were significantly associated with worse RFS after BCG treatment in the European cohort15,21. ARID1B were mutually exclusive isoforms with ARID1A, and ARID1B mutations were less frequently reported compared with ARID1A mutations. Wang et al. have found high expression of ARID1B was significantly associated with worse prognosis in patients with BCa, and patients with low expression of ARID1B could benefit more from adjuvant chemotherapy34. In our study, we found that ARID1B mutations rather than ARID1A mutations were more connected with the RFS after BCG treatment in the Chinese population, but the sample size was limited and further studies were needed.
Previous studies have established many risk stratification criteria based on transcriptomic profiling. They stratified patients into several molecular subtypes to predict the response to treatments. For example, Hedgaard et al. have divided NMIBC patients into three major classes with basal- and luminal-like characteristics and different clinical outcomes using a 17-gene classifier35. However, these molecular subtypes mainly reflected the intrinsic characteristics of tumor cells but not the signals from tumor surroundings and cell-cell interactions. BCG functioned mainly by activating the immune system rather than killing the tumor cells directly. Thus it was essential to assess the landscape of immune cell infiltration and inflammation in the TME to predict the efficacy of BCG. Damrauer et al. have established an immune infiltration signature as a predictive biomarker for BCG response, and they have found a high immune score was connected with better RFS after BCG treatment36. In this study, using mIF assay, we found significant differences in the types, numbers, and spatial distribution of immune cells. Immune cells infiltrating in the tumor parenchymal area were predominantly M1 macrophages and CD56dim NK cells. TILs, CD8 + T cells, CD56bright NK cells, M2 macrophages, and Tex cells were significantly more infiltrated in the stroma area. The correlation between the stroma and parenchymal areas of the tumor was higher for M2 macrophages, CD56dim NK cells, and Tex cells, which indicated the ability to penetrate into parenchymal areas of the tumor of the different immune cells was different. Further analysis revealed that TILs in the tumor parenchymal region were significantly correlated with RFS, and CD56dim NK cells, as well as macrophages in the tumor mesenchymal region were also correlated with RFS, although lacking statistical significance. Lymphocytes were the first immune cell subset which were studied and considered to play a vital role in the BCG mediated anti-tumor immune activation. They can exert anti-tumor function by both killing tumors directly or activating innate immunity by releasing several cytokines37. The number of T cells were found to elevate in the urine and bladder mucosa in patients after BCG treatment, especially CD4 + T cells. The depletion of CD4 + T cells or CD8 + T cells could both influence the efficacy of BCG treatment in mouse models38. Studies also found BCG could not only increase the quantity of CD4 + T cells but also increase the CD4/FoxP3 ratio in the TME. However, the transcriptome profile of CD4 + T cells after BCG treatment seemed the same as those already in the TME, which suggested BCG could only recruit immune cells, but their characters were mainly dependent on the original TME39.
NK cells also played a significant role in BCG mediated antitumor immunity37. According to the density of CD56 markers on the membrane, NK cells can be divided into two subsets, including CD56dim NK cells and CD56bright NK cells. CD56dim NK cells accounted for more than 90% of the NK cells population, which were mainly cytotoxic and expressed IL-2 receptor (IL-2R) with moderate affinity and stronger killing activity. And CD56dim NK cells produced a large number of cytokines, which were considered immature, mainly played an immunomodulatory role and highly expressed IL-2R26. In our study, we found CD56dim NK cells possessed greater penetration capabilities and accessed the tumor parenchymal region more easily compared to CD56bright NK cells, but CD56bright NK cells were more connected with the RFS after BCG treatment. Previous studies have reported the population of CD56bright NK cells in the peripheral blood mononuclear cells would increase after exposure to BCG. However, these BCG-activated CD56bright NK cells retained many characteristics of mature CD56dim NK cells. They could fight against tumor cells directly by degranulation and mediating antibody-dependent cell-mediated cytotoxicity. These unique subpopulation of NK cells could be only observed in patients treated with BCG but not those treated with mitomycin C. Combined with our findings, we hypothesized that these special CD56bright NK cells played the crucial role in BCG-mediated antitumor immunity and influenced the patients’ prognosis40.
Macrophages were a bridge between innate immunity and adaptive immunity, which could not only kill and phagocytose pathogens and tumor cells, but also present antigens, secrete cytokines, and activate T cells. Macrophages could be simply grouped into M1 and M2, two subtypes according to their functions and characteristics. M1 macrophages had proinflammatory characteristics, and could defend tumor by producing reactive oxygen/nitrogen species and pro-inflammatory cytokines, which could recruit and activate other immune cells. However, M2 macrophages mainly suppressed immunity and contributed to the migration and invasion of tumors. Studies have found higher frequency of TAMs infiltration was associated with poor prognosis in various cancers41. Previous studies demonstrated higher TAM counts were connected with worse RFS and increased risk of BCG failure42,43. Furthermore, Lima et al. found high infiltration of CD163+ macrophages (M2 subtype), especially in the stroma area but not in the tumor parenchymal region, could predict a worse prognosis after BCG treatment44. In our study, consistently, we also found M2 macrophages were more concentrated in the stroma area and more infiltration of macrophages in the stroma area of tumor was connected with worse RFS after BCG treatment in patients with NMIBC. However, the specific mechanisms need to be further explored in the future.
As we have mentioned above, BCG might only recruit immune cells into the TME, but the characteristics of these newly recruited immune cells mainly depended on the original TME. Therefore, we next explored the interaction of the molecular landscape of tumors and the infiltration of immune cells. We found significant associations between KDM6A and TERT mutations and the infiltration of immune cells. Jie Yin et al. found KDM6A mutations could regulate NK cells survival and activity in mice without any tumors45. Studies also found inhibition and knockdown of KDM6A reduced NK cell proliferation and IFN-γ production46. Enhancer of Zeste Homolog 2 (EZH2), which could be counteracted by KDM6A and was often overexpressed in muscle-invasive bladder cancer, was associated with a worse prognosis. Ramakrishnan et al. found LoF mutations in KDM6A could make patients with muscle-invasive bladder cancer more susceptible to EZH2 inhibition therapy and activate the NK cells at the same time47. Therefore, genomic alterations might influence the patients’ prognosis not only by influencing the innate characteristics of tumors, but also by remodeling the immune microenvironment.
There still existed some limitations in this study. First of all, the number of patients was relatively small, especially in some subgroups when subgroup analyses were performed, so some conclusions needed to be interpreted with caution. Besides, this is a single-center study, and muti-center study with more cohorts was needed in the future to get more valid conclusions. Although mIF assays could discriminate different kinds of immune cells crudely, more detailed categorization should be acquired by more advanced methods such as single cell sequencing. Finally, the follow up period was relatively short, and some patients interrupted BCG treatment owing to COVID-19 epidemic. These limitations should be carefully considered and overcome in future studies.
In conclusion, this study comprehensively depicted the genomic and TME profiles in NMIBC and identified potential predictive biomarkers for BCG treatment. Although the sample size is currently small and not sufficient to draw strong conclusions, we will continue our efforts to conduct more in-depth research. Our goal is to translate our findings into clinical applications that facilitate the stratification of patients and better guide clinical decision-making in the management of NMIBC.
Data availability
The data generated in this study are available upon request from the corresponding author.
References
Babjuk, M. et al. European Association of Urology Guidelines on Non-muscle-invasive Bladder Cancer (Ta, T1, and Carcinoma in Situ). Eur. Urol. 81, 75–94 (2022).
Antoni, S. et al. Bladder cancer incidence and mortality: a global overview and recent trends. Eur. Urol. 71, 96–108 (2017).
Lobo, N. et al. Epidemiology, screening, and prevention of bladder cancer. Eur. Urol. Oncol. 5, 628–639 (2022).
Li, Z., Zhou, Z., Cui, Y. & Zhang, Y. Systematic review and meta-analysis of randomized controlled trials of perioperative outcomes and prognosis of transurethral en-bloc resection vs. conventional transurethral resection for non-muscle-invasive bladder cancer. Int. J. Surg. 104, 106777 (2022).
Lenis, A. T., Lec, P. M., Chamie, K. & Mshs, M. D. Bladder cancer: a review. Jama 324, 1980–1991 (2020).
Sylvester, R. J., van der Meijden, A. P., Witjes, J. A. & Kurth, K. Bacillus calmette-guerin versus chemotherapy for the intravesical treatment of patients with carcinoma in situ of the bladder: a meta-analysis of the published results of randomized clinical trials. J. Urol. 174, 86–91 (2005).
Malmström, P. U. et al. An individual patient data meta-analysis of the long-term outcome of randomised studies comparing intravesical mitomycin C versus bacillus Calmette-Guérin for non-muscle-invasive bladder cancer. Eur. Urol. 56, 247–256 (2009).
Chamie, K. et al. Recurrence of high-risk bladder cancer: a population-based analysis. Cancer 119, 3219–3227 (2013).
Strandgaard, T. et al. Elevated T-cell exhaustion and urinary tumor DNA levels are associated with Bacillus Calmette-Guérin Failure in patients with non-muscle-invasive bladder cancer. Eur. Urol. 82, 646–656 (2022).
Batista, R. et al. TERT promoter mutation as a potential predictive biomarker in BCG-Treated bladder cancer patients. Int. J. Mol. Sci. 21, 947 (2020).
Facchinetti, F. et al. Resistance to selective FGFR inhibitors in FGFR-driven urothelial cancer. Cancer Discov. 13, 1998–2011 (2023).
Guercio, B. J. et al. Clinical and genomic landscape of FGFR3-altered urothelial carcinoma and treatment outcomes with Erdafitinib: a real-world experience. Clin. Cancer Res. 29, 4586–4595 (2023).
Mayr, R. et al. Prognostic and predictive value of fibroblast growth factor receptor alterations in high-grade non-muscle-invasive bladder cancer treated with and without Bacillus Calmette-Guérin Immunotherapy. Eur. Urol. 81, 606–614 (2022).
Suriano, F. et al. Tumor associated macrophages polarization dictates the efficacy of BCG instillation in non-muscle invasive urothelial bladder cancer. J. Exp. Clin. Cancer Res. 32, 87 (2013).
Pietzak, E. J. et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur. Urol. 72, 952–959 (2017).
Chen, X. et al. Blood tumor mutation burden can predict the clinical response to immune checkpoint inhibitors in advanced non-small cell lung cancer patients. Cancer Immunol. Immunother. 70, 3513–3524 (2021).
Liu, Z. et al. HER2 copy number as predictor of disease-free survival in HER2-positive resectable gastric adenocarcinoma. J. Cancer Res. Clin. Oncol. 147, 1315–1324 (2021).
Wu, D. et al. The landscape of kinase ___domain duplication in Chinese lung cancer patients. Ann. Transl. Med. 8, 1642 (2020).
Stack, E. C., Wang, C., Roman, K. A. & Hoyt, C. C. Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70, 46–58 (2014).
Katoh, M. & Nakagama, H. FGF receptors: cancer biology and therapeutics. Med. Res. Rev. 34, 280–300 (2014).
Bacon, J. V. W. et al. Somatic features of response and relapse in non-muscle-invasive bladder cancer treated with Bacillus Calmette-guérin immunotherapy. Eur. Urol. Oncol. 5, 677–686 (2022).
Rachakonda, P. S. et al. TERT promoter mutations in bladder cancer affect patient survival and disease recurrence through modification by a common polymorphism. Proc. Natl Acad. Sci. USA 110, 17426–17431 (2013).
Hayashi, Y. et al. TERT C228T mutation in non-malignant bladder urothelium is associated with intravesical recurrence for patients with non-muscle invasive bladder cancer. Mol. Oncol. 14, 2375–2383 (2020).
Chen, X. et al. Significance of KDM6A mutation in bladder cancer immune escape. BMC Cancer 21, 635 (2021).
Telli, M. L. et al. Homologous Recombination Deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin. Cancer Res. 22, 3764–3773 (2016).
Morvan, M. G. & Lanier, L. L. NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer 16, 7–19 (2016).
Chávez-Galán, L., Olleros, M. L., Vesin, D. & Garcia, I. Much More than M1 and M2 Macrophages, There are also CD169(+) and TCR(+) Macrophages. Front. Immunol. 6, 263 (2015).
Zhao, Y. et al. Tumor Infiltrating Lymphocyte (TIL) Therapy for solid tumor treatment: progressions and challenges. Cancers 14, 4160 (2022).
Leão, R. et al. Mechanisms of human telomerase reverse transcriptase (hTERT) regulation: clinical impacts in cancer. J. Biomed. Sci. 25, 22 (2018).
Zvereva, M. et al. Activating Telomerase TERT promoter mutations and their application for the detection of bladder cancer. Int. J. Mol. Sci. 21, 6034 (2020).
Allory, Y. et al. Telomerase reverse transcriptase promoter mutations in bladder cancer: high frequency across stages, detection in urine, and lack of association with outcome. Eur. Urol. 65, 360–366 (2014).
Ascione, C. M. et al. Role of FGFR3 in bladder cancer: treatment landscape and future challenges. Cancer Treat. Rev. 115, 102530 (2023).
Conde, M. & Frew, I. J. Therapeutic significance of ARID1A mutation in bladder cancer. Neoplasia 31, 100814 (2022).
Wang, B. et al. Expression of ARID1B is associated with poor outcomes and predicts the benefit from adjuvant chemotherapy in bladder urothelial carcinoma. J. Cancer 8, 3490–3497 (2017).
Hedegaard, J. et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 30, 27–42 (2016).
Damrauer, J. S. et al. Identification of a novel inflamed tumor microenvironment signature as a predictive biomarker of Bacillus Calmette-Guérin immunotherapy in non-muscle-invasive bladder cancer. Clin. Cancer Res. 27, 4599–4609 (2021).
Redelman-Sidi, G., Glickman, M. S. & Bochner, B. H. The mechanism of action of BCG therapy for bladder cancer-a current perspective. Nat. Rev. Urol. 11, 153–162 (2014).
Ratliff, T. L., Ritchey, J. K., Yuan, J. J., Andriole, G. L. & Catalona, W. J. T-cell subsets required for intravesical BCG immunotherapy for bladder cancer. J. Urol. 150, 1018–1023 (1993).
Kates, M. et al. Intravesical BCG Induces CD4(+) T-Cell expansion in an immune competent model of bladder cancer. Cancer Immunol. Res. 5, 594–603 (2017).
García-Cuesta, E. M. et al. Characterization of a human anti-tumoral NK cell population expanded after BCG treatment of leukocytes. Oncoimmunology 6, e1293212 (2017).
Sharifi, L. et al. A review on the role of M2 macrophages in bladder cancer; pathophysiology and targeting. Int. Immunopharmacol. 76, 105880 (2019).
Ajili, F., Kourda, N., Darouiche, A., Chebil, M. & Boubaker, S. Prognostic value of tumor-associated macrophages count in human non-muscle-invasive bladder cancer treated by BCG immunotherapy. Ultrastruct. Pathol. 37, 56–61 (2013).
Takayama, H. et al. Increased infiltration of tumor associated macrophages is associated with poor prognosis of bladder carcinoma in situ after intravesical bacillus Calmette-Guerin instillation. J. Urol. 181, 1894–1900 (2009).
Lima, L. et al. The predominance of M2-polarized macrophages in the stroma of low-hypoxic bladder tumors is associated with BCG immunotherapy failure. Urol. Oncol., 32, 449–457 (2014).
Yin, J. et al. Ezh2 regulates differentiation and function of natural killer cells through histone methyltransferase activity. Proc. Natl Acad. Sci. USA 112, 15988–15993 (2015).
Cribbs, A. et al. Inhibition of histone H3K27 demethylases selectively modulates inflammatory phenotypes of natural killer cells. J. Biol. Chem. 293, 2422–2437 (2018).
Ramakrishnan, S. et al. Inhibition of EZH2 induces NK cell-mediated differentiation and death in muscle-invasive bladder cancer. Cell Death Differ. 26, 2100–2114 (2019).
Acknowledgements
This research is supported by grants from Hubei Urology Minimally Invasive Therapy Clinical Medical Center (SCZ2023015 and SCZ2023017) and grants from Natural Science Foundation of Hubei Province (ZRMS2020002466). Authors would like to thank the patients who participated in our study. We also appreciate BioRender software (https://biorender.com) for helping to create Fig. 1A.
Author information
Authors and Affiliations
Contributions
Qi-dong Xia, Jian-Xuan Sun, and Zhi-Peng Yao collected samples, enrolled the patients and recorded clinical information. Qi-dong Xia, Jian-Xuan Sun, Jun-Lin Lu and Chen-Qian Liu wrote the manuscript. Jin-Zhou Xu, Si-Han Zhang, Ye An and Meng-Yao Xu revised the manuscript. Xing-Yu Zhong, Na Zeng, Hao-Dong He, Si-Yang Ma, Yi Lu and Bing Li analyzed the data and drew the figures. Jia Hu, Yao-Bing Chen, Zheng Liu and Shao-gang Wang participated in the design of the study and revised the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xia, QD., Sun, JX., Yao, ZP. et al. The role of TERT C228T and KDM6A alterations and TME in NMIBC treated with BCG. npj Precis. Onc. 8, 216 (2024). https://doi.org/10.1038/s41698-024-00725-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-024-00725-4