Abstract
The development and implementation of microbial chassis cells have profound impacts on circular economy. Non-model bacterium Zymomonas mobilis is an excellent chassis owing to its extraordinary industrial characteristics. Here, the genome-scale metabolic model iZM516 is improved and updated by integrating enzyme constraints to simulate the dynamics of flux distribution and guide pathway design. We show that the innate dominant ethanol pathway of Z. mobilis restricts the titer and rate of these biochemicals. A dominant-metabolism compromised intermediate-chassis (DMCI) strategy is then developed through introducing low toxicity but cofactor imbalanced 2,3-butanediol pathway, and a recombinant D-lactate producer is constructed to produce more than 140.92 g/L and 104.6 g/L D-lactate (yield > 0.97 g/g) from glucose and corncob residue hydrolysate, respectively. Additionally, techno-economic analysis (TEA) and life cycle assessment (LCA) demonstrate the commercialization feasibility and greenhouse gas reduction capability of lignocellulosic D-lactate. This work thus establishes a paradigm for engineering recalcitrant microorganisms as biorefinery chassis.
Similar content being viewed by others
Introduction
The improvement on sustainable waste management and resource utilization efficiency can help reduce pollution and strengthen sustainable biochemical production to achieve the goal of circular economy1. Non-food materials such as lignocellulose, glycerol, molasses, and waste gas serve as sustainable feedstocks of biomanufacturing2,3,4. The construction of efficient microbial cell factories is crucial for the sustainable conversion of low-cost non-food materials into high-value biochemicals5.
The rapid progress in systems biology and synthetic biology has greatly accelerated the development of efficient genome-engineering toolkits and effective microbial cell factories for diverse biochemicals production5,6. However, most achievements in this field have been accomplished using model microorganisms such as Escherichia coli, Saccharomyces cerevisiae, Corynebacterium glutamicum, and Bacillus subtilis because of their clear genetic background, comprehensive genome-editing tools, as well as abundant biological parts and devices7,8,9,10.
Given the limited number of model species and inherent drawbacks associated with their use in industrial production, significant efforts have been focused on identifying and developing non-model microorganisms with excellent characteristics for biomanufacturing, such as Vibrio natriegens (fast growth)11, Acinetobacter baylyi (high natural transformation efficiency)12, Lactococcus lactis (lactate production)13, and Halomonas bluephagenesis (PHA production)14. In addition, some non-model microorganisms possess diversified industrial characteristics that make them suitable for engineering as a biorefinery chassis, such as robustness against severe environmental conditions as well as toxic inhibitors, biosynthetic intermediates and end-products, broad spectrum of substrate utilization and biochemical production capabilities, high yield of target products with few by-products, innate immunity from phage infection and stable genome structure15,16,17. However, compared to model microorganisms, the lack of abundant information and efficient genome-engineering toolkits for non-model microorganisms limits their development and applications despite the continuous development of numerous tools for non-model species with microbial cell factories being constructed at an unparalleled speed for industrial production. Efficient and effective toolkits and pipelines for enzyme selection, rational pathway design, and genome engineering are still in their infancy. Tremendous efforts are needed to expedite the development of diverse biorefinery chassis and their derivative cell factories for sustainable circular bioeconomy. More importantly, non-model microorganisms require more systematic research, including the utilization of multiple feedstocks and production of diverse products, to become the biorefinery chassis while being developed into specific cell factories.
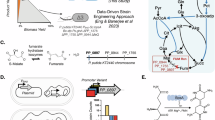
Zymomonas mobilis is a non-model polyploid ethanologenic Gram-negative bacterium with many industrial merits and unique physiological properties18, which is the only known microorganism that can utilize the Entner-Doudoroff (ED) pathway under anaerobic conditions19 and possesses excellent characteristics such as high sugar uptake rate, high ethanol yield and ethanol tolerance20. Significant efforts have been continuously made to achieve accurate genome sequence and annotation21,22, as well as significant amounts of omics datasets21,22,23, which have been used for the prediction and expansion of biological parts such as RBS and promoters24,25. In addition, efficient genome-editing tools based on heterologous CRISPR-Cas12a, endogenous Type I-F CRISPR-Cas, and associated repair pathways, such as microhomology-mediated end joining (MMEJ) have been developed26,27,28. Several microbial cell factories have been constructed for the production of various biochemicals, including acetaldehyde, D-lactate, L-alanine, L-serine, acetoin, 2,3-butanediol (2,3-BDO), isobutanol, L-malate, succinic acid, poly-3-hydroxybutyrate (PHB), farnesene, and ethylene29,30,31,32,33,34,35,36,37,38,39,40 (Fig. 1). Moreover, an improved genome-scale metabolic model (GEM) of iZM516 for Z. mobilis was recently constructed, which contains the most reactions, metabolites, and genes among all other reported models. This model was used to design the metabolic pathways for the production of succinate and 1,4-butanediol (1,4-BDO)41.
The blue font in the pathway represents the key enzymes. The solid line represents a one-step reaction, while the dotted line indicates a multi-step reaction. The thick blue solid line represents the native recalcitrant ethanol pathway. 2,3-BDO 2,3-butanediol, DMAPP dimethylallyl diphosphate, DXP 1-deoxy-D-xylulose-5-phosphate, ED Entner-Doudoroff pathway, FPP farnesyl diphosphate, G3P glyceraldehyde-3-phosphate, G6P glucose-6-phosphate, GPP geranyl diphosphate, IPP isopentenyl diphosphate, MEP methylerythritol 4-phosphate pathway, PHB poly-3-hydroxybutyrate, PPP pentose phosphate pathway, TCA tricarboxylic acid cycle.
Despite these accomplishments, the primary obstacle in making Z. mobilis a chassis for non-food feedstock biorefinery19,42 lies in circumventing its dominant ethanol production pathway from pyruvate comprised of efficient pyruvate decarboxylase (PDC) and alcohol dehydrogenases (ADHs). A central-carbon metabolism control-valve strategy has been explored to redirect the carbon flux to the 2,3-BDO, lactate, or isobutanol pathways with ca 65% overall yield from glucose43. Similarly, efforts have been made to shift ethanol metabolism towards lactate production by replacing the strong promoter of pdc with an inducible promoter Ptet31. However, none of these studies have succeeded in completely eliminating Z. mobilis’ dominant ethanol pathway, thereby limiting its potential as a biorefinery chassis for the production of biochemicals other than ethanol with a high titer, yield, and rate.
In this work, we improve the GEM iZM516 of Z. mobilis ZM4 by integrating enzyme constraints to obtain eciZM547 for the rational design of metabolic pathways for C2-C5 biochemical production. We design and construct metabolic pathways for the production of 1,3-propanediol (1,3-PDO) from glycerol, butanediol from glucose, xylonic acid, ethylene glycol, glycolic acid, and 1,4-butanediol from xylose using Z. mobilis as the chassis. We then develop a metabolic strategy to bypass dominant metabolism by first constructing a metabolism-comprised intermediate chassis instead of directly engineering the chassis for the target biochemicals. Subsequently, we construct a D-lactate producer successfully. Finally, we evaluate the economic feasibility of D-lactate production using corncob residue hydrolysate (CRH) through techno-economic analysis (TEA) and life cycle assessment (LCA). We envision that the dominant-metabolism compromised intermediate-chassis will be a valuable strategy for efficiently guiding biochemical production in many microorganisms for circular economy.
Results
Integration of enzyme constraints to improve the genome-scale metabolic model
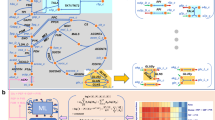
High-quality GEMs play critical roles in the rational design of microbial cell factories in the classical Design-Build-Test-Learn cycle of synthetic biology studies41. A high-quality GEM iZM516 of Z. mobilis ZM4 was constructed recently with the MEMOTE evaluation score of 91% among all published models, which contains 1389 reactions, 1437 metabolites, 516 genes, and 3 cell compartments41. Recently, numerous ecModels have been applied to model microorganisms such as E. coli, S. cerevisiae, C. glutamicum, and B. subtilis with superior potential to identify rate-limiting enzymes, simulate overflow metabolism, and elucidate the trade-off between biomass yield and enzyme usage efficiency44,45.
The previous model of iZM516 built upon the ModelSeed database differs from iZM4_47846 in terms of Gene-Protein-Reaction (GPR) relationships by the BiGG database47. The unique genes and reactions from iZM4_478 were manually curated and incorporated into iZM516 to obtain the revised iZM547 in this study that contains 2501 reactions, 1455 metabolites, and 547 genes. However, it is important to note that iZM547 only considers stoichiometric constraints and does not reflect the cellular status or provide targets that limit flux through specific product synthesis pathways. With the accumulation of enzyme kinetic data, the predictive accuracy of models can be further enhanced by integrating enzyme constraints that reflect limitations related to protein resources during cell growth. Hence, we applied ECMpy244 and Kcat values provided by AutoPACMEN48, which is the closest to the experimental results and more accurate to other methods such as DLkcat49, TurNup50, and UniKP51, for developing an enzyme-constrained model (ecModel) (Supplementary Fig. 1). The resulting eciZM547_AutoPACMEN_mean (subsequently abbreviated as eciZM547) is the enzyme-constrained metabolic network model closest to the experimental results (Supplementary Fig. 1), and ultimately selected for the subsequent analyses.
A relatively comprehensive comparison between iZM516 and iZM4_478 has already been conducted in previous work41. The results indicate that iZM516 exhibits superior simulation accuracy regarding strain behavior according to the report41. As shown in Fig. 2, small discrepancies between iZM516 and eciZM547 were observed, mainly manifested in the existence of a maximum in the enzyme-constrained model eciZM547 when the value of glucose uptake exceeds around 71 mmol·gDW−1·h−1. This indicates a shift from glucose-limited growth to proteome-limited growth. The maximum predicted growth rate and ethanol production rate were 0.50 h−1 and 134.76 mmol·gDW−1·h−1, respectively (Fig. 2). Therefore, eciZM547 exhibited better accuracy than the previous model, which highly overestimated the maximum growth rate and ethanol production rate.
a Predictions for specific cell growth rates. b Predictions for ethanol production fluxes. iZM4_478 is a model published recently41. iZM516 is a revised model version constructed in this study on the basis of the published one. eciZM547 is the enzyme-constrained version on the basis of revised iZM516. The bullet points with different color and shape were experimental data from published literature52,82,84,85.
Additionally, iZM516 indicates that most carbon sources are directed towards acetate based on growth criteria when glucose was served as the sole carbon source with an uptake rate of 10 mmol·gDW−1·h−1 under aerobic conditions, whereas eciZM547 calculates that carbon sources flow into both acetate and acetoin, aligning more closely with our previous data36. To further investigate the aforementioned simulation results, we conducted the 13C-metabolic flux analysis (MFA) of ZM4 under aerobic condition, and subsequently calculated the distributions of metabolic flux (Supplementary Fig. 2a). Consistent with the predictions made by eciZM547, a greater influx of carbon sources into both acetate and acetoin was observed compared to the anaerobic condition reported before52. In addition, in the classical revised model iZM516, the growth rate increased linearly with the substrate uptake. In contrast, the enzyme constraint model of eciZM547 narrowed the solution space, resulting in predictions that are closer to actual experimental conditions in Z. mobilis (Fig. 2).
To investigate the potential of endogenous C2-C5 biochemicals synthesis pathway in Z. mobilis ZM4, we calculated and evaluated the metabolic pathways, flux rates, and theoretical conversion rates of biochemicals through simulation and analyses using the iZM516 and eciZM547 models, respectively (Table 1).
Besides ethanol, lactate, isobutanol, 2,3-BDO, and xylitol also had the same rates under anaerobic or aerobic conditions (Table 1), revealing the advantages of Z. mobilis in producing these biochemicals anaerobically, which is crucial for commodity biochemical production since production anaerobically can save the cost around 30–50%. The yield of 1,4-BDO decreases under enzyme constraints (Table 1). However, as the only microorganism using the ED pathway under anaerobic conditions, the conversion rate of 1,4-BDO remained consistent regardless of anaerobic or aerobic conditions as predicted by either iZM516 or eciZM547 models (Table 1).
Although the rate of succinate production under aerobic condition with the supplementation of CO2 was higher than that under anaerobic condition, it remains feasible to develop a cell factory of Z. mobilis for anaerobic succinate production at a rate of 0.9 (Table 1). The significant disparity in production rates between anaerobic and aerobic processes for itaconic acid and L-alanine indicates that Z. mobilis is more effective for anaerobic production, as suggested by eciZM547 simulation results (Table 1). Moreover, the low rates of ethylene glycol with glucose and xylose as carbon sources under aerobic or even less under anaerobic conditions imply that Z. mobilis is not an appropriate chassis for ethylene glycerol production (Table 1). In contrast to ethylene glycol, the rate of 1,3-PDO when utilizing glucose as a carbon source is relatively low. However, a higher rate of 1,3-PDO derived from glycerol suggests its feasibility for production using Z. mobilis (Table 1).
Construction of microbial cell factories for C2-C5 biochemical production
A series of recombinant strains were then constructed for biochemical production based on model-guided pathway design, including succinate from glucose, 1,3-PDO from glycerol, xylonic acid, ethylene glycerol, glycolic acid, and 1,4-BDO from xylose (Fig. 3a).
a Model-guided pathway design strategy and metabolic pathway of biochemicals production from glucose, xylose, or glycerol. b Summary of succinic acid production in recombinant Z. mobilis with different strategies. c The titer of ethylene glycerol, xylonic acid, glycolic acid, and 1,4-butanediol in recombinant strains from xylose through Dahms pathway. d The titer of 1,3-PDO from glycerol by overexpressing different dhaB. The solid represents a one-step reaction, while the dotted line indicates a multi-step reaction. Metabolites: 2D3DXA: 2-dehydro-3-deoxy-D-xylonate, 5HKG: 5-hydroxy-α-ketoglutarate, 6PG: 6-phospho-gluconate, AcCoA: acetyl-CoA, EG: ethylene glycol, GA: glycolate, G3P: glyceraldehyde-3-phosphate, G6P: glucose-6-phosphate, HBA: hydroxy-butyraldehyde, KdcA: keto-acid decarboxylase, KDPG: 2-keto-3-dehydro-6-phosphogluconate, KGAS: ketoglutaric semialdehyde, OAA: oxaloacetate, PEP: phosphoenolpyruvate, XA: xylonic acid. Enzymes: Adh: alcohol dehydrogenase, AldA: aldehyde dehydrogenase, DhaB: glycerol dehydratase, Frd: fumarate reductase, FumC, fumarate hydratase, Mdh, malate dehydrogenase, Pdc: pyruvate decarboxylase, Ppc, phosphoenolpyruvate carboxylase, Xdh: D-xylose dehydrogenase, XylX: 2-dehydro3-deoxy-D-xylonate dehydratase, YagE: 2-dehydro-3-deoxy-D-xylonate aldolase, YagF: D-xylonate dehydratase, YqhD: aldehyde reductase. Genes: CbdhaB from Clostridium butyricum, CfdhaB from Citrobacter freundii, Kppdu and kpdhaB were from Klebsiella pneumoniae. Brown font represents carbon source. Blue, green, orange background colors represent C2, C3, C4 compounds, respectively. Data are presented as mean ± s.e.m. (n = 3 biologically independent samples). Statistical analysis was performed using a two-tailed Student t-test. ****p < 0.0001, ***p < 0.001 versus wild-type strain. Source data are provided as a Source Data file.
Succinic acid has been designated as one of the top 12 building block chemicals by the U.S. Department of Energy for its unique advantages in chemical, food, agricultural, and plastic industries53, and microbial cell factories using E. coli and other microorganisms have been developed for succinic acid production54. Previous model simulations employing iZM516 have indicated that succinic acid production under anaerobic condition using Z. mobilis exhibits significant economic advantages41. The model-guided metabolic pathway was subsequently optimized through the co-expression of genes involved in the reductive TCA cycle and the individual expression of malate dehydrogenase (encoded by mdh) from C. glutamicum to enhance succinic acid production (Supplementary Fig. 3a). Phosphoenolpyruvate carboxylase (encoded by ppc), fumarate hydratase (encoded by fumC), and fumarate reductase (encoded by frd) from different sources were screened and tested using the strain SA3 with mdh overexpression as the parental strain (Supplementary Fig. 3b–d). Although there was no significant increase in succinic acid production in these recombinant strains (Fig. 3b), enhanced CO2 supply can significantly improve succinic acid production, as predicted by the model (Table 1, Fig. 3b). Further overexpression of mdh and ppc from C. glutamicum, fumC and frd from S. cerevisiae in Z. mobilis with the supplementation of NaHCO3 can generate 560 mg/L succinic acid (Fig. 3b). Despite attempts to divert carbon from ethanol to succinic acid using the CRISPRi system, no significant improvement was achieved owing to the robust metabolic characteristic of the ethanol pathway and redox imbalance present in Z. mobilis (Supplementary Fig. 4).
Xylose is the second most abundant sugar, accounting for 18–30% of lignocellulose components. Efficient utilization of xylose is crucial for economical biochemical production from renewable biomass55. Z. mobilis has been engineered to produce ethanol from xylose by introducing the xylose assimilation and pentose phosphate pathway genes56,57. Xylose dehydrogenase (Xdh) catalyzes the first step of the Dahms pathway from xylose to xylonic acid (Fig. 3a). The codon-optimized xdh gene from Paraburkholderia xenovorans58 driven by the strong promoter Ppdc in pXA1 was constructed to produce 16.78 ± 1.58 g/L xylonic acid with CaCO3 supplementation in RMG2X2 (Fig. 3c). The introduction of the heterologous Dahms pathway could effectively convert xylonic acid into glycolaldehyde and the biosynthesis of ethylene glycol in Z. mobilis based on model prediction using eciZM547. The strong promoter of Peno was used to control the expression of the genes of yagF, yagE, and yqhD from E. coli (Supplementary Fig. 5). Following CaCO3 supplementation in the recombinant strain EG2 (Fig. 3c), the final ethylene glycerol titer reached 3.26 ± 0.07 g/L. Furthermore, pyruvate generated from the Dahms pathway can be further converted to ethanol (Fig. 3a). By replacing yqhD with aldA, it is possible to achieve a production of 1.5 g/L glycolic acid in the recombinant strain GA (Fig. 3c). In addition, 1,4-butanediol production from xylose can be achieved to 1.2 g/L in the recombinant strain BDO1 by modifying the Dahms pathway and introducing genes including yagF, xylX, yqhD, and kdcA (Fig. 3c).
Glycerol is a promising renewable waste feedstock in biotechnology for the production of high-value products such as 1,3-PDO, ethanol, lactate, and succinic acid59. 1,3-PDO has various applications in food, cosmetics, pharmaceuticals, and biomaterials, as it serves as a monomer for the production of polyesters such as polypropylene terephthalate (PTT)60. In this study, we tested the feasibility of producing 1,3-PDO using glycerol in Z. mobilis by introducing glycerol dehydratase based on eciZM547 model prediction. By comparing different sources of dhaB and co-expressing the aldehyde reductase gene yqhD, the optimized recombinant strain could achieve 4.1 g/L 1,3-PDO from glycerol (Fig. 3d). The model prediction results also indicated that the Embden-Meyerhof-Parnas (EMP) pathway has a greater advantage in 1,3-PDO synthesis than the ED pathway of Z. mobilis (Supplementary Fig. 6). Therefore, we attempted to complement the truncated EMP pathway of Z. mobilis by co-expressing phosphofructokinase (encoded by pfk from Borrelia burgdorferi), fructose bisphosphate aldolase (encoded by fba from Z. mobilis), and triosephosphate isomerase (encoded by tpi from Z. mobilis) to increase glycerol biosynthesis for 1,3-PDO production (Supplementary Fig. 7). The results showed that while introducing the EMP pathway resulted in a slight reduction in glucose consumption and ethanol synthesis of the recombinant strain, the ethanol yield remained unchanged. Notably, however, the glycerol yield increased from 0.28 g/L to 0.8 g/L (Supplementary Fig. 7). Therefore, completion of the EMP pathway proves advantageous for enhancing glycerol production and further utilization of carbon source.
Development of dominant-metabolism compromised intermediate-chassis strategy to reconfigure ethanologen Z. mobilis into a D-lactate producer
Despite the successful construction of several cell factories in this study aimed at producing C2-C5 biochemicals using glucose, xylose, or glycerol as carbon sources, the titers of these biochemicals remain insufficient for industrialization. This limitation is primarily attributed to the dominant ethanol pathway, which diverts carbon flux to ethanol production. We were unable to directly replace the ethanol pathway with the D-lactate pathway by replacing the pdc gene with D-ldh. This challenge arises because both pathways utilize pyruvate as the substrate, and are redox and cofactor balanced (Fig. 4a). This result is consistent with a previous report, which indicated that the complete replacement of ethanol with the biosynthesis pathway using inducible promoters to regulate pdc expression led to a maximum conversion rate of only 70%43.
a Construction and application of the 2,3-BDO intermediate-chassis for D-lactate production in Z. mobilis. b Process of constructing D-lactate producer of Z. mobilis using the 2,3-BDO intermediate-chassis. c Flask fermentation results of glucose consumption and D-lactate production of the recombinant D-lactate producer ZMB6 in different concentrations of glucose. d Fermentor results of glucose consumption and D-lactate production of the recombinant strain ZMB6 using corncob residue hydrolysate (CRH). Adh: alcohol dehydrogenase, AldC: acetolactate decarboxylase, Als: acetolactate synthase, Bdh: butanediol dehydrogenase, LmldhA (Ldh): lactate dehydrogenase from L. mesenteroides, Pdc (encoding by ZMO1360): pyruvate decarboxylase. Data are presented as mean ± s.e.m. (n = 2 biologically independent samples). Source data are provided as a Source Data file.
The heterologous 2,3-BDO pathway was then introduced first into Z. mobilis, which is redox imbalanced with only one NADH comsumption35 (Fig. 4a). Therefore, microaerobic conditions are essential for optimal cell growth to maintain redox balance by oxidizing the extra NADH through respiration with Ndh, which also helps maintain the optimal NADH level for cofactor balancing (Fig. 4a). In addition, the growth of Z. mobilis is not inhibited by elevated concentrations of 2,3-BDO, and Z. mobilis can tolerate to above 100 g/L 2,3-BDO indicating its potential for 2,3-BDO production at high titers35.
Then, the model eciZM547 was used to simulate the result of replacing ethanol pathway with D-lactate pathway in the 2,3-BDO producer background. When glucose was served as the sole carbon source at an uptake rate of 40 mmol·gDW−1·h−1, the quantity of ammonia as the sole nitrogen source was unlimited with biomass as the objective. The results indicated that carbon flux will be shifted toward D-lactate pathway upon knocking out ethanol pathway gene ZMO1360 (pdc). Furthermore, when both ethanol and D-lactate metabolic pathways were knocked out, the enzyme-constrained metabolic model was able to utilize the 2,3-BDO pathway as an alternative for normal cell growth, which is consistent with subsequent experimental results. However, meaningful results cannot be obtained when using eciZM547 to simulate the replacement of ethanol pathway with biochemicals such as isobutanol and 1,3-PDO due to the imbalance in NADH/ NAD+ and ATP/ADP ratios.
Hence, the 2,3-BDO producer of Z. mobilis can thus be regarded as the dominant ethanol metabolism-comprised strain under anaerobic condition. This strain was then used in the construction of a D-lactate producer by knocking out pdc gene, using the 2,3-BDO producer as the intermediate chassis (Fig. 4a). As illustrated in Fig. 4b, the wild-type strain ZM4 produces a significant amount of ethanol at a yield of 0.48 g/g. A synthetic operon for high 2,3-BDO production was introduced into the genome of ZM4 to obtain the recombination strain ZMB1, which can produce ethanol, 2,3-BDO, and acetoin at a yield of 0.12, 0.11, and 0.28 (g/g), respectively. To further improve the yield of 2,3-BDO, an additional copy of bdh was integrated at the ZMO0038 chromosomal locus, resulting in the decrease of acetoin and the increase of 2,3-BDO in the recombinant strain ZMB2. Subsequently, the pdc gene was knocked out in ZMB2 to obtain ZMB3 strain, which does not produce ethanol and converts almost all carbon into 2,3-BDO at a yield of 0.42 g/g with only a small amount of acetoin being produced. Therefore, a dominant-metabolism compromised intermediate chassis ZMB3 with 2,3-BDO production was constructed.
To determine the metabolic flux distribution of ZMB3, labeling data from [1,2–13C] tracer experiments with biomass, 2,3-BDO, acetoin production rates were simultaneously fitted to a single flux map of eciZM547 including central carbon metabolism as well as the biosynthesis of 2,3-BDO and acetoin (Supplementary Data 1). During exponential growth, over 96% glucose flux was converted to 2,3-BDO synthetic pathway from ethanol via ED pathway, and the remaining central metabolic reactions, including the TCA cycle and amino acid biosynthesis, had low activity with minimal flux to fulfill biosynthetic needs, which was highly consistent with the results predicted by our model. Additionally, ZMB3 exhibited better fermentation characteristics under aerobic condition compared to ZM4 (Supplementary Table 1), which may be attributed to the enhanced redox balance of ZMB3 under aerobic condition. Therefore, the recombinant strain ZMB3 can be regarded as an excellent aerobic dominant-metabolism compromised intermediate chassis aimed at synthesizing other biochemicals.
Consequently, bdh was replaced by LmldhA gene encoding D-lactate dehydrogenase to obtain ZMB4 for the production of D-lactate and 2,3-BDO at yields of 0.74 and 0.15 g/g, respectively. To further enhance D-lactate yield, another copy of LmldhA was integrated into the chromosomal locus ZMO1759 to obtain strain ZMB5. Finally, a recombinant strain ZMB6 was constructed with three copies of D-lactate dehydrogenase engineered into 2,3-BDO intermediate chassis (Fig. 4b). To assess the fermentation performance and D-lactate production of ZMB6, shake flask fermentation was carried out using RM medium with varying glucose concentrations. Nearly all glucose was converted into D-lactate with a titer of 140.92 g/L and a yield of 99% by the recombinant strain ZMB6 (Fig. 4c, Supplementary Table 2).
Approximately 40–70% of total costs are allocated to fermentation substrates such as sugars in microbial lactate production61. The potential of ZMB6 using non-food feedstock of corncob residue hydrolysate as a substrate was then evaluated. The results demonstrated a high conversion rate of glucose to D-lactate (Fig. 4d). Due to the advantages of abundance, low price, and ability to mitigate conflicts with food supplies, it is an attractive and promising approach for lactate production using the non-food feedstocks. In a scale-up fermenter with corncob residue hydrolysate, ZMB6 also demonstrated excellent glucose conversion capacity with a D-lactate titer of 104.6 g/L at a yield exceeding 0.97 g/g within 45 h (Fig. 4d), along with an optical purity of 99.1% (Supplementary Tables 2 and 3). These findings indicated the significant advantages in commercial applications.
TEA and LCA of D-lactic acid cell factory
The economic feasibility of utilizing lignocellulosic residues from corncob waste for D-lactate production by recombinant Z. mobilis was evaluated (Fig. 5a). The designed production scenario demonstrated the potential to generate over 31,100 tons of D-lactate annually, representing approximately 1.7% of the global lactic acid market, thus establishing a significant market share for the emerging bioeconomy. The comprehensive depiction of assumptions and carbon flow diagram are provided in Supplementary Table 4 and Supplementary Fig. 10. Additionally, the total capital investment (TCI) and total operating cost (TOC) associated with the proposed processes were calculated and listed in Supplementary Tables 5 and 6. Our analysis reveals that with the advantages of lower feedstocks price, higher yield, and scale-up effect in this study, the minimum selling price (MSP) for D-lactate amounted to USD 0.35 kg−1, remarkably lower than the current market price for lactic acid, set at USD 3.15 kg−1, serving as a benchmark for comparison62,63. When considering the cost of corn stover pretreatment as part of the capital investment (represents approximately 12.89% of the total cost for equipment and installation64), the MSP of lactic acid is estimated to be around USD 0.37 kg−1. The competitive advantage of MSP primarily derives from utilizing corncob residues, typically regarded as industrial waste, with a feedstock cost of merely $38.7 per tonne. This is substantially lower than alternative carbon sources like corn dextrose or molasses (Supplementary Table 7), leading to a significant reduction in raw material costs and TOC. The titer, productivity, and yield of D-lactate of this study are 104.6 g/L, 2.32 g/L/h, and 97%, respectively, which are higher than the values reported in previous studies (Supplementary Table 7). The comparisons of the economic feasibility (TCI, TOC, MSP) of lactate from previous literature reports (Supplementary Tables 7 and 8) reveal that the MSP identified in this study aligns with prior findings and falls within the lower range of reported MSP, which fluctuate based on production capacity, feedstock types, and process design, thereby indicating the economic feasibility of D-lactate production using the recombinant strain ZMB6 developed in this study.
a Schematic diagram of the industrial production process of D-lactate. b Single-point sensitivity analysis of the minimum selling price to produce 31,100 tons/year of D-lactate. c Minimum selling price (MSP) of lactic acid in various prospective scenarios. d Contribution analysis of the cradle-to-gate global warming potential to produce 1 kg of D-lactate. e Global warming potential comparison of producing 1 kg of lactic acid and fossil-based organic acid. D-LA: D-lactate.
As depicted in Fig. 5b, a single-point sensitivity analysis was conducted to assess seven cost-driving forces based on reasonable ranges, which illustrated the maximum and minimum values of the MSP of D-lactate derived from corncob residues. The analysis demonstrated that the MSP fluctuated from USD 0.29 to 0.45 kg−1. Importantly, the high sensitivity to annual plant capacity is aligned with the supply of corncob residue, thereby influencing the TCI, TOC, and MSP. Doubling the corncob residue supply from 5000 to 10,000 kg/h leads to a decrease in MSP to USD 0.29 kg−1. In addition, D-lactate productivity and titer stand out as the most critical production factors influencing overall costs. Reduced productivity and titer can result in a notable increase in MSP, largely attributed to the substantial correlation between TCI and annual plant capacity. However, the impact of productivity and titer on MSP decreases above 1 g/L/h and 50 g/L, respectively (Supplementary Fig. 9), indicating a reduced impact at certain levels of productivity and titer.
A comparison using combined sensitivity analysis can provide further insights into how improving fermentation parameters affects MSP (Fig. 5c). The analysis includes the base case (baseline in the single-point sensitivity analysis), short-term case, and long-term case. Rational predictions for lactic acid productivity, titer, and yield were made based on the optimization of genetic engineering. The selection of long-term factor values was determined by the highest value observed in the single-point sensitivity analysis. The MSP for the short-term and long-term scenario can be USD 0.31 kg−1 and USD 0.28 kg−1, which is reduced by 11% and 20% compared to the base case, respectively. Although the MSP of the base case has shown impressive economic performance, improvements in fermentation-related parameters would further enhance its economic viability.
In addition to economic considerations, a preliminary LCA was also conducted, including results and process simulation using Aspen Plus v14, to demonstrate the global warming potential (GWP) associated with the production of 1 kg D-lactate. The GWP of producing 1 kg D-lactic acid from the corncob residue hydrolysate was determined to be 0.49 kg CO2-eq (Fig. 5d). A contribution analysis was further performed to identify environmental hotspots throughout the process. The utilization of cellulolytic enzyme (Cellic® CTec3, Novozyme, Denmark) was the most significant driver, accounting for 57.5% of all categories, followed by the conversion process at 39.8%. The proposed pathway for D-lactate production has the potential to further reduce greenhouse gas (GHG) emissions by integrating alternative renewable energy sources, such as solar, wind, and geothermal energy.
Upon comparing production data from various non-food feedstocks, including lignocellulosic biomass, vine shoots, corn stover, sugarcane bagasse, and brown leaves, with the corncob residue hydrolysate used in this study, it was noteworthy that the biorefinery GWP demonstrated a reduction of up to 22.51 tons of GHG emissions per ton of D-lactate produced65,66,67,68 (Fig. 5e). Moreover, the utilization of this pathway could result in a reduction of GHG emissions by up to 98.16% when compared to fossil-based organic acids (Fig. 5e). Thus, D-lactate production with Z. mobilis from corncob residue hydrolysate processes not only offers a practical method for reusing biomass in an industrial setting but also has the potential to mitigate local GHG emissions and ultimately to reduce the overall carbon footprint.
Discussion
The success of circular economy requires the identification and development of efficient chassis cells with diverse industrial characteristics, enabling us to engineer them as biorefinery cell factories for the economic production of a broad spectrum of biochemicals using non-food substrates such as lignocellulosic biomass, molasses, and CO269,70. Model microorganisms have been mostly studied and developed due to their clear genetic background with rich knowledge, efficient genome-engineering toolkits, and abundant biological parts and devices. However, model species often lack industrial characteristics such as vulnerability to extreme environmental conditions and toxic inhibitors. In addition, the ease of genome engineering for model species could also lead to proneness of phage infection and genome instability71,72. Therefore, there is growing interest in exploring non-model species with excellent industrial features for diverse biochemical production using a wide range of substrates72. Taking the recalcitrant polyploid ethnologic bacterium Z. mobilis as an example, we have developed a strategy to help develop non-model species biorefinery chassis.
An improved GEM model of eciZM547 was first constructed in this study by incorporating enzyme constraints, which provides a more accurate prediction of metabolic pathway than the previous iZM516 to guide rational design of metabolic pathways for creating efficient cell factories. For example, eciZM547 verified that 1,3-PDO production via the EMP pathway anaerobically in Z. mobilis is superior to that under aerobic condition. Guided by the improved GEM, a series of cell factories were successfully constructed using Z. mobilis as the chassis. These cell factories are capable of producing various biochemicals, including succinic acid from glucose, ethylene glycol, glycolic acid, and 1,4-butanediol from xylose, and 1,3-PDO from glycerol (Fig. 6).
More importantly, the GEM model simulation also suggested that Z. mobilis is well-suited for the economic production of ethanol, lactic acid, isobutanol, 2,3-BDO, and xylitol under anaerobic conditions, providing a direction for bioproducts selection in industrial applications (Table 1). Under aerobic conditions, a more significant yield increase was observed for succinic acid and itaconic acid, two compounds related to the TCA cycle. Therefore, further modifications are necessary to achieve efficient production under aerobic conditions (Table 1). With the development of artificial intelligence (AI)-driven approaches, more accurate and extensive GEMs will be developed to intelligently simulate various metabolic activities within microbial chassis cells to expedite the development of efficient microbial cell factories for industrial applications44.
To overcome the challenge of recalcitrant microorganisms with dominant native pathways, we developed a paradigm of dominant-metabolism compromising intermediate-chassis strategy with low toxicity and cofactor relative imbalanced to reconfigure the ethanologenic bacterium Z. mobilis into a D-lactate producer. We constructed an intermediate chassis of Z. mobilis that compromises its dominant ethanol pathway while producing 2,3-BDO, which was subsequently employed to completely reconfigure the recalcitrant ethanologenic bacterium Z. mobilis into a D-lactate producer. The recombinant strain ZMB6 can produce pure D-lactate from glucose and corncob residue hydrolysate with a titer of 140.92 g/L and 104.6 g/L at a yield exceeding 0.97 g/g, respectively. TEA and LCA also confirmed that D-lactate produced from corncob residue hydrolysate can effectively mitigate the environmental impact in industrial applications, and is economically feasible at a price of USD 0.35 kg−1. It is worth noting that although the yield of D-lactic acid increases with prolonged fermentation time, productivity is significantly reduced as a trade-off. Therefore, it is crucial to further improve overall economy viability by increasing product titer, rate, and yield (Supplementary Fig. 9).
In addition to Z. mobilis, S. cerevisiae is another model ethanologenic microorganism with excellent industrial merits. However, inactivation of PDCs strains is notoriously known for inability to grow in the presence of excess glucose medium due to NADH/NAD+ imbalance under aerobic condition73. Similar to Z. mobilis, recent research has determined that the introduction of the 2,3-BDO pathway directly into S. cerevisiae results in a coupled anaerobic 2,3-BDO and glycerol production through the ecGEM prediction in ethanol block-out strains74. A previous study also showed that the introduction of 2,3-BDO and lactic acid pathway significantly improved NADH-consuming73. Hence, we propose that a dominant-metabolism compromising intermediate-chassis strategy remains applicable to S. cerevisiae for anaerobic biochemicals production, and a more comprehensive GEM in S. cerevisiae can efficiently guide biochemicals production without ethanol constraints.
The unique physiological and genetic features of Z. mobilis evolved to adapt to the unique environment, make it a robust chassis for pyruvate-derived extracellular biochemical production such as the innate ethanol biosynthesis pathway or the D-lactate production pathway constructed in this study. The successful reconfiguration of the ethanologenic bacterium Z. mobilis into several cell factories for C2-C5 biochemical production using glucose, xylose, or glycerol as substrates in this study (Fig. 6) demonstrates its feasibility to develop Z. mobilis as efficient cell factories for the commercialization of other biochemicals such as L-alanine, isobutanol, and 2,3-BDO using pyruvate as the direct precursor32,35,36.
However, it is disadvantageous to construct cell factories for producing energy-consuming biochemicals using Z. mobilis since there is only one molar of ATP produced from one molar of glucose because of its unique anaerobic ED pathway and truncated EMP and TCA. For example, compared to the classical EMP pathway, the only known anaerobic ED pathway provides efficient substrate utilization but limits its ability to synthesize compounds such as L-serine and 1,3-PDO33 using glyceraldehyde-3-phosphate as precursor. Additionally, incomplete TCA results in high energy utilization efficiency with declined biomass, which hinders the economic production of intracellular biochemicals39.
Although metabolic engineering, random mutagenesis, and adaptive laboratory evolution can be applied to reconfigure Z. mobilis for enhanced energy production and biomass generation, designing and predicting these modifications remains a significant challenge. This difficulty arises from a significant knowledge gap in gene functions, physiological and information metabolism, as well as regulatory signals and networks. Therefore, more microbial chassis with different advantages are needed, and it is crucial to understand the strengths and weaknesses of both model and non-model microorganisms when considering them for biotechnology applications. Additionally, it is crucial to leverage their inherent advantages while introducing metabolic or regulatory pathways in the design of biorefinery cell factories.
In addition, it is also important to integrate AI technology and big data analysis with microbial physiological, metabolic, and omics datasets of diverse biochemical producers under different conditions to construct high-accuracy mathematical models75. This will help predict and enhance the adaptability of metabolic pathways to specific chassis cells, improve product biosynthesis ability, and enhance strain robustness. With the rapid development of mass spectrometry, high-resolution microscopy, as well as efficient gene sequencing, editing, and synthesis techniques, more microorganisms with unique industrial features will be identified and characterized, and the corresponding genome information and genome-engineering toolkits can then be developed rapidly to help engineer them as biorefinery cell factories. Furthermore, the accumulation of physiological and omics datasets along with an increased understanding of the underlying mechanisms of cell organization, regulation, and proliferation, the increased knowledge of biological parts, devices and systems, and effective systems and synthetic biology approaches will facilitate rational design, synthesis assembly, and delivery of artificial genomes and synthetic organisms as the ultimate universal chassis for circular bioeconomy development.
Methods
Strains and media
Z. mobilis ZM4 was employed for the construction of biochemicals production strain. E. coli DH5α and Trans110 were used as a base strain for plasmid construction and demethylation. Plasmids, strains and oligonucleotides used in this study are listed in Supplementary Data 2.
E. coli was cultured at 37 °C, 250 rpm using Luria-Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl), and Rich medium (RMG5, 50 g/L glucose, 10 g/L yeast extract, 2 g/L KH2PO4) was used for Z. mobilis cultivation at 30 °C, 100 rpm. RMG2X2 (20 g/L glucose, 20 g/L xylose, 10 g/L yeast extract, 2 g/L KH2PO4) was used for cultivation of the xylose utilization recombinant strains. 100 µg/mL spectinomycin was added for both E. coli and Z. mobilis when appropriate.
Genetic manipulation
The acquisition of fragments and linear vectors, as well as the plasmids construction process, were carried out according to previously reported method76. Specifically, the insert(s) and vector were ligated through T5 exonuclease kit (NEB, USA) with a molar ratio of 3:1. The ligation product was used for transformation into E. coli Dh5α competent cells. The transformants were confirmed by colony PCR using specific primers of 15A-Fwd/Rev to confirm the insert size, correct plasmids were further verified by Sanger sequencing (TsingKe, Wuhan, China). Genetic modification of Z. mobilis was based on native Type I-F CRISPR-Cas system by editing plasmid pL2R27. For gene insertions, the gene expression cassette flanking 600 bp upstream and 600 bp downstream of editing sites was assembled into the pL2R linearized vector. The correct plasmid was then transferred into Z. mobilis competent cells via electroporation77. The CRISPRi system application for inhibiting pdc and ldh expression was then carried out34. The targeting gRNA sequence was annealed using two single-stranded oligonucleotides and ligated into linearized pEZ-sgr which carries a minimal CRISPR array.
C2-C5 biochemicals metabolic pathway construction
Heterologous gene sequences using for C2-C5 biochemicals production in this study were listed in Supplementary Data 3, these genes were synthesized from TsingKe (Wuhan, China) after codon-optimized. The promoter used were cloned from Z. mobilis ZM4 genome. For constructing the succinate synthesis pathway in Z. mobilis. The strong promoter Peno was used to drive mdh (encoding malate dehydrogenase) genes from different sources on plasmid to evaluate the efficiency (Supplementary Fig. 3a). The expression cassette with the higher titer was integrated into the ZMO0028 locus of Z. mobilis genome to obtain SA3 strain. Based on SA3, different ppc (encoding phosphoenolpyruvate carboxylase) genes driven by Peno were tested in Z. mobilis (Supplementary Fig. 3b). The better efficiency of ppc from C. glutamicum was inserted into the genome of SA2 at ZMO0038 locus to obtain SA3 strain. Similarly, the other genes fum (encoding fumarate hydratase) and frd (fumarate reductase) cloned from different sources were overexpressed on plasmid and transformed to SA3 for testing (Supplementary Fig. 3d, e). Then the efficient synthetic operon of fum-frd driven by Ptet was constructed and transformed into SA3 for succinic acid production.
Similarly, a synthetic operon was constructed to overexpress different source of genes dhaB (encoding glycerol dehydratase) driven by strong promoter Ppdc, and gdrAB (encoding activating factor), yqhD (encoding aldehyde reductase) driven by strong promoter Peno on plasmid for 1,3-PDO production. The sequence of RBS used for gdrAB and yqhD connecting is 5’ ATTAAAGAGGAGAAA 3’. For xylonic acid production, Ppdc promoter was used for xdh overexpressed to obtain plasmid pXA1. ethylene glycol, glycolate, and 1,4-butanediol production, the genes yagF (encoding xylonate dehydratase), yagE (encoding 2-keto-3-deoxy D-xylonate aldolase), and yqhD (encoding aldehyde reductase) were cloned from E. coli. These genes were overexpressed by Peno promoter and inserted into pXA1 plasmid to obtain pEG2 for ethylene glycol production. Gene yqhD of pEG2 was replaced with aldA to obtain pGA1 plasmid for glycolate production. For achieving 1,4-BDO production from xylose in Z. mobilis, the gene yagF of pEG2 was replaced with xylX to obtain pBDO1 plasmid. Then the pBDO1 plasmid was transformed into ZMQ3 strain36 for 1,4-BDO production. The gene LmldhA driven by constitutive promoter PadhB (encoding D-lactate dehydrogenase) for lactic acid production was chosen and used in this work31,36.
Electroporation transformation and recombinant strain selection
The plasmids were transformed into electrocompetent cells of Z. mobilis by electroporation (Bio-Rad Gene Pulser, 0.1-cm gap cuvettes, 1.6 kV, 200 ohms, 25 μF). The electrocompetent cells were transferred and recovered in RMG5 medium at 30 °C for 3 h36. For recombinant strain selection, transformants were cultured on RM agar plate containing 100 µg/mL spectinomycin (RMSp). Potential mutants were screened by colony PCR with the specific primer pair. Then the current recombinant strains were further cultivated in RMSp medium for preservation and further flask evaluation. For curing of editing plasmid, transformants with correct PCR results were cultivated in RM media without antibiotics at 30 °C and passaged for five generations.
Construction of dominant-metabolism compromised intermediate chassis
Z. mobilis ZM4 was used as the initial strain and conducted three rounds of genome editing (Fig. 5b). First, we replaced the ZMO1650 of ZM4 with 2,3-BDO expression cassette including Bsals driven by inducible promoter Ptet and a synthetic operon Aldc-Bdh driven by Pgap promoter, resulting in ZMB1, which enabled the heterologous expression of the 2,3-BDO pathway. Then, the ZMO0038 of ZMB1 was replaced by bdh driven by Pgap promoter, generating ZMB2 for further optimizing the metabolic flux of the 2,3-BDO pathway. Finally, the ZMO1360 (pdc) of ZMB2 was deleted by editing plasmid pB3 to obtain dominant-metabolism compromised intermediate-chassis ZMB3, completely redirecting the central carbon flow from the ethanol pathway to the 2,3-BDO pathway.
In order to further redirect carbon flux towards D-lactic acid production, we replaced the ZMO0038-bdh gene of ZMB3 with Dldh, resulting in ZMB4, which achieved the heterologous expression of the D-lactic acid pathway and redirected carbon flow from the 2,3-BDO pathway to the D-lactic acid pathway. Subsequently, we replaced the ZMO1759 gene of ZMB4 with Dldh to obtain ZMB5, which increased the copy number of lactate dehydrogenase and further competed for metabolic carbon flow, consequently enhancing the flux of the D-lactic acid metabolic pathway. Finally, we replaced the ZMO1650-2,3-BDO expression cassette of ZMB5 with Dldh to obtain ZMB6.
Batch fermentation in shaking flasks
Recombinant strains were pre-cultured overnight with OD600nm value of 1.0–2.0 in RMG5 medium for the seed culture preparation, which was inoculated in 50 mL shake flasks containing 40 mL corresponding medium with an initial OD600 nm value of 0.1 after being collected and washed twice. During fermentation, cell growth in terms of the absorbance value (OD600 nm) was determined spectrophotometrically at 600 nm (UV-1800, AOE, China) at different time points. Samples were centrifuged at 17,000 × g for 2 min, collected supernatants were passed through 0.22-μm filters and stored at −80 °C for subsequent HPLC analysis if needed.
For biochemicals production from xylose, the seed culture was transferred into 50 mL shake flasks containing 40 mL RMG2X2 medium with an initial OD600nm value of 0.1 at 30 °C, 100 rpm. 10 g/L CaCO3 was supplemented to control the pH during fermentation. Samples from different time periods were collected and stored at −80 °C for subsequent HPLC analysis.
For 1,3-PDO production from glycerol, the seed culture was transferred into 50 mL shake flasks containing 40 mL RMG5 medium with an initial OD600nm value of 0.1 at 30 °C on a rotary shaker at 100 rpm. 10 g/L glycerol and 5 mg/L vitamin B12 was supplemented into the media.
For D-lactic acid production from CRH in shake flasks, the seed culture was transferred into medium with an OD600nm value of 0.5 at 33 °C on a rotary shaker at 100 rpm. 2 g/L yeast extract was supplemented into the media.
Production of D-lactate from corncob residue hydrolysate
Corncob residue hydrolysate with approximately 107 g/L glucose (pH 4.50) used in this study was provided by Wuhan Ruijiakang Biotechnology Co., Ltd. (Hubei, China). As for batch fermentation in 5-L fermentor, 50 g/L CaCO3 and 2 g/L yeast extract was supplemented into medium. Strain ZMB6 at an initial OD600nm value of 0.5 was cultured at 33 °C, 100 rpm for 72 h for the measurements of glucose and D-lactate.
13C metabolic flux analysis
13C metabolic flux analysis (MFA) of amino acid in the wild-type ZM4 and ZMB3 strains was carried out according to previously reported method78,79. In particular, [1,2–13C] glucose (99.1 atom% 13C) purchased from Sigma-Aldrich (St. Louis, MO) was used as the sole carbon source for cell growth. Briefly, Cells cultured in MM medium (20 g/L glucose, 1 g/L K2HPO4, 1 g/L KH2PO4, 1 g/L (NH4)2SO4, 0.5 g/L NaCl, 0.42 g/L MgCl2·6 H2O, 0.001 g/L calcium pantothenate) were collected at the exponential phase and hydrolysate by 6 M HCl at 100 °C for 18 h. The supernatant of hydrolysate was freeze-dried by low temperature concentration cold trap. Dried exudates were subsequently dissolved with 100 μL tetrahydrofuran followed by the addition of 100 μL N-tere-Butyldimethylsilyl-N-methyltrifluoroacetamide (TBDMS) at 70 °C for 1 h to generate derivatives. After centrifugation for 5 min, 1-μL derived sample was quantified by GC-MS (Gas chromatograph Trace, Thermo; Triple Quadrupole Mass Spectrometer), fitted with a TG-5MS column (30 mm × 0.25 mm × 0.25 μm, Thermo; USA) with 1.2 mL/min flow rate of carried gas. The detector procedure of GC was set as follows: the initial temperature was held at 150 °C for 2 min and was programmed to raise to 280 °C at 3 °C per min, and further increase to 300 °C at 20 °C per min and hold for another 5 min. Solvent delay was set as 5 min, and the range of mass to charge ratio (m/z) in MS was set from 60 to 50078. The software WuFlux-Ms Tool was applied to analyze and correct amino acid MS data (fragments of [M-57] +, [M-159] +, [M-85] +, and [f302]).
13C-MFA was calculated by using the INCA software, which is based on the elementary metabolic unit framework. A metabolic reaction network of Z. mobilis central metabolism, including ED and EMP glycolytic reactions, non-oxidative PPP reactions, TCA cycle reaction, lumped amino acid biosynthesis, was constructed from model eciZM547. Data of labeling amino acids were substituted into the model for calculating metabolic flux of central metabolism. Several reactions about acetate, acetoin and 2,3-BDO syntheses including Acetaldehyde + NAD+ → Acetate + NADH, 2 Acetaldehyde→Acetoin, Pyruvate → acetolactate + CO2, Acetolactate → Acetoin + CO2, Acetoin + NADH ↔ 2,3-BDO + NAD+, Acetoin + NAD+ → Acetyl-CoA + NADH, Acetoin + ATP → Acetoin. ext, 2,3-BDO → 2,3-BDO. ext were added to the metabolic network model. Cell growth rate, glucose consumption rate and fermentation production rate were characterized for exponential phase growth at 30 °C in MM medium under 10% medium volume condition for using as MFA constraints (Supplementary Table 1). The measured data of labeling amino acid was substituted into the model for calculating metabolic flux (Supplementary Data 1). OD600 measurements were converted to dry weight52. The flux estimations were performed by using Metlab R2012b (The Mathworks Inc.)
Analytical methods
The concentration of glucose and ethanol in the supernatant was analyzed by HPLC LC-20AD (Shimadzu, Japan) with a RezexTM, RFQ-Fast Acid H+ (8%) (Phenomenex, CA, USA) at 80 °C. Elution was performed with 5 mM H2SO4 at 0.6 mL/min36. Lactic acid, succinic acid, 1,3-PDO, 1,4-BDO, glycerol, xylose and xylonic acid concentration were determined by HPLC equipped with a Bio-Rad Aminex HPX-87H column (Bio-Rad, Hercules, CA, U.S.A.) and a refractive index detector54,60,80. The column temperature was set at 60 °C and 5 mM H2SO4 solution was used as the mobile phase with a flow rate of 0.5 mL/min.
Since xylonic acid and xylose have similar retention times with refractive index detector, xylonic acid has determined using the UV detector when xylose was present by HPLC-2030 Plus (Shimadzu, Kyoto, Japan) equipped with a Bio-Rad Aminex HPX-87H column (Bio-Rad, Hercules, CA, U.S.A.) at 60 °C. 5 mM H2SO4 solution was used as the mobile phase with a flow rate of 0.5 mL/min. Xylose concentrations were estimated by subtraction of the xylonic acid peak from the combined peak of xylose and xylonic acid when xylonic acid was present58.
Construction of enzyme-constrained metabolic network model
The metabolic model of this study is based on iZM516. By comparing with iZM4_478, this study added the unique genes and related reactions of iZM4_478 relative to iZM516, tentatively designated as iZM547, and the detailed changes were included Supplementary Data 4 and 5. The construction of the enzyme-constrained model is mainly completed using the process ECMpy2.
Data collection, molecular weight (MW) and Subnit number data were collected from the Interaction information in the UniProt database based on Gene ID. Four methods (AutoPACMEN, DLkcat, TurNup, and UniKP) were used to determine Kcat values49,50,51. AutoPACMEN was mainly determined based on the Kcat values of various species in the Brenda and SABIO-RK databases using EC numbers. Data of Z. mobilis were directly accessed, otherwise the average or median Kcat values of all species under the selected EC number will be used as the Kcat value for the relevant reaction. If there is no Kcat value, the default Kcat value was filled based on the average or median Kcat values of all different EC numbers. The protein abundance of Z. mobilis was obtained by selecting PXD030417 protein data from the PRIDE database, and the original data was completed through the online service of MaxQuant and PAXdb81. The protein-to-cell ratio ptot is set to 0.60538,82. The approximated average saturation of enzyme σ was set as 0.5 in default. Enzyme mass fraction f was calculated according to Eq. 1:
where Ai and Aj represented the abundance of the i-th protein (p_num represents proteins expressed in the model) and j-th protein (g_num represents proteins expressed in the whole proteome).
First, reversible reactions in iZM547 were divided into pairs of irreversible reactions, and reactions catalyzed by multiple isoenzymes were split into different reactions (append num in reaction ID, e.g., rxn00001_reverse_num1). Next, the MW of each enzyme was calculated. According to Eq. 2, the total sum of proteins in the complex was used for reactions catalyzed by enzyme complexes. Finally, an enzymatic constraint (Eq. 3) was introduced into the model with function trans_model2enz_json_model_split_isoenzyme. Then, the biomass calculation was calibrated in enzyme-constrained model by identifying the reaction with the enzyme cost and substituting its Kcat values with the highest value found in the Brenda or SABIO-RK database.
where m is the number of different subunits in the enzyme complex and Nj is the number of j-th subunits in the complex.
Process simulation
Schematics of the D-lactate production from corncob residue hydrolysate process is presented in Supplementary Fig. 8. In this study, lactic acid production process was designed using Aspen Plus® software, which was also used to calculate mass and energy balance. The D-lactate production process includes five areas: glucose production (A100), D-lactate production (A200), D-lactate purification (A300), wastewater treatment (A400), and utilities (A500). Detailed process descriptions can be found in the Supplementary Method 1. The major assumptions and details of the process design in Aspen Plus v14 can be found in Supplementary Table 4. The mass and energy data collected from Aspen Plus v14 are presented in Supplementary Tables 9–13 and Supplementary Data 6.
Techno-economic analysis
TEA is an integrated process that can be used to assess the feasibility of lactic acid production from corncob residue based on an economic and technical point of view. The TCI, TOC, and MSP were estimated using an in-house model developed by the National Renewable Energy Laboratory (NREL)83. The mass balance of the lactic acid production process conducted using Aspen Plus v14 is presented in Supplementary Fig. 10. An annual lactic acid production capacity of 31,100 tons for the biorefinery was assumed along with influencing factors such as equipment size, raw material expenditures, and other associated costs. The TEA is an “nth-plant” model that can estimate the investment necessary for the pre-commercial process as well83. The key assumptions and basic parameters for the process scale-up are shown in Supplementary Table 14, which are sourced from NREL’s reports and studies83.
The equipment and raw materials costs used in the TCI and TOC estimation were obtained from published literature and official reports (Supplementary Table 15)83. The costs of equipment necessarily change with implementation size and obey a power law of the original prices65. The inside battery limit (ISBL) is commonly used in TEA for basic related to warehouse investments, site development, and additional piping. In this study, the ISBL includes equipment and installation costs for A100 to A300. The equipment and raw materials costs will be processed in an Excel spreadsheet using established formulas to calculate the TCI and TOC of the proposed processes (Supplementary Tables 5, 6). Using the discounted cash flow method, the MSP of lactic acid can be determined at the point where the net present value (NPV) equals zero and the internal rate of return (IRR) reaches 10%83. All costs are converted to 2023 US dollars to eliminate the influence of the rate of inflation on expenditure according to the Plant Cost Index from Chemical Engineering Magazine, the Industrial Inorganic Chemical Index from SRI Consulting, and the US Department of Labor Bureau of Labor Statistics83.
Life cycle assessment
The main goal of the LCA was to measure the carbon footprint of manufacturing lactic acid using corncob residue. The system boundary is “from cradle to gate”, raw material consumption and production processes were considered. In LCA investigations, the functional unit for chemicals is commonly defined as the mass of the product in kilogram. In order to compare the results to other similar research, this study selected 1 kg of lactic acid as the functional unit to serve as a benchmark for the inputs and outputs. The lactic acid production pathway in this study obtained crucial raw materials and energy inputs from Aspen Plus v14. Additionally, the disposal capability for corncob residue was determined to be 5000 kg/h. Supplementary Table 16 presents the data of materials and energy used in the process of producing lactic acid. The Environmental Protection Agency (EPA) produced version 2.1 of the tool for reduction and assessment of chemicals and other environmental impacts (TRACI), which provides characterization parameters that estimate the potential effects of inputs and discharges on certain manufacturing processes. This study evaluated the GWP of the lactic acid production process. The background data utilized in the life cycle assessment is based on Ecoinvent v3.6.
Statistical analysis
The number of biological replicates is specified in figure and table legends. Data analysis was performed using GraphPad Prism statistical software (version 8.0.1, GraphPad, CA, USA). Independent unpaired t-test was used for two-tailed comparison. p ≤ 0.05 was considered statistically significant.
Data availability
Data supporting the findings of this work are available within the paper and its Supplementary Information files. A reporting summary for this article is available as a Supplementary Information file. Source data are provided with this paper.
Code availability
The code was implemented in Python 3.9, and the code relies on the COBRApy and ECMpy packages. Full code and models of eciZM547 are available on GitHub [https://github.com/AaroncrowAries/eciZM547].
References
Roebroek, C. T. J., Duveiller, G., Seneviratne, S. I., Davin, E. L. & Cescatti, A. Releasing global forests from human management: how much more carbon could be stored? Science 380, 749–753 (2023).
Zhao, X., Liu, C. & Bai, F. Making the biochemical conversion of lignocellulose more robust. Trends Biotechnol. 42, 418–430 (2024).
Singh, N. et al. Global status of lignocellulosic biorefinery: challenges and perspectives. Bioresour. Technol. 344, 126415 (2022).
Martín, M., Taifouris, M. & Galán, G. Lignocellulosic biorefineries: a multiscale approach for resource exploitation. Bioresour. Technol. 385, 129397 (2023).
Choi, K. R. et al. Systems metabolic engineering strategies: integrating systems and synthetic biology with metabolic engineering. Trends Biotechnol. 37, 817–837 (2019).
Shi, Z. et al. Data-driven synthetic cell factories development for industrial biomanufacturing. Biodes. Res. 2022, 9898461 (2022).
Gao, D. et al. De novo biosynthesis of vindoline and catharanthine in Saccharomyces cerevisiae. Biodes. Res. 2022, 0002 (2022).
Cho, J. S., Kim, G. B., Eun, H., Moon, C. W. & Lee, S. Y. Designing microbial cell factories for the production of chemicals. JACS Au 2, 1781–1799 (2022).
Yilmaz, S., Nyerges, A., van der Oost, J., Church, G. M. & Claassens, N. J. Towards next-generation cell factories by rational genome-scale engineering. Nat. Catal. 5, 751–765 (2022).
Liu, L., Helal, S. E. & Peng, N. CRISPR-Cas-based engineering of probiotics. Biodes. Res. 5, 0017 (2023).
Hoff, J. et al. Vibrio natriegens: an ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 22, 4394–4408 (2020).
Suárez, G. A., Renda, B. A., Dasgupta, A. & Barrick, J. E. Reduced mutation rate and increased transformability of transposon-free Acinetobacter baylyi ADP1-ISx. Appl. Environ. Microbiol. 83, e01025–01017 (2017).
Abdel-Rahman, M. A., Tashiro, Y. & Sonomoto, K. Recent advances in lactic acid production by microbial fermentation processes. Biotechnol. Adv. 31, 877–902 (2013).
Ling, C. et al. Engineering NADH/NAD+ ratio in Halomonas bluephagenesis for enhanced production of polyhydroxyalkanoates (PHA). Metab. Eng. 49, 275–286 (2018).
Sun, M. L. et al. Building Yarrowia lipolytica cell factories for advanced biomanufacturing: challenges and solutions. J. Agric. Food Chem. 72, 94–107 (2024).
Yang, Y. et al. Progress and perspective on lignocellulosic hydrolysate inhibitor tolerance improvement in Zymomonas mobilis. Bioresour. Bioprocess. 5, 6 (2018).
Yan, X., He, Q., Geng, B. & Yang, S. Microbial cell factories in the bioeconomy era: from discovery to creation. Biodes. Res. 6, 0052 (2024).
Fuchino, K., Wasser, D. & Soppa, J. Genome copy number quantification revealed that the ethanologenic alpha-proteobacterium Zymomonas mobilis is polyploid. Front. Microbiol. 12, 705895 (2021).
Wang, X. et al. Advances and prospects in metabolic engineering of Zymomonas mobilis. Metab. Eng. 50, 57–73 (2018).
Xia, J., Yang, Y., Liu, C., Yang, S. & Bai, F. Engineering Zymomonas mobilis for robust cellulosic ethanol production. Trends Biotechnol. 37, 960–972 (2019).
Yang, S. et al. Improved genome annotation for Zymomonas mobilis. Nat. Biotechnol. 27, 893–894 (2009).
Yang, S. et al. Complete genome sequence and the expression pattern of plasmids of the model ethanologen Zymomonas mobilis ZM4 and its xylose-utilizing derivatives 8b and 2032. Biotechnol. Biofuels 11, 125 (2018).
Cao, L. et al. Deciphering molecular mechanism underlying self-flocculation of Zymomonas mobilis for robust production. Appl. Environ. Microbiol. 88, e0239821 (2022).
Yang, Y. et al. Prediction and characterization of promoters and ribosomal binding sites of Zymomonas mobilis in system biology era. Biotechnol. Biofuels 12, 52 (2019).
Yang, Y. et al. Identification and characterization of ethanol-inducible promoters of Zymomonas mobilis based on omics data and dual reporter-gene system. Biotechnol. Appl. Biochem. 67, 158–165 (2020).
Shen, W. et al. Establishment and application of a CRISPR-Cas12a assisted genome-editing system in Zymomonas mobilis. Micro. Cell Fact. 18, 162 (2019).
Zheng, Y. et al. Characterization and repurposing of the endogenous type I-F CRISPR-Cas system of Zymomonas mobilis for genome engineering. Nucleic Acids Res. 47, 11461–11475 (2019).
Wang, X. et al. CRISPR-mediated host genomic DNA damage is efficiently repaired through microhomology-mediated end joining in Zymomonas mobilis. J. Genet. Genomics 48, 115–122 (2021).
Yang, S. et al. Zymomonas mobilis as a model system for production of biofuels and biochemicals. Micro. Biotechnol. 9, 699–717 (2016).
Vriesekoop, F. & Pamment, N. B. Acetaldehyde stimulation of the growth of Zymomonas mobilis subjected to ethanol and other environmental stresses: effect of other metabolic electron acceptors and evidence for a mechanism. Fermentation 7, 80 (2021).
Hu, M. et al. Metabolic engineering of Zymomonas mobilis for co-production of D-lactic acid and ethanol using waste feedstocks of molasses and corncob residue hydrolysate. Front. Bioeng. Biotechnol. 11, 1135484 (2023).
Uhlenbusch, I., Sahm, H. & Sprenger, G. A. Expression of an L-alanine dehydrogenase gene in Zymomonas mobilis and excretion of L-alanine. Appl. Environ. Microbiol. 57, 1360–1366 (1991).
Wang, Z. et al. Metabolic engineering of an industrial bacterium Zymomonas mobilis for anaerobic l-serine production. Synth. Syst. Biotechnol. 9, 349–358 (2024).
Bao, W., Shen, W., Peng, Q., Du, J. & Yang, S. Metabolic engineering of Zymomonas mobilis for acetoin production by carbon redistribution and cofactor balance. Fermentation 9, 113 (2023).
Yang, S. et al. Metabolic engineering of Zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol. Biofuels 9, 189 (2016).
Qiu, M. et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol. Biofuels 13, 15 (2020).
Khandelwal, R., Srivastava, P. & Bisaria, V. S. Expression of Escherichia coli malic enzyme gene in Zymomonas mobilis for production of malic acid. J. Biotechnol. 351, 23–29 (2022).
Lee, K. Y., Park, J. M., Kim, T. Y., Yun, H. & Lee, S. Y. The genome-scale metabolic network analysis of Zymomonas mobilis ZM4 explains physiological features and suggests ethanol and succinic acid production strategies. Micro. Cell Fact. 9, 94 (2010).
Li, Y. et al. Metabolic engineering of Zymomonas mobilis for continuous co-production of bioethanol and poly-3-hydroxybutyrate (PHB). Green Chem. 24, 2588–2601 (2022).
He, Y. et al. Metabolic engineering of Zymomonas moblis for ethylene production from straw hydrolysate. Appl. Microbiol. Biotechnol. 105, 1709–1720 (2021).
Wu, Y. et al. Construction and application of high-quality genome-scale metabolic model of Zymomonas mobilis to guide rational design of microbial cell factories. Synth. Syst. Biotechnol. 8, 498–508 (2023).
He, M. et al. Zymomonas mobilis: a novel platform for future biorefineries. Biotechnol. Biofuels 7, 101 (2014).
Liu, Y. et al. Regulated redirection of central carbon flux enhances anaerobic production of bioproducts in Zymomonas mobilis. Metab. Eng. 61, 261–274 (2020).
Mao, Z. et al. ECMpy 2.0: a Python package for automated construction and analysis of enzyme-constrained models. Synth. Syst. Biotechnol. 9, 494–502 (2024).
Sánchez, B. J. et al. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017).
Ong, W. K. et al. Model-driven analysis of mutant fitness experiments improves genome-scale metabolic models of Zymomonas mobilis ZM4. PLoS Comput. Biol. 16, e1008137 (2020).
Norsigian, C. J. et al. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 48, D402–d406 (2020).
Bekiaris, P. S. & Klamt, S. Automatic construction of metabolic models with enzyme constraints. BMC Bioinformatics 21, 19 (2020).
Li, F. et al. Deep learning-based kcat prediction enables improved enzyme-constrained model reconstruction. Nat. Catal. 5, 662–672 (2022).
Kroll, A., Rousset, Y., Hu, X., Liebrand, N. A. & Lercher, M. J. Turnover number predictions for kinetically uncharacterized enzymes using machine and deep learning. Nat. Commun. 14, 4139 (2023).
Yu, H., Deng, H., He, J., Keasling, J. D. & Luo, X. UniKP: a unified framework for the prediction of enzyme kinetic parameters. Nat. Commun. 14, 8211 (2023).
Jacobson, T. B. et al. 2H and 13C metabolic flux analysis elucidates in vivo thermodynamics of the ED pathway in Zymomonas mobilis. Metab. Eng. 54, 301–316 (2019).
Zhu, X. et al. Metabolic evolution of two reducing equivalent-conserving pathways for high-yield succinate production in Escherichia coli. Metab. Eng. 24, 87–96 (2014).
Cui, Z. et al. Reconfiguration of the reductive TCA cycle enables high-level succinic acid production by Yarrowia lipolytica. Nat. Commun. 14, 8480 (2023).
Mosier, N. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour. Technol. 96, 673–686 (2005).
Zhang, M., Eddy, C., Deanda, K., Finkelstein, M. & Picataggio, S. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267, 240–243 (1995).
Lou, J. et al. Development and characterization of efficient xylose utilization strains of Zymomonas mobilis. Biotechnol. Biofuels 14, 231 (2021).
Herrera, C. R. J. et al. Engineering Zymomonas mobilis for the production of xylonic acid from sugarcane bagasse hydrolysate. Microorganisms 9, 1372 (2021).
Xin, B. et al. Coordination of metabolic pathways: enhanced carbon conservation in 1,3-propanediol production by coupling with optically pure lactate biosynthesis. Metab. Eng. 41, 102–114 (2017).
Li, Z. et al. Systems metabolic engineering of Corynebacterium glutamicum for high-level production of 1,3-propanediol from glucose and xylose. Metab. Eng. 70, 79–88 (2022).
Rawoof, S. A. A. et al. Production of optically pure lactic acid by microbial fermentation: a review. Environ. Chem. Lett. 19, 539–556 (2021).
Gezae Daful, A. & Görgens, J. F. Techno-economic analysis and environmental impact assessment of lignocellulosic lactic acid production. Chem. Eng. Sci. 162, 53–65 (2017).
Fei, Q. et al. Biological valorization of natural gas for the production of lactic acid: techno-economic analysis and life cycle assessment. Biochem. Eng. J. 158, 107500 (2020).
Humbird, D. et al. Process Design and Economics for Biochemical Conversion of Lignocellulosic Biomass to Ethanol: Dilute-acid Pretreatment and Enzymatic Hydrolysis of Corn Stover (National Renewable Energy Lab (NREL), 2011).
Vatavuk, W. M. Updating the CE plant cost index. Chem. Eng. 109, 62–70 (2002).
Li, Y. et al. Sustainable lactic acid production from lignocellulosic biomass. ACS Sustain Chem. Eng. 9, 1341–1351 (2021).
Pachón, E. R., Mandade, P. & Gnansounou, E. Conversion of vine shoots into bioethanol and chemicals: prospective LCA of biorefinery concept. Bioresour. Technol. 303, 122946 (2020).
Ögmundarson, Ó., Sukumara, S., Laurent, A. & Fantke, P. Environmental hotspots of lactic acid production systems. GCB Bioenergy 12, 19–38 (2020).
Zhong, W., Li, H. & Wang, Y. Design and construction of artificial biological systems for one-carbon utilization. Biodes. Res. 5, 0021 (2023).
Gao, J. et al. Engineering co-utilization of glucose and xylose for chemical overproduction from lignocellulose. Nat. Chem. Biol. 19, 1524–1531 (2023).
Mantynen, S., Laanto, E., Oksanen, H. M., Poranen, M. M. & Diaz-Munoz, S. L. Black box of phage-bacterium interactions: exploring alternative phage infection strategies. Open Biol. 11, 210188 (2021).
Fatma, Z., Schultz, J. C. & Zhao, H. Recent advances in domesticating non‐model microorganisms. Biotechnol. Prog. 36, e3008 (2020).
Yao, Z. et al. A highly efficient transcriptome-based biosynthesis of non-ethanol chemicals in Crabtree negative Saccharomyces cerevisiae. Biotechnol. Biofuels 16, 37 (2023).
Sjöberg, G., Reķēna, A., Fornstad, M., Lahtvee, P.-J. & van Maris, A. J. A. Evaluation of enzyme-constrained genome-scale model through metabolic engineering of anaerobic co-production of 2,3-butanediol and glycerol by Saccharomyces cerevisiae. Metab. Eng. 82, 49–59 (2024).
Shi, Z. et al. Data-driven synthetic cell factories development for industrial biomanufacturing. Biodes. Res. 2022, 1–12 (2022).
Yan, X. et al. Cysteine supplementation enhanced inhibitor tolerance of Zymomonas mobilis for economic lignocellulosic bioethanol production. Bioresour. Technol. 349, 126878 (2022).
Okamoto, T. & Nakamura, K. Simple and highly efficient transformation method for Zymomonas mobilis: electroporation. Biosci. Biotechnol. Biochem. 56, 833 (1992).
Hu, S. et al. Transcription factor DegU-mediated multi-pathway regulation on lichenysin biosynthesis in Bacillus licheniformis. Metab. Eng. 74, 108–120 (2022).
He, P. et al. 13C-metabolic flux analysis reveals the metabolic flux redistribution for enhanced production of poly-γ-glutamic acid in dlt over-expressed Bacillus licheniformis. Front. Microbiol. 10, 105 (2019).
Wu, M. Y., Sung, L. Y., Li, H., Huang, C. H. & Hu, Y. C. Combining CRISPR and CRISPRi systems for metabolic engineering of E. coli and 1,4-BDO biosynthesis. ACS Synth. Biol. 6, 2350–2361 (2017).
Huang, Q., Szklarczyk, D., Wang, M., Simonovic, M. & von Mering, C. PaxDb 5.0: curated protein quantification data suggests adaptive proteome changes in yeasts. Mol. Cell Proteomics 22, 100640 (2023).
Widiastuti, H. et al. Genome-scale modeling and in silico analysis of ethanologenic bacteria Zymomonas mobilis. Biotechnol. Bioeng. 108, 655–665 (2011).
Zhang, C. et al. An upcycling bioprocess for sustainable aviation fuel production from food waste-derived greenhouse gases: life cycle assessment and techno-economic analysis. Chem. Eng. J. 486, 150242 (2024).
Fuhrer, T., Fischer, E. & Sauer, U. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187, 1581–1590 (2005).
Kremer, T. A., LaSarre, B., Posto, A. L. & McKinlay, J. B. N2 gas is an effective fertilizer for bioethanol production by Zymomonas mobilis. Proc. Natl Acad. Sci. USA 112, 2222–2226 (2015).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2022YFA0911800 to S.Y.), the International Science and Technology Cooperation base of Hubei Province (No. SH2318 to S.Y.), the Innovation Base for Introducing Talents of Discipline of Hubei Province (No. 2019BJH021 to S.Y.), the Technological innovation Plan Project of Hubei Province (No. 2024BCB025 to Q.H.), the Technology Achievement Transformation Project of Wuhan Science and Technology Innovation Bureau (No. 2024030803010187 to S.Y.), and Science Fund for Distinguished Young Scholars of Shanxi Province (No. 2022JC09 to Q.F.). Funding was also supported by the State Key Laboratory of Biocatalysis and Enzyme Engineering.
Author information
Authors and Affiliations
Contributions
S.Y. designed and supervised the experiments with inputs from all authors. X.Y. and W.B. constructed plasmids and recombinant strains with the help from Z.H., Q.P., M.H., and B.G. Y.W. performed the model construction and optimization with help from Z.M., Q.Y., Q.F., and H.M. X.Y., P.H. performed the 13C metabolic flux analysis with the help from S.C. C.Z. and Q.F. performed the techno-economic analysis and life cycle assessment of D-lactate production from corncob residue. X.Y., W.B., Y.W., Z.H., Q.H., Q.F., and S.Y. analyzed the data. X.Y., Q.H., and S.Y. wrote the manuscript with the help from Q.F. and H.M. All authors helped revise, read, and approve the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have patent applications associated with this study. S.Y., X.Y., Q.H. have filed two patents (CN202310246847.7 and US18401511) for protecting strains for EG production. S.Y., W.B., Q.P. have filed two patents (CN202410112699.4 and US18777118) for protecting strains for D-lactate production. Other authors claim no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yan, X., Bao, W., Wu, Y. et al. Paradigm of engineering recalcitrant non-model microorganism with dominant metabolic pathway as a biorefinery chassis. Nat Commun 15, 10441 (2024). https://doi.org/10.1038/s41467-024-54897-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-54897-5
This article is cited by
-
Economic and sustainable revolution to facilitate one-carbon biomanufacturing
Nature Communications (2025)