Abstract
Our understanding of how global changes in cellular metabolism contribute to human kidney disease remains incompletely understood. Here we show that nicotinamide adenine dinucleotide (NAD+) deficiency drives mitochondrial dysfunction causing inflammation and kidney disease development. Using unbiased global metabolomics in healthy and diseased human kidneys, we identify NAD+ deficiency as a disease signature. Furthermore using models of cisplatin- or ischaemia-reperfusion induced kidney injury in male mice we observed NAD+ depletion Supplemental nicotinamide riboside or nicotinamide mononucleotide restores NAD+ levels and improved kidney function. We find that cisplatin exposure causes cytosolic leakage of mitochondrial RNA (mtRNA) and activation of the cytosolic pattern recognition receptor retinoic acid-inducible gene I (RIG-I), both of which can be ameliorated by restoring NAD+. Male mice with RIG-I knock-out (KO) are protected from cisplatin-induced kidney disease. In summary, we demonstrate that the cytosolic release of mtRNA and RIG-I activation is an NAD+-sensitive mechanism contributing to kidney disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Gene expression data in this paper are deposited to GSE207587. The RNA-seq data for large-scale human kidney samples are available in GSE115098. Source data are provided with this paper.
References
Doke, T. et al. Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis. J. Clin. Investig. 131, e141801 (2021).
Qiu, C. et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat. Med. 24, 1721–1731 (2018).
Sheng, X. et al. Mapping the genetic architecture of human traits to cell types in the kidney identifies mechanisms of disease and potential treatments. Nat. Genet. 53, 1322–1333 (2021).
Liu, B. C., Tang, T. T., Lv, L. L. & Lan, H. Y. Renal tubule injury: a driving force toward chronic kidney disease. Kidney Int. 93, 568–579 (2018).
Nadour, Z. et al. Validation of a liquid chromatography coupled to tandem mass spectrometry method for simultaneous quantification of tryptophan and 10 key metabolites of the kynurenine pathway in plasma and urine: application to a cohort of acute kidney injury patients. Clin. Chim. Acta; Int. J. Clin. Chem. 534, 115–127 (2022).
Xu, J., Kitada, M. & Koya, D. NAD(+) homeostasis in diabetic kidney disease. Front. Med. 8, 703076 (2021).
Ralto, K. M., Rhee, E. P. & Parikh, S. M. NAD(+) homeostasis in renal health and disease. Nat. Rev. Nephrol. 16, 99–111 (2020).
Tran, M. T. et al. PGC1α drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature 531, 528–532 (2016).
Jia, Y. et al. Nicotinamide mononucleotide attenuates renal interstitial fibrosis after AKI by suppressing tubular DNA damage and senescence. Front. Physiol. 12, 649547 (2021).
Morevati, M. et al. Effect of NAD+ boosting on kidney ischemia-reperfusion injury. PLoS ONE 16, e0252554 (2021).
Myakala, K. et al. NAD metabolism modulates mitochondrial function and inflammation and prevents progression of diabetic kidney disease. https://www.biorxiv.org/content/10.1101/2021.12.05.471273v2 (2021).
Zheng, M. et al. Nicotinamide reduces renal interstitial fibrosis by suppressing tubular injury and inflammation. J. Cell. Mol. Med. 23, 3995–4004 (2019).
Liu, X. et al. Impaired nicotinamide adenine dinucleotide biosynthesis in the kidney of chronic kidney disease. Front. Physiol. 12, 723690 (2021).
Poyan Mehr, A. et al. De novo NAD(+) biosynthetic impairment in acute kidney injury in humans. Nat. Med. 24, 1351–1359 (2018).
Raines, N. H. et al. Niacinamide may be associated with improved outcomes in COVID-19-related acute kidney injury: an observational study. Kidney360 2, 33–41 (2021).
Simic, P. et al. Nicotinamide riboside with pterostilbene (NRPT) increases NAD(+) in patients with acute kidney injury (AKI): a randomized, double-blind, placebo-controlled, stepwise safety study of escalating doses of NRPT in patients with AKI. BMC Nephrol. 21, 342 (2020).
Yoshino, J., Baur, J. A. & Imai, S. I. NAD(+) intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528 (2018).
Martin, D. R., Lewington, A. J., Hammerman, M. R. & Padanilam, B. J. Inhibition of poly(ADP-ribose) polymerase attenuates ischemic renal injury in rats. Am. J. Physiol. Regulatory, Integr. Comp. Physiol. 279, R1834–R1840 (2000).
Doke, T. et al. Single-cell analysis identifies the interaction of altered renal tubules with basophils orchestrating kidney fibrosis. Nat. Immunol. 23, 947–959 (2022).
Tigano, M., Vargas, D. C., Tremblay-Belzile, S., Fu, Y. & Sfeir, A. Nuclear sensing of breaks in mitochondrial DNA enhances immune surveillance. Nature 591, 477–481 (2021).
Dhir, A. et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018).
Guan, Y. et al. Nicotinamide mononucleotide, an NAD(+) precursor, rescues age-associated susceptibility to AKI in a sirtuin 1-dependent manner. J. Am. Soc. Nephrology 28, 2337–2352 (2017).
Rabb, H. et al. Inflammation in AKI: current understanding, key questions, and knowledge gaps. J. Am. Soc. Nephrology 27, 371–379 (2016).
Kirita, Y., Wu, H., Uchimura, K., Wilson, P. C. & Humphreys, B. D. Cell profiling of mouse acute kidney injury reveals conserved cellular responses to injury. Proc. Natl Acad. Sci. USA 117, 15874–15883 (2020).
Hasenberg, A. et al. Catchup: a mouse model for imaging-based tracking and modulation of neutrophil granulocytes. Nat. Methods 12, 445–452 (2015).
Rehwinkel, J. & Gack, M. U. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 20, 537–551 (2020).
Desjardins, P., Frost, E. & Morais, R. Ethidium bromide-induced loss of mitochondrial DNA from primary chicken embryo fibroblasts. Mol. Cell. Biol. 5, 1163–1169 (1985).
Kleih, M. et al. Direct impact of cisplatin on mitochondria induces ROS production that dictates cell fate of ovarian cancer cells. Cell Death Dis. 10, 851 (2019).
Redza-Dutordoir, M. & Averill-Bates, D. A. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta 1863, 2977–2992 (2016).
Hou, F. et al. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146, 448–461 (2011).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Maekawa, H. et al. Mitochondrial damage causes inflammation via cGAS-STING signaling in acute kidney injury. Cell Rep. 29, 1261–1273.e1266 (2019).
Abedini, A. et al. Spatially resolved human kidney multi-omics single cell atlas highlights the key role of the fibrotic microenvironment in kidney disease progression. https://www.biorxiv.org/content/10.1101/2022.10.24.513598v1 (2022).
McReynolds, M. R. et al. NAD(+) flux is maintained in aged mice despite lower tissue concentrations. Cell Syst. 12, 1160–1172.e1164 (2021).
Liu, L. et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080.e1065 (2018).
Peng, J. et al. Clinical implications of a new DDX58 pathogenic variant that causes lupus nephritis due to RIG-I hyperactivation. J. Am. Soc. Nephrol. 34, 258–272 (2022).
Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Doke, T. et al. Genome-wide association studies identify the role of caspase-9 in kidney disease. Sci. Adv. 7, eabi8051 (2021).
Steen, C. B., Liu, C. L., Alizadeh, A. A. & Newman, A. M. Profiling cell type abundance and expression in bulk tissues with CIBERSORTx. Methods Mol. Biol. 2117, 135–157 (2020).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Mukherjee, S. et al. SIRT3 is required for liver regeneration but not for the beneficial effect of nicotinamide riboside. JCI Insight 6, e147193 (2021).
Acknowledgements
This work has been supported by the National Institute of Health, in the Susztak laboratory grant nos. NIH R01 DK087635, DK076077 and DK105821 and in the Baur laboratory by grant nos. DK098656 and HL165792. Additional funding and research materials were provided by Metro International Biotech through a sponsored research agreement to J.A.B. and K.S. We thank the Molecular Pathology and Imaging Core (grant no. P30-DK050306) and the Diabetes Research Center (grant nos. P30-DK19525 and S10 OD025098) at University of Pennsylvania for their services.
Author information
Authors and Affiliations
Contributions
This study was led by K.S. with assistance from J.A.B. These authors jointly supervised this work. T.D. performed experiments with assistance from S.M., D.M., K.C., J.G.D., B.C., A.A. and P.D. T.D., J.A.B. and K.S. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
Work in the Susztak laboratory is supported by Gilead, GSK, Boehringer, Regeneron, Novo Nordisk, Novartis, Calico, Astra Zeneca, Genentech, Ventus and Maze Biotech. J.A.B. is consultant to Pfizer and Cytokinetics, an inventor on a patent for using NAD+ precursors in liver injury and has received research funding and materials from Elysium Health and Metro International Biotech, both of which have an interest in NAD+ precursors. The other authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Hiroshi Itoh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Isabella Samuelson, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
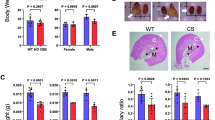
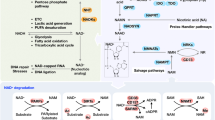
Extended Data Fig. 1 Changes in de novo NAD+ synthesis pathway in human and mouse kidneys.
a, Simplified de novo NAD+ synthesis pathway. The colour indicates metabolites significantly higher (red) or lower (blue) in injured kidneys of human and mice. b, Relative quantification of tryptophan, kynurenine, and quinolinate in human kidneys (Healthy control n = 25. KD n = 25). *P < 0.05. NS, not significant. c, Relative quantification of tryptophan, kynurenine, and quinolinate in mouse kidneys (PBS n = 4. Cis n = 4). **P < 0.01, ***P < 0.001. d, Heatmap showing the expression of genes involved in NAD+ metabolism in human kidneys. Color indicates higher (red) or lower (blue) expression. e, Heatmap showing the expression of genes involved in NAD+ metabolism in mouse kidneys. Color indicates higher (red) or lower (blue) expression. Data are presented as mean ± s.e.m. and were analysed using a two-tailed Student’s t-test.
Extended Data Fig. 2 Changes in de novo NAD+ synthesis pathway in age-matched human kidneys.
a, Demographic and clinical data of age-matched human kidney samples. b, Relative quantification of NAD+, NR, NAM, and quinolinate in human kidneys (Healthy control n = 25. KD n = 25). *P < 0.05. Data are presented as mean ± s.e.m. and were analyzed using a two-tailed Student’s t-test.
Extended Data Fig. 3 The expression levels of genes involved in cytosolic DNA and RNA sensing in kidneys.
a, TPM values of cGAS, Aim2, Tlr9, Zbp1, and Ddx58 in kidneys. b, Relative transcript levels of Ddx58, Isg15, Irf7, and Ifitm3 in in the kidneys of experimental groups (PBS n = 4. Cis + PBS n = 8. Cis + NMN n = 8. Cis + NR n = 8). Upper: Data were normalized using Gapdh. Lower; Data were normalized using Actb. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 4 NMN and NR supplementation improved kidney function and lowered inflammation in IRI mouse KD model.
a, The experiment designs. NAD+ precursors (NMN or NR) or vehicle (PBS) were injected i.p. for 4 consecutive days. First dose was injected 2 h before IRI. Kidneys were collected 3 days after IRI. b, Kidney NAD+ levels in experimental groups. *P < 0.05. c, Representative images of hematoxylin and eosin staining and semi-quantitative analysis of tubule injury in experimental groups. Scale bars: 20 μm. *P < 0.05. d, Serum creatinine and blood urea nitrogen (BUN) levels in experimental groups. **P < 0.01. e, Relative expression levels of Lcn2 and Havcr1 in the kidneys of mice in experimental groups. **P < 0.01. f, Relative expression levels of Ddx58, Isg15, Irf7, ifitm3, Cxcl10, and Cxcl16 in the kidneys of mice in experimental groups. *P < 0.05, **P < 0.01. b-f, PBS n = 4. Cis + PBS n = 8. Cis + NMN n = 8. Cis + NR n = 8. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 5 The effect of NAD+ depletion by NAMPT inhibitor (FK866).
a, Experimental design. Renal tubule cells were cultured and treated with NAMPT inhibitor, FK866 (100 nM) for indicated days. NAD+ levels in renal tubule cells and changes in live cell numbers of experimental groups on day1 (D1), day2 (D2), and day3 (D3). b, Relative transcript levels of Bax, Ddx58, Isg15, Irf7, ifitm3, Cxcl10, and Cxcl16 in renal tubule cells of experimental groups on D2. Gene expression levels were normalized to Gapdh. (A, B) PBS n = 3. FK866 + PBS n = 3. FK866 + NR n = 3. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 6 NAD+ precursors (NMN or NR) treatment restored mitochondria respiration capacity, lowered apoptosis, and improved energy production.
a, Cytotoxicity assay. The data is as represented as fold change (FC) normalized to control PBS group (n = 8 in each group). b, Relative transcript levels of Bax in renal tubule cells of experimental groups (n = 3 in each group). Gene expression levels were normalized to Gapdh. c, The result of oxygen consumption rate (OCR) in cultured renal tubule cells of experimental groups PBS n = 6. Cis + PBS n = 6. Cis + NMN n = 5. Cis + NR n = 5. *P < 0.05. The data was normalized to total protein levels. d, ATP levels in renal tubule cells in experimental groups (n = 3 in each group). *P < 0.05. The data was normalized to total protein levels. e, Live cell numbers of cisplatin-treated renal tubule cells in indicated experiment groups (n = 3 in each group). veh; vehicle control. NS, not significant. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 7 MitoTEMPO and BAX inhibitor reduced RIG-I cytosolic RNA sensing pathway induction in renal tubule cells.
a, (Left) The experimental design of the MitoTEMPO study. (Right) Relative transcript levels of Ddx58, isg15, and Irf7 in experimental groups (n = 3 in each group). Gene expression levels were normalized using Gapdh. veh; vehicle control. *P < 0.05. b, (Left) The experimental design of the BAX inhibitor study. (Right) Relative transcript levels of Ddx58, isg15, and Irf7 in experimental groups (n = 3 in each group). Gene expression levels were normalized using Gapdh. veh; vehicle control. *P < 0.05. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 8 RIG-I depletion protected from kidney injury, cell death, and inflammation.
a, Western blot quantification of RIG-I, MAVS, cleaved caspase-3 (cCASP3) in mice kidneys in indicated groups (n = 3 in each group). *P < 0.05, ***P < 0.001. NS, not significant. b, Experimental design. Renal tubule cells were isolated from WT and RIG-I KO mice. Relative transcript levels of Ifih1, cGAS, Isg15 and Irf7 in renal tubule cells of experimental groups (n = 3 in each group). Gene expression levels were normalized to Gapdh. *P < 0.05. NS, not significant. c, Kidney NAD+ levels in experimental groups (WT + PBS n = 4. RIG-I KO + PBS n = 4. WT + Cis n = 6. RIG-I KO + Cis n = 6). *P < 0.05. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 9 The effect of MDA5 and cGAS deletion on expression of inflammatory molecules in cisplatin treated renal tubule cells.
a, Experimental design. Renal tubule cells were isolated from WT and MDA5 KO mice. Western blot image of MDA5 in renal tubule cells. Relative transcript levels of Ddx58, cGAS, Isg15, Irf7, and Ifitm3 in experimental groups (n = 3 in each group). Gene expression levels were normalized to Gapdh. NS, not significant. b, Experimental design. Renal tubule cells were isolated from cGAS flox/flox mice, and infected with Adenovirus-GFP (Ade-GFP) or Adenovirus-Cre-GFP (Ade-Cre-GFP). Western blot image of cGAS in renal tubule cells. Relative transcript levels of Ddx58, Ifih1, Isg15, Irf7, and Ifitm3 in experimental groups (n = 3 in each group). Gene expression levels were normalized to Gapdh. *P < 0.05. NS, not significant. Data are presented as mean ± s.e.m. and were analyzed using a one-way ANOVA followed by Tukey post hoc test for multigroup comparison.
Extended Data Fig. 10 The correlation between the degree of renal fibrosis and expression of RIG-I and cytosolic RNA sensing pathway genes.
a, Correlation between the degree of kidney fibrosis and normalized transcription levels of DDX58, ISG15, and IRF7 in human kidney samples. b, Correlation between transcription levels of DDX58 and ISG15, IRF7, CXCL10, and CXCL16 in human kidney samples. c, Correlation of relative transcript levels of PPARGC1A with kidney NAD+ levels and eGFR. P value was calculated by Pearson’s correlation.
Supplementary information
Supplementary Information
Supplementary figs. 1 and 2 and uncropped image for supplementary fig. 2b.
Supplementary Tables
Supplementary tables 1–12.
Source data
Source Data Figs. 3–5 and 7 and Extended Data Fig. 9.
Unprocessed western blots.
Source Data Figs. 1–7.
Statistics source data for main figures.
Source Data Extended Data Figs. 2–9.
Statistics source data for extended data figures.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Doke, T., Mukherjee, S., Mukhi, D. et al. NAD+ precursor supplementation prevents mtRNA/RIG-I-dependent inflammation during kidney injury. Nat Metab 5, 414–430 (2023). https://doi.org/10.1038/s42255-023-00761-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-023-00761-7
This article is cited by
-
JOSD2 alleviates acute kidney injury through deubiquitinating SIRT7 and negativity regulating SIRT7-NF-κB inflammatory pathway in renal tubular epithelial cells
Acta Pharmacologica Sinica (2025)
-
SLC29A1 and SLC29A2 are human nicotinamide cell membrane transporters
Nature Communications (2025)
-
Targeting a key disulfide linkage to regulate RIG-I condensation and cytosolic RNA-sensing
Nature Cell Biology (2025)
-
IRBITs, signaling molecules of great functional diversity
Pflügers Archiv - European Journal of Physiology (2025)
-
AZD6738 Attenuates LPS-Induced Corneal Inflammation and Fibrosis by Modulating Macrophage Function and Polarization
Inflammation (2025)