Abstract
The period length of the circadian clock is important for organisms to maintain homeostasis. CHRONO has been shown to play an important role in regulating the period of the mammalian clock and has recently been implicated to function in several important physiological aspects. However, the mechanisms underlying transcriptional regulation by CHRONO to determine the circadian period remain elusive. Here, we report that CHRONO acts as a transcriptional repressor on JUN/FOS complexes independent of the classical BMAL1/CLOCK complexes. CHRONO can regulate BMAL1 expression by differentially regulating REV-ERBα and REV-ERBβ. We further developed a mathematical model and the model simulations strongly agreed with our findings that CHRONO acts on both REV-ERBα and BMAL1 to regulate the circadian period. The model also sheds light on the physiological significance of the fact that CHRONO cycles in most of the mammalian tissues. Thus, our experimental and computational approaches reveal the molecular mechanisms of regulating the circadian period by an important circadian component, CHRONO.
Similar content being viewed by others
Introduction
Disruptions in circadian rhythms, misalignments, and loss of entrainment significantly impact endocrine, metabolic, and sleep/wake cycles, potentially leading to various health issues, including metabolic disorders1,2,3, neurological diseases4, immune dysfunction5,6, and challenges associated with aging7,8. The period of the endogenous circadian clock is known to influence both the circadian phase and the phase angle of entrainment9. Animal studies have indicated a correlation between the period length of the endogenous clock and the phase angle following entrainment10,11. Specifically, organisms with shorter circadian periods tend to entrain earlier, while those with longer periods entrain later to external environmental cycles, which are typically 24 h12,13. It has been demonstrated that additional light exposure is necessary for humans to adapt to environments exceeding a 24 h cycle14. Furthermore, research indicates that women’s circadian rhythms are generally shorter than those of men, aligning with earlier phases of melatonin secretion and sleep cycles in females15,16. Therefore, investigating the mechanisms that govern circadian clock periods is essential for understanding their broader physiological implications.
A group of essential clock genes forms a highly conserved cell-autonomous transcription-translation feedback loop (TTFL). This includes core regulatory factors such as BMAL1, CLOCK (along with its homolog NPAS2), PER1, PER2, PER3, CRY1, and CRY217. Additionally, auxiliary factors involved in this regulatory network include factors such as REV-ERBα/β, RORs, DBP, NFIL3, CSNK1d/e, DEC1/2, FBXL3/2118. Recent research has identified CHRONO as an important core clock gene in addition to the classical clock genes19,20,21. This gene is broadly expressed across multiple tissues and organs, and its knockout in mice leads to an extension of the circadian period19,20,22. Notably, CHRONO has been found to interact with the C-terminus of BMAL1, disrupting the association between BMAL1 and CBP/P300, which subsequently impairs BMAL1’s functional activity21,23.
Collectively, the molecular functions attributed to CHRONO include: (i) direct interaction with BMAL1 to regulate the activity of the BMAL1/CLOCK complexes21,23; (ii) exhibiting the highest number of oscillating tissues among clock genes, indicating its critical role in circadian regulation24; and (iii) causing a prolonged circadian period upon knockout19,20. Additionally, we have recently characterized the nuclear localization signal (NLS) peptide of CHRONO, demonstrating that its entry into the nucleus is crucial for maintaining normal cellular circadian rhythms25. Despite these insights, the exact mechanisms by which CHRONO determines the circadian period remain poorly understood.
In this study, we employed genetic manipulation techniques to knock out and overexpress CHRONO, allowing us to assess its influence on the circadian period. Through these experiments, we identified a novel cis-binding element site for CHRONO, which it utilizes to repress the transcriptional activity of JUN/FOS complexes. We confirmed that CHRONO binding to this element is independent of BMAL1. Our model incorporated that CHRONO differentially regulates REV-ERBα and REV-ERBβ, thereby influencing the circadian clock period. Finally, we developed mathematical models to elucidate the mechanisms by which CHRONO regulates the circadian clock and to highlight the significance of CHRONO oscillation across various tissues.
Results
RNA-sequencing supports a distinct role of CHRONO in regulating transcription
We have employed CRISPR/Cas9 technology to achieve a knockout of the CHRONO gene in U2OS cells25. The resultant CHRONO knockout (KO) cells exhibited a significantly prolonged circadian period in comparison to wild-type U2OS cells (Fig. 1A, B). Furthermore, we extended our investigation to NIH-3T3 cells containing a Bmal1::Luciferase reporter. The luminescence traces from the Chrono knockout NIH-3T3 cells demonstrated a notable increase in the circadian period relative to those of the wild-type cells (Fig. 1C, D). In order to investigate the role of CHRONO in affecting cellular physiology, we carried out RNA-sequencing using WT and CHRONO−/− U2OS cells. Systematically, the most highly affected genes that were significantly changed with log2(fold change) > 1 or log2(fold change) < (−1) were plotted with a volcano plot (Fig. 1E & S1A). From the clustering (Fig. 1F) and principal component (Fig. 1G) analyses, we confirmed that WT and CHRONO−/− cells are distinct at the transcriptional level, indicating that CHRONO plays an important role in regulating transcription. In addition, KEGG and gene ontology (GO) pathway analyses of these differentially expressed genes revealed the enrichment of several pathways including signaling and cancer pathways (Fig. S1B, C). Next, we planned to further investigate the function of CHRONO.
ACHRONO−/− U2OS cells were generated using the CRISPR/Cas9 method. Representative subtracted bioluminescence data are shown for WT and CHRONO−/− cell lines. The black dashed line represents the error bar of the WT cells, and the red dashed line represents that of the CHRONO−/− cells. B The circadian period of CHRONO−/− U2OS cells was significantly prolonged (n = 6). Data are presented as the mean ± SD. *** P < 0.001. C Chrono−/− NIH3T3 cells were generated using the CRISPR/Cas9 method. Representative subtracted bioluminescence data are shown for WT and Chrono−/− cell lines. The black dashed line represents the error bar of the WT NIH3T3 cells, and the red dashed line represents tht of the Chrono−/− cells. D The circadian period of Chrono−/− NIH3T3 cells was significantly prolonged (n = 6). Data are presented as the mean ± SD. *** P < 0.001. E Volcano plot showing the differentially expressed genes between the WT and CHRONO−/− groups, p value < 0.05 and | log2(fold change) | >1. Clustering (F) and PCA (G) analysis of RNA-sequencing samples from WT and CHRONO−/− U2OS cells. Data are presented as the individual biological replicates or as an integration of all independent biological replicates (n = 2).
Overexpression of CHRONO caused dysregulation of BMAL1 and REV-REBα, but not CLOCK and REV-REBβ
Using lentivirus, we successfully made two U2OS cell lines that stably overexpress CHRONO with Myc tags. Unlike the CHRONO KO U2OS cells which lengthened the circadian period, CHRONO overexpression led to arrhythmic BMAL1 expression when we tested the circadian rhythms of the U2OS-CHR-1 and U2OS-CHR-2 cell lines (Fig. 2A, B). Overexpression of CHRONO not only disrupted the circadian rhythm of luminescence under the Bmal1 promoter, but also increased the level of luminescence (Fig. 2A). We therefore hypothesized that overexpression of CHRONO could upregulate the expression of BMAL1 proteins. According to the luminescence trace of the U2OS-WT control (Fig. 2A), we harvested time course samples of U2OS-WT and U2OS-CHR starting at 4 h before the trough of WT luminescence (arrow indicates the trough time in Fig. 2A). The relevant clock proteins and their transcript levels were then detected by Western blotting (WB) and quantitative real-time PCR (qPCR), respectively. Time point 0 was designated as 4 h before the trough (see the arrow in Fig. 2A). Clearly, CHRONO protein levels were highly upregulated in U2OS-CHR cells (Fig. 2C, D). BMAL1 was upregulated and REV-ERBα was downregulated continuously over the time course (Fig. 2C, D). Interestingly, the mobility shift of BMAL1 protein indicated that BMAL1 was in a phosphorylated state in CHRONO overexpressing cells, which was confirmed by WB using antibodies against phosphorylated BMAL1 at Ser42 (p-BMAL1) (Fig. 2D). Figure 2C, D clearly show that BMAL1 and p-BMAL1 are upregulated in CHRONO overexpressing cells, while REV-ERBα was significantly downregulated over time. However, the protein levels of both CLOCK and REV-ERBβ were unaffected. With the overexpression of CHRONO transcripts (Fig. 2E), the transcript levels of BMAL1 in U2OS-CHR cells were significantly upregulated over time (Fig. 2F), and those of REV-ERBα were significantly downregulated over time (Fig. 2G). The expression of other classical circadian genes over time was also determined by qPCR (Fig. S2). We further harvested cells from U2OS-WT, U2OS-CHRONO−/−, U2OS-CHR-1 and U2OS-CHR-2 at the trough of WT luminescence (arrow in Fig. 2A). WB results showed that REV-ERBα was dramatically reduced, whereas BMAL1 was upregulated in CHRONO overexpressing cells (Fig. 2H). Meanwhile, REV-ERBα was upregulated in CHRONO−/− cells (Fig. 2H). The transcript level of BMAL1 showed the same regulation as the protein level (Fig. 2I). On the other hand, REV-ERBα was upregulated in CHRONO−/− cells and downregulated in both U2OS-CHR-1 and U2OS-CHR-2 cells (Fig. 2J). In summary, REV-ERBα but not REV-ERBβ was downregulated in U2OS cells when CHRONO was overexpressed.
A Representative raw bioluminescence data are shown for WT and two CHRONO- overexpression cell lines derived from U2OS cells expressing a Bmal1::Luc reporter. B Fourier analysis of the luminescence traces of each cell line in panel A. PCA analysis (C) Time point 0 was designated as 4 h before the trough (see the arrow in panel A) of PBMAL1::Luc oscillation, and then cells were collected every 4 h, designated as 0, 4, 8, 12, 16 and 20. Different target proteins were detected by WB using the indicated antibodies. D Different target proteins in U2OS-CHR cells were detected during the time course, as done in (C). * labels the non-specific bands of CHRONO and CLOCK in (C, D). E The relative expression levels of CHRONO transcripts in WT and U2OS-CHR cells during the time course for both WT and U2OS-CHR cells. The relative expression levels of BMAL1 (F) and REV-ERBα (G) were determined by RT-qPCR during the time course for both WT and U2OS-CHR cells. H Western results of REV-ERBα, REV-ERBβ, BMAL1, p-BMAL1 from WT, CHRONO KO, and overexpressing cells, collected as indicated by the arrow in (A). I The mRNA level of BMAL1 of the WT, CHRONO KO, and overexpressing cells. J The mRNA level of REV-ERBα of the WT, CHRONO KO, and overexpressing cells. Experiments were carried out at least three independent times. Data are presented as the mean ± SD. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
CHRONO repressed the expression of REV-ERBα through the TRE box
Given our observation that REV-ERBα expression was strongly inhibited upon CHRONO overexpression (Fig. 2), we hypothesized the presence of a specific cis-element exists within the REV-ERBα promoter region. Myc antibodies did not precipitate any detectable chromatins from U2OS-WT cells (Ctrl in Fig. 3A–C). Therefore, we used ChIP-seq to localize the DNA binding sites for CHRONO on REV-ERBα and REV-ERBβ genes in U2OS-CHR cells. Indeed, the promoter of REV-ERBα contains a TRE box, 5’-TGA(C/G)TCA-3’, which is mainly activated by heterodimers of the AP-1 family proteins (Fig. 3A, for sequence of this binding site see Fig. S3A, named REV-ERBα-B3). In contrast, the one binding peak in the promoter of REV-ERBβ contains only E-box (Fig. 3B, for sequence of this binding site see Fig. S3B). Among the binding peaks, the promoter region of the UBC gene contains only the TRE box, but not the E-box (Fig. 3C, for sequence of this binding site see Fig. S3C). ChIP-PCR was used to confirm that CHRONO can bind to the TRE box on the UBC locus (Fig. 3C). We further knocked out BMAL1 in the U2OS-CHR cells and we confirmed that in the absence of BMAL1 (Fig. S3D), CHRONO can associate with the unique upstream regions of UBC and REV-ERBα, but not with the E-box motifs upstream of PER2 and REV-ERBβ (Fig. 3D, primers see Table S1, and quantification of this panel see Fig. S4).
A The UCSC genome browser view of CHRONO occupancy at the REV-ERBα locus in both U2OS-WT and U2OS-CHR cells. The binding peak that contains the TRE box was labeled. Notice there are three binding peaks for CHRONO. B The UCSC genome browser view of CHRONO occupancy at the REV-ERBβ locus in both U2OS-WT and U2OS-CHR cells. The only binding peak at the REV-ERBβ locus contains an E-box. C The UBC locus contains only the TRE element but not the E-box. ChIP-PCR confirmed that CHRONO can bind to the TRE box on the UBC locus. D CHRONO still binds to UBC and REV-ERBα -B3, but not PER2, REV-ERBβ loci when BMAL1 was knocked out. ChIP-PCR confirmed that CHRONO binds to the UBC locus. E JUN/FOS was present in the immuno-precipitation product when CHRONO was precipitated. “*” labels the non-specific bands of FOS proteins. F Co-IP confirmed the interaction between CHRONO and the JUN/FOS complex. G CHRONO suppressed the luciferase activity which is under the control of a UBC promoter containing only the TRE element. H CHRONO suppressed the luciferase activity which is under the control of a REV-ERBα promoter containing a TRE element. I JUN/FOS did not activate the expression when the TRE element was mutated. Data are presented as the mean ± SD. (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Indeed, when we performed motif analysis using the top 500 peaks of the ChIP-seq results, we found that the most enriched motif is the TRE element followed by the E-box element (Fig. S3E). JUN and FOS proteins are the most studied AP-1 factors. Therefore, we further hypothesized that the JUN/FOS complexes can activate the expression of REV-ERBα through the TRE box while CHRONO represses the JUN/FOS complexes through binding with them. We then immunoprecipitated Myc-labeled CHRONO from U2OS-CHR cells, and we identified both JUN and FOS in the precipitated product (Fig. 3E). Furthermore, Co-IP experiments confirmed the interaction between CHRONO and the JUN/FOS heterocomplex when we co-expressed 5m6h-CHR (5×Myc-6×His tagged CHRONO) with FH-tagged (Flag-HA tagged) JUN and FOS in 293 T cells (Fig. 3F). We then constructed a luciferase reporter PUBC-TRE::luc and tested the effect of CHRONO on this construct. As predicted, JUN/FOS activated the expression of luciferase that is under the control of the TRE DNA element, while CHRONO repressed the function of the JUN/FOS complex (Fig. 3G). A luciferase reporter PREV-ERBα -B3::luc containing only the TRE box from the REV-ERBα promoter was constructed. Again, JUN/FOS activated luciferase expression and CHRONO significantly suppressed the function of JUN/FOS complex (Fig. 3H). When the TRE elements were mutated, the activating effect of JUN/FOS was abolished, and the inhibitory effect of CHRONO disappeared (Fig. 3I).
Modeling insights into the mechanism that CHRONO regulates the circadian period
Building upon a previously published mathematical model26 and our observations that CHRONO binds to the TRE box to repress the transcription activation of REV-ERBα (Fig. 3), we first constructed a modified model (see Methods and Supplementary Text S1 for a complete model description) in which CHRONO is incorporated into the transcriptional repressions on both BMAL1/CLOCK and JUN/FOS complexes. This updated model takes into account the differences between REV-ERBα and REV-ERBβ since the former gene contains different cis-regulatory elements besides E-box (Fig. 3A, B). CHRONO binds to the heterocomplex JUN/FOS to repress the transcription of REV-ERBα (Figs. 3H & 4A). Briefly, the model does not show differences among PER1/2/3, CRY1/2, and RORα/β/γ. Based on a parameter set (Table S2), our modified model (Model I) simulates well the circadian oscillations of the key component BMAL1, which shows rhythmicity with a period of about 24 h in the presence of CHRONO (Fig. 4B). The changed period of CHRONO−/− simulation (Fig. 4B) partially recapitulates the phenotype of the CHRONO deletion cell lines, which is approximately 1–2 h longer than WT cells (Fig. 1). Under the conditions of CHRONO−/− simulation, REV-ERBα is upregulated (Fig. 4C) and REV-ERBβ remains unaffected (Fig. 4D). Second, by overexpressing the amount of CHRONO (ksCHRONO is 6.835 in CHRONO overexpression condition compared to 0.091 in WT condition, see Supplementary Text S1), the model predicts an increased amount of BMAL1 expression (Fig. 4E, and see experimental results in Fig. 2D). Notably, under this single perturbation, the rhythm of BMAL1 expression is disturbed due to the repressed expression of REV-ERBα (Fig. 4F, and see experimental results in Fig. 2D). To further confirm the role of CHRONO in regulating the circadian period, we knocked out the CHRONO gene in PC9 cells, a human lung adenocarcinoma cell line. The PC9-CHRONO−/− cells showed a prolonged circadian period (Fig. 4G, H), consistent with what was shown in the model (Fig. 4B). By comparing the model predictions with our experimental observations in U2OS, NIH3T3 and PC9 cell lines, this model supports that CHRONO affects both BMAL1/CLOCK and JUN/FOS complexes. In summary, our early experimental observation that CHRONO overexpression upregulates the expression of BMAL1 is faithfully captured by our model.
A Diagram of the network of interacting components of the circadian model I incorporating the CHRONO/REV-ERBα loop. B When CHRONO is knocked out in the model, the period of BMAL1 expression is prolonged. C The expression level of REV-ERBα in the situation of CHRONO knockout is increased in the model simulation results. D The expression level of REV-ERBβ in the situation of CHRONO knockout is not affected. E When CHRONO is overexpressed, BMAL1 loses the rhythmicity of its expression. F Overexpression of CHRONO strongly represses the expression of REV-ERBα. The model recapitulates the experimentally observed differential period response due to lack of CHRONO. Solid black lines represent the control WT condition, while dash-dotted lines represent CHRONO−/−. The red lines represent the simulated CHRONO expression in WT (B) and U2OS-CHR cells (E). G The luminescence trace of PC9-CHRONO−/− cell line, which harbors a luciferase reporter driven by the Bmal1 promoter (Bmal1::Luc). H CHRONO knockout in PC9 cell line showed prolonged period, which agreed with model predictions. Data are presented as the mean ± SD.
Periodic sensitivity analysis indicated that the degradation of BMAL1 may be affected by the deficiency of CHRONO
In our Model I, the CHRONO knockout prolongs the period (Fig. 4B, simulated periods of Fig. 4B–D see Table S3) but not as much as our experimental observations (Fig. 1B, D), suggesting that there may be other sets of parameters regulating the period. We then carried out the periodic sensitivity analysis with respect to the period dependence of the CHRONO-related parameter in Model I. This sensitivity analysis was performed on the basis of the parameter set obtained previously (Table S2). Two types of sensitivity are noticeable: the first is that the period at the boundaries varies by a factor greater than 2 h, and the second is that the window for oscillations is particularly narrow (Table S5). In this analysis we found the highest sensitivity of the parameter kdn of BMAL1, which controls the degradation of the BMAL1 protein. It is sensitive in both types of sensitivity: the period at the boundaries varies by more than 2 h, and the oscillation window is narrow (Table S5). In a narrow range of the boundaries of kdn of BMAL1, the lower the degradation rate, the longer the period, as shown in the bifurcation diagrams (Fig. 5A). Other parameters of CHRONO, such as ksCHRONO and v0Chrono, can affect the period, but not as sensitive as the kdn of BMAL1 does (Fig.5B–D). When we incorporated the slower degradation rate of BMAL1 into Model I (Fig.4A), we found that the slower degradation rate further prolonged the period by 0.48 h (Fig. 5E, BMAL1 degradation rate is 0.068 in CHRONO knockout cells compared to 0.0877 in WT cells). This simulation indicated that the degradation rate of BMAL1 proteins would affect the period of the rhythmicity.
Bifurcation diagrams showing the variation of the period as a function of kdn of BMAL1, ksCHRONO, v0Chrono and vsChrono in Model I. The curves (A–D) were obtained for sensitive parameter after sensitivity analysis. In all panels, the red dot on each curve refers to the basal parameter value listed in Table S2, the blue dot refers to the boundaries of the oscillatory ___domain listed in Table S5. E Under the dedicated degradation rates of BMAL1 (kdnB = 0.0877), CHRONO−/− prolonged the period by 0.39 h, when degradation rates of BMAL1 is slower (kdnB = 0.068), CHRONO−/− would further lengthen the period by 0.48 h.
Updated computational model further incorporated the nuclear localization of BMAL1
We further measured the degradation rate of BMAL1 in WT and CHRONO−/− cells. In the presence of cyclohexmide which stops the synthesis of new proteins, BMAL1 appears to be degraded more slowly in the CHRONO−/− cell (Fig. S5). Our recent findings have observed that there was more BMAL1 in the cytoplasm of CHRONO−/− cells25, thus we hypothesized that the cytoplasmic/nuclear distribution of BMAL1 might be influenced by the nuclear import rate of BMAL1 proteins. We then further updated Model I by separating BMAL1 into the cytoplasmic BMAL1 and the nuclear BMAL1 (Model II, Fig. 6A, see Supplementary Text S1). We first performed a bifurcation analysis for the CHRONO-related parameters in Model II (Fig. S6). The window of ksCHRONO for oscillations is very narrow, ranging from 0.0 to about 0.3. When ksCHRONO goes down to zero, mimicking the knockout of CHRONO gene, the period is lengthened less than an hour; when ksCHRONO goes high above the boundary, mimicking the overexpression of CHRONO, the rhythmicity disappears (Fig. 6B). Furthermore, a period sensitivity analysis was performed in Model II (Table S6). Indeed, we found that the rate constant for the nuclear import of BMAL1 (kim,B) influences on the period (Fig. 6C). Within a certain range, the smaller the kim,B is, the longer the apparent period (Fig. 6C). A lower nuclear import rate may result in more BMAL1 in the cytoplasm, affecting the overall rate of BMAL1 degradation. The rate constant for the nuclear export of BMAL1 (kex,B) may affect the period, but in our simulation, the best fit parameter stays in the lowest rate of kex,B (Fig. 6D, see the basal parameters in Table S4). Using this updated Model II, as predicted by Fig. 5E, the circadian oscillation of BMAL1, REV-ERBα, and REV-ERBβ exhibited longer period in the absence of CHRONO (Fig. S7). Taken together, the combination of differential regulation of REV-ERBs and a slower degradation rate of BMAL1 recapitulates the lengthened phenotype of the CHRONO deletion cell lines.
A The schematic of Model II, which considers that CHRONO affects the cytoplasmic/nucleic localization of BMAL1. The dynamic changes of BMAL1 in the cytoplasm and nucleus are described separately in two ordinary differential equations. Further details can be found in Supplementary Text S1. B The bifurcation diagram shows the variation of the period as a function of ksCHRONO. The bifurcation diagram shows the variation of the period as a function of kim,B (C) and kex,B (D) in Model II. In all panels, the red dot on each curve refers to the basal parameter value listed in Table S4, the blue dot refers to the boundaries of the oscillatory ___domain listed in Table S6.
Modeling insights into the significance of CHRONO
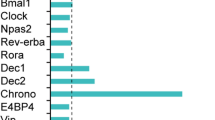
Since CHRONO was detected as the most rhythmically expressed gene in various tissues (52 out of 64 examined tissues) in Baboons24, we wondered whether it is most rhythmically expressed in mice. Though Chrono is not the most cycling gene in mice using a publicly available dataset (Ciart is another name for Chrono)27, it is rhythmically expressed in most tissues that were examined (Fig. 7A). The peak phase time of Chrono in different mouse tissues was very close to ZT12, which is exactly anti-phase to that of both Bmal1 (another name for Bmal1 is Arntl) and Clock/Npas2 (Fig. 7B). In consideration of other clock genes, the peak time of each gene in various tissues is restricted in a roughly 6-hour window. Therefore, Chrono is cycling broadly in both mice and Baboons, indicating the significance of the gene in the physiological processes. According to the model (Fig. 6A), our simulated results indicate that the amplitude of the BMAL1 oscillation is larger in the CHRONO knockout conditions under a certain range of BMAL1 degradation rates: from 0.021 to 0.148 (Fig. 7C, D). Therefore, we speculate that the existence of CHRONO may be important for maintaining a stable circadian clock in various tissues.
AChrono is rhythmically expressed in most tissues of the mouse. The detailed expression data of all examined genes were downloaded from a publicly available dataset GSE54652. B The phase distribution of different clock genes in the publicly available dataset. C, D The expression and amplitude of BMAL1 recapitulates over a certain range of BMAL1 degradation rates (kdn of BMAL1 = 0.034–0.148).
Discussion
In this study, we identify that the JUN/FOS complexes can bind to the promoter region of REV-ERBα to activate its expression, while CHRONO binds to the complexes to repress the transcription activation through the TRE box. Motif analysis of the promoter regions of REV-ERBα and REV-ERBβ supports our experimental observations of differential regulation between them when CHRONO was overexpressed. We further incorporate our observations that CHRONO can simultaneously regulate both BMAL1/CLOCK and JUN/FOS complexes into a mathematical model. Our findings that CHRONO KO extended the period over an hour further lead us to consider that CHRONO may regulate the circadian period in part by facilitating the nuclear localization of BMAL1. Furthermore, our mathematical model shed insights into the robustness effects of CHRONO that oscillates in the most tissues in both Baboons24 and mice (Fig. 7). Overall, our observations and simulations reveal the detailed mechanism by which CHRONO regulates the period of the circadian clock.
With regard to the prolonged period in CHRONO−/− and the disturbed BMAL1 promoter rhythmicity in CHRONO-overexpressing cells, WB and qPCR analysis indicate that this is determined by the changes in both BMAL1 and REV-ERBα. Our ChIP-seq results provide evidence to explain how CHRONO affects the period of the circadian clock. We have identified REV-ERBα, but not REV-ERBβ, as the key binding factor for CHRONO that contributes to the period effect. When BMAL1 is absent, CHRONO still associates with the TRE box upstream of UBC and REV-ERBα-B3, but not with the E-box upstream of PER2 and REV-ERBβ (Fig. 3D). These findings are supported by our modeling efforts that a novel cis-regulatory element, TRE box, links CHRONO to differentially regulate REV-ERBα and REV-ERBβ.
When we incorporated the differential regulation by CHRONO into a mathematical model, the simulations show that the period of CHRONO−/− cells is ~0.5 h longer than that of WT cells, suggesting that another parameter of CHRONO may affect the period of the circadian clock. The sensitivity analysis showed that the rate of BMAL1 degradation had a stronger effect on period length (Fig. 5A). We modified the mathematical model by adjusting the degradation rate of BMAL1, and the period length of CHRONO−/− cells is further prolonged (Fig. 5E). As previously reported, nuclear translocation of BMAL1 enhances proteolysis of both BMAL1 and CLOCK via ubiquitin-dependent and -independent pathways28,29. Our observations that BMAL1 was degraded more slowly in CHRONO−/− cells (Fig. S5) indicated the nuclear transportation of BMAL1 was affected in CHRONO−/− cells. In the computational simulations, a lower nuclear import rate of BMAL1 results in a longer period. Based on this observation, we conclude that the period effect of CHRONO results from two aspects: the differential regulation on REV-ERBα and REV-ERBβ, and the changes in BMAL1 degradation. Recently, we found that CHRONO interacts with several nuclear transporters for its nuclear localization25. How CHRONO affects the cellular localization of BMAL1 requires further investigation, although CHRONO may form meta-complexes with importins together with BMAL1 and CLOCK proteins to facilitate their nuclear entry.
A previous study reported that overexpression of REV-ERBα leads to loss of BMAL1 reporter oscillation, emphasizing the importance of the REV-ERBα – BMAL1 loop30. Other mathematical models have also confirmed the critical role of the REV-ERBα – BMAL1 loop in generating circadian oscillators in various tissues31. Our experimental data showed that overexpression of CHRONO constitutively inhibits the expression of REV-ERBα, resulting in a de-repression of BMAL1 (Fig. 2). This is in agreement with the important role of the REV-ERBα – BMAL1 loop. Although the BMAL1 promoter was disturbed to higher activity by CHRONO overexpression (Fig. 2), the circadian rhythmicity may still be maintained. Several studies have proved that even constitutive expression of BMAL1 (or BMAL2) can sustain circadian rhythmicity32,33,34. We will continue to investigate the effect of CHRONO overexpression on the cellular circadian clock. There are several limitations of our model. First, the mismatched period may be explained by other missing components such as phosphorylated clock proteins, HAT, HDAC, etc.35. Second, the degradation rate of PER or CRY proteins may also affect the circadian period31,36. Nevertheless, our model reproduces our experimental data under both CHRONO knockout and overexpression conditions.
In addition to the importance of the period for entrainment, the amplitude of endogenous clocks is also closely related to entrainment. The ratio between the strength of the external cues and the amplitude of the endogenous oscillations affects the entrainment phase, which is a critical parameter for good entrainment. A well entrained organism maintains both aligned phases with the external cue and aligned phases between the central and peripheral clocks. Reduced circadian amplitude of the Per gene in Clock mutant mice allows increased efficacy of phase resetting37. Consistently, the amplitude of the circadian clock has also been implicated in the entrainment of the human circadian rhythm, such that a reduced amplitude makes the clock more sensitive to the entraining cues38,39. CHRONO is the most recently discovered core clock component, cycling mostly in the studied tissues in both Baboons24 and mice (Fig. 7). For the circadian clocks in peripheral tissues to entrain faster than the central pacemaker, the amplitude of the peripheral clocks must be smaller12. Using our mathematical model, we observed that CHRONO can reduce the amplitude of BMAL1 expression over a range of BMAL1 degradation rates (Fig. 7C, D). The reduced amplitude in the presence of CHRONO may provide an explanation for the importance of CHRONO in various tissues. In summary, we have provided both experimental and computational evidence to understand the importance of CHRONO.
Methods
CHRONO knock out cells
CRISPR/Cas9 system was used to generate the CHRONO knockout cells. The sgRNA1 (CCGCTGCAGGCATCGATCGA) targeted to CHRONO and sgRNA2 (CTGTCCCGGGGTCACCATGGCAG) targeted to Chrono were designed using the website http://crispr.mit.edu. The sgRNA sequences were cloned into the pX459 vector (pSpCas9{BB}-2A-Puro was a gift from Feng Zhang, Addgene plasmid No. 48139). The constructed plasmid of each individual gene was transfected into NIH-3T3-B1-B10 cells40 or U2OS-C2641 cells to knock out the gene. After screening the monoclonal cell lines, the targeted sequence was amplified and analyzed by subsequent sequencing to confirm the knockout. The Chrono knockout NIH3T3 cells have been described previously by Wu et al.42 and the CHRONO knockout U2OS cells have been described by Zhou et al.25. The sequences of the knockout alleles were confirmed by Sanger sequencing.
BMAL1 knockout in U2OS-CHR cells
CRISPR/Cas9 was used to knock out BMAL1 in U2OS-CHR cells. The procedure was similar to the process of generating CHRONO knockout cells. Briefly, the sgRNA, GTTTCTCGGCACGCGATAGA, targeting the exon 12 was picked to knockout the BMAL1 gene. U2OS-C26 cells41 were transfected with constructed plasmids to knock out the BMAL1 gene. The monoclonal cell line U2OS-CHR-BMAL1−/− had both alleles being modified, leading to pre-mature stop of the protein synthesis. The knockout of BMAl1 was confirmed with western blot, as presented in the Supplementary Fig. S3D.
Western blotting and Co-immunoprecipitation (Co-IP)
Rabbit antibody against Myc-tag (#2276, Cell Signaling Technology, Danvers, USA), mouse antibody against GAPDH (60004-1-Ig, Proteintech, Wuhan, China), rabbit antibody against REV-ERBα (#13418, Cell Signaling Technology, Denver, USA), rabbit antibody against REV-ERBβ (#13906-1-AP, Proteintech, Wuhan, China), rabbit antibody against CLOCK (#5157, Cell Signaling Technology, Denver, USA), rabbit antibody against BMAL1 (#14020, Cell Signaling Technology, Denver, USA), rabbit antibody against phosphor-BMAL1 (Ser42) (#13936, Cell Signaling Technology, Denver, USA), rabbit antibody against CHRONO (#PA5-55643, Invitrogen, California, USA), rabbit antibody against JUN (#9165, Cell Signaling Technology, Denver, USA), rabbit antibody against FOS (#2250, Cell Signaling Technology, Denver, USA), rabbit antibody against Luciferase (#27986-1-AP, Proteintech, Wuhan, China) were subjected to Western blotting according to the manufacturer’s protocol. For Western blotting, cells were harvested at the indicated time points after dexamethasone synchronization and lysed with the radio immunoprecipitation assay buffer (RIPA buffer, #P0013B, Beyotime, Shanghai, China) at 4 °C for 30 min, centrifuged at 12,000 rpm, and the soluble fraction was collected in a new tube for SDS-PAGE, followed by immunoblotting analysis with the specific antibodies.
For Co-IP, the desired plasmids were transfected into cells using PEI. After 48 h, cells were lysed in Western and IP cell lysis buffer (#P0013, Beyotime, Shanghai, China) at 4 °C for 30 min. The supernatant of the lysate was prepared after centrifugation at 4 °C, 12,000 rpm for 20 min and immunoprecipitated with corresponding antibody and mouse IgG with gentle rotation at 4 °C overnight, then incubated with the Protein G magnetic beads (#9006S, Cell Signaling Technology, Danvers, USA) solution at 4 °C for 2 h. The the beads were washed with pre-cold PBS for five times with gentle rotation. Samples were resuspended in the SDS loading dye, boiled for 10 min, and resolved by SDS-PAGE and followed by immunoblotting analysis with the specific antibodies.
Luciferase reporter assay
The firefly luciferase reporter pGL3-PUBC::Fluc and pGL3-PREV-ERBα-B3::Fluc were constructed by inserting the triple TRE element (TGA G/C TCA) site of UBC (Chr 12: 124917220 - 124917267) or the CHRONO binding locus of REV-ERBα containing TRE site (Chr 17: 40099129 - 40099189) into pGL3-luc vector. PREV-ERBα-B3-mut::Fluc was constructed by removing the TRE element. JUN and FOS were cloned into pcDNA3.1 with N-terminal Flag-HA tag. Approximately 5 × 105 HEK-293T cells were seeded into each well of a 60 mm dish 1 day prior to transfection. When cells reached 70% ~ 90% confluence, 1 µg of the firefly luciferase reporter, 100 ng of the renilla luciferase reporter pCMV::Rluc, and 2 µg of JUN, FOS or CHR were co-transfected into HEK-293T using ExFect Transfection Reagent (#T101, Vazyme Biotech, Nanjing, China). The constant amount of DNA (7.1 µg/well) was adjusted with pcDNA3.1(+)/Hygro vector. 24 h after transfection, the cells were washed three times with ice-cold PBS (SH30256.01, Hyclone/GE, USA), then processed with a TransDetect Double-Luciferase Reporter Assay Kit (#FR201, Transgen Biotech, Beijing, China) and measured using SpectraMax Paradigm Multi-Mode Microplate Reader (Molecular Devices, Silicon Valley, USA). Each construct was assayed at least for three independent times.
Bioluminescence recoding and data analysis
2 × 105 corresponding cells were seeded in 35 mm dishes the day before synchronization. On the next day, cells which reached around 80% confluency in each dish were synchronized with 2 h treatment of 200 nM dexamethasone (dissolved in DMSO, dimethyl sulfoxide) and recorded in a LumiCycle as described previously23. Bioluminescence data were analyzed with the LumiCycle analysis program (Actimetrics, Florida, USA) to obtain circadian parameters such as period and amplitude.
ChIP-seq experiment and analysis
ChIP-DNA was prepared using the SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) (#9003, Cell Signaling Technology, Danvers, USA) according to the manufacturer’s protocol. Briefly, For CHRONO-ChIP, approximately 1 × 107 U2OS-WT and U2OS-CHR cells were cross-linked in 15 cm culture dishes with 1% formaldehyde for 10 min at room temperature and quenched with glycine. After two washes with ice-cold PBS, the cells were scraped off for nuclear lysis buffer containing DTT and PIC (protease inhibitor cocktail), kept on ice for 10 min and mixed every 3 min by inverting the tube. The nuclei were digested with micrococcal nuclease and then sonicated 3 sets of 20 s pulses at 60 W (Sonifier450D, USA). After centrifugation at 9400 × g for 10 min at 4 °C, the cross-linked chromatin was prepared in the supernatant, 10 µg of fragmented chromatin was incubated with anti-Myc antibody overnight at 4 °C. 40 µL of magnetic beads Protein G was added to the chromatin and incubated for a further 4 h. The beads were then washed three times with low salt wash buffer and once with high salt wash buffer. Co-immunoprecipitated DNA fragments were eluted with ChIP elution buffer and then reverse cross-linked by incubation with NaCl and Proteinase K at 65 °C for 2 h. Finally, DNA was purified using DNA Purification Buffers and Spin Columns (#14209, Cell Signaling Technology, Danvers, USA).
cDNA synthesis and q-PCR analysis
Total cellular RNA extraction was performed with Trizol reagent (#ET101, Transgene, Beijing, China) according to the manufacturer’s instructions. Next, cDNA was synthesized using the HiScript III RT SuperMix for qPCR (+gDNA wiper) (#R323, Vazyme Biotech, Nanjing, China) and was subjected to qPCR using AceQ Universal SYBR qPCR Master Mix (#Q511-02, Vazyme Biotech, Nanjing, China) on a real-time fluorescence quantitative PCR instrument (Thermo Scientific, QuantStudio™6 Flex, USA). The value of each cDNA was calculated using the ΔΔCt method and normalized to the value of the house-keeping gene control GAPDH. Data were plotted as a fold of change. The sequences of primers were shown in Table S7.
RNA-seq
Total RNA was extracted from cells using the Trizol reagent following the manufacturer’s instructions. Three replicate RNA-seq libraries were prepared. A total of 9 libraries were sent to BGI Genomics for sequencing using the BGISEQ-500 sequencer. The sequencing data were filtered using SOAPnuke (v1.5.6) by removing reads containing sequencing adapters, removing reads with low-quality base ratios (base quality less than or equal to 15) was greater than 20% and unknown base ratios (‘N’ base) greater than 5%. The sequencing reads were also checked for quality using FastQC v0.10.1. The clean reads were stored in FASTQ format. Bowtie2 (2.3.4.3) was applied to align the clean reads to the gene set, including known and novel, coding and non-coding transcripts were included. Gene expression levels were calculated using RSEM (v1.3.1). Essentially, differential expression analysis was performed using the DESeq2 (v1.4.5) (or DEGseq or PoissonDis) with Q value ≤ 0.05 (or FDR ≤ 0.001). To gain insight into the change in phenotype, GO (http://www.geneontology.org/) and KEGG (https://www.kegg.jp/) enrichment analysis of annotated differently expressed genes was performed by Phyper (https://en.wikipedia.org/wiki/Hypergeometric_distribution) based on Hypergeometric test. The significant levels of terms and pathways were corrected by Q value with a strict threshold (Q value ≤ 0.05)
Model for the CHRONO-dependent regulation of BMAL1 and REV-ERBα (Model I)
Model I, as shown in Fig. 4A, is derived from the model proposed in 201826. CHRONO is another key inhibitor in this loop, binding directly to BMAL1 to inhibit the transcriptional activation of downstream genes by BMAL1/CLOCK. Meanwhile, CHRONO can also bind to the AP-1 element of REV-ERBα with JUN and FOS to repress its expression. All modeling assumptions and equations are outlined in Supplementary Text S1. The parameters were estimated by optimizing the ODE system via an evolutionary strategy, as employed by Foteinou et al.26. The period sensitivity analysis and Hopf bifurcation analysis were carried out based on the method described by Leloup et al.43. Further details can be found in Supplementary Text S1.
Model for the CHRONO related BMAL1 localization (Model II)
Model II, updated from Model I, considers that CHRONO affects the cytoplasmic/nucleic localization of BMAL1. BMAL1 is divided into cytoplasmic BMAL1 and nuclear BMAL1. The dynamic changes of BMAL1 in the cytoplasm and nucleus are described separately in two ordinary differential equations. Further details can be found in Table S2, S4–S6 and Fig. S6 in Supplementary Materials.
Statistical analysis
Data of the period length, gene expression, luciferase activity, and quantification of WB images were presented as the mean ± SD. They were obtained from three (or more) independent experiments. An unpaired Student’s t-test was applied to compare the mean between two independent groups. P < 0.05 was considered statistically significant. For multi-group comparison, P-values were derived from one-way ANOVA with Tukey’s correction for multiple comparisons. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. Data were analyzed using Graph Pad Prism Version 6.01 software (San Diego, CA, USA).
Data Availability
The RNA-seq and ChIP-seq data that support the findings of this study have been deposited in the Gene Expression Omnibus under the accession code GSE247830 and GSE237793. Custom codes for compiling the mathematical models can be find in https://github.com/ganlanxin/clock.git.
References
Begemann, K. et al. Endocrine regulation of circadian rhythms. npj Biol Timing Sleep. 2, 10 (2025).
Bishehsari, F., Voigt, R. M. & Keshavarzian, A. Circadian rhythms and the gut microbiota: from the metabolic syndrome to cancer. Nat Rev Endocrinol. 16, 731–739 (2020).
Reinke, H. & Asher, G. Crosstalk between metabolism and circadian clocks. Nat Rev Mol Cell Biol. 20, 227–241 (2019).
Tu, H. Q. et al. Rhythmic cilia changes support SCN neuron coherence in circadian clock. Science. 380, 972–979 (2023).
Curtis, A. M., Bellet, M. M., Sassone-Corsi, P. & O’Neill, L. A. Circadian clock proteins and immunity. Immunity. 40, 178–186 (2014).
Aiello, I. et al. Circadian disruption promotes tumor-immune microenvironment remodeling favoring tumor cell proliferation. Sci Adv. 6, eaaz4530 (2020).
Cederroth, C. R. et al. Medicine in the Fourth Dimension. Cell Metab. 30, 238–250 (2019).
Blacher, E. et al. Aging disrupts circadian gene regulation and function in macrophages. Nat Immunol. 23, 229–236 (2022).
Roenneberg, T. & Merrow, M. The Circadian Clock and Human Health. Curr Biol. 26, R432–R443 (2016).
Aschoff, J. & Pohl, H. Phase relations between a circadian rhythm and its zeitgeber within the range of entrainment. Naturwissenschaften. 65, 80–84 (1978).
Bordyugov, G. et al. Tuning the phase of circadian entrainment. J R Soc Interface. 12, 20150282 (2015).
Duffy, J. F. & Czeisler, C. A. Age-related change in the relationship between circadian period, circadian phase, and diurnal preference in humans. Neurosci Lett. 318, 117–120 (2002).
Mendoza, J. Brain circadian clocks timing the 24h rhythms of behavior. npj Biol Timing Sleep. 2, 13 (2025).
Gronfier, C., Wright, K. P. Jr, Kronauer, R. E. & Czeisler, C. A. Entrainment of the human circadian pacemaker to longer-than-24-h days. Proc Natl Acad Sci USA. 104, 9081–9086 (2007).
Duffy, J. F. et al. Sex difference in the near-24-hour intrinsic period of the human circadian timing system. Proc Natl Acad Sci USA. 108, 15602–15608 (2011).
Santhi, N. et al. Sex differences in the circadian regulation of sleep and waking cognition in humans. Proc Natl Acad Sci USA. 113, E2730–E2739 (2016).
Koike, N. et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 338, 349–354 (2012).
Reppert, S. M. & Weaver, D. R. Coordination of circadian timing in mammals. Nature. 418, 935–941 (2002).
Annayev, Y. et al. Gene model 129 (Gm129) encodes a novel transcriptional repressor that modulates circadian gene expression. J Biol Chem. 289, 5013–5024 (2014).
Anafi, R. C. et al. Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 12, e1001840 (2014).
Goriki, A. et al. A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 12, e1001839 (2014).
Ono, D. et al. CHRONO and DEC1/DEC2 compensate for lack of CRY1/CRY2 in expression of coherent circadian rhythm but not in generation of circadian oscillation in the neonatal mouse SCN. Sci Rep. 11, 19240 (2021).
Yang, Y., Li, N., Qiu, J., Ge, H. & Qin, X. Identification of the Repressive Domain of the Negative Circadian Clock Component CHRONO. Int J Mol Sci. 21, 2469 (2020).
Mure, L. S. et al. Diurnal transcriptome atlas of a primate across major neural and peripheral tissues. Science. 359, eaao0318 (2018).
Zhou, Q. et al. The nuclear transportation of CHRONO regulates the circadian rhythm. J Biol Chem. 300, 107917 (2024).
Foteinou, P. T. et al. Computational and experimental insights into the circadian effects of SIRT1. Proc Natl Acad Sci USA. 115, 11643–11648 (2018).
Zhang, R., Lahens, N. F., Ballance, H. I., Hughes, M. E. & Hogenesch, J. B. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci USA. 111, 16219–16224 (2014).
Kwon, I. et al. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol. 26, 7318–7330 (2006).
Zheng, X. et al. RAE1 promotes BMAL1 shuttling and regulates degradation and activity of CLOCK: BMAL1 heterodimer. Cell Death Dis. 10, 62 (2019).
Relógio, A. et al. Tuning the mammalian circadian clock: robust synergy of two loops. PLoS Comput Biol. 7, e1002309 (2011).
Pett, J. P., Kondoff, M., Bordyugov, G., Kramer, A. & Herzel, H. Co-existing feedback loops generate tissue-specific circadian rhythms. Life Sci Alliance. 1, e201800078 (2018).
Padlom, A. et al. Level of constitutively expressed BMAL1 affects the robustness of circadian oscillations. Sci Rep. 12, 19519 (2022).
Shi, S. et al. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 20, 316–321 (2010).
Liu, A. C. et al. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 4, e1000023 (2008).
Vanselow, K. et al. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 20, 2660–2672 (2006).
Gabriel, C. H. et al. Circadian period is compensated for repressor protein turnover rates in single cells. Proc Natl Acad Sci USA. 121, e2404738121 (2024).
Vitaterna, M. H. et al. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci USA. 103, 9327–9332 (2006).
Jewett, M. E., Kronauer, R. E. & Czeisler, C. A. Light-induced suppression of endogenous circadian amplitude in humans. Nature. 350, 59–62 (1991).
Brown, S. A. et al. Molecular insights into human daily behavior. Proc Natl Acad Sci USA. 105, 1602–1607 (2008).
Guo, G. et al. Autokinase Activity of Casein Kinase 1 δ/ε Governs the Period of Mammalian Circadian Rhythms. J Biol Rhythms. 34, 482–496 (2019).
Baggs, J. E. et al. Network features of the mammalian circadian clock. PLoS Biol. 7, e52 (2009).
Wu, Y. et al. Systematic Studies of the Circadian Clock Genes Impact on Temperature Compensation and Cell Proliferation Using CRISPR Tools. Biology ((Basel)). 10, 1204 (2021).
Leloup, J. C. & Goldbeter, A. Modeling the mammalian circadian clock: sensitivity analysis and multiplicity of oscillatory mechanisms. J Theor Biol. 230, 541–562 (2004).
Acknowledgements
We are grateful to the staff for providing technical support with using the facility of Institute of Physical Science and Information Technology at Anhui University. This work was financially supported by the grants from the National Natural Science Foundation of China (32071158 to X.Q., 12171003 and 11971240 to D.L.), Anhui Provincial Natural Science Foundation (2008085MC68 to X.Q.). The funders had no role in study design, data collection and analysis.
Author information
Authors and Affiliations
Contributions
X.Q. and D.L. conceived and designed research; Q.Z., R.W. and S.L. performed research; L.G., X.Q. and D.L. wrote and ran computational simulations; all authors were involved in analyzing data; Q.Z., D.L. and X.Q. wrote and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, Q., Gao, L., Wang, R. et al. CHRONO regulates the circadian clock via the REV-ERBα – BMAL1 loop. npj Biol Timing Sleep 2, 22 (2025). https://doi.org/10.1038/s44323-025-00041-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44323-025-00041-5