Abstract
Dynamic regulation of metabolic activities in astrocytes is critical to meeting the demands of other brain cells. During neuronal stress, lipids are transferred from neurons to astrocytes, where they are stored in lipid droplets (LDs). However, it is not clear whether and how neuron-derived lipids trigger metabolic adaptation in astrocytes. Here, we uncover an endolysosomal function that mediates neuron-astrocyte transcellular lipid signaling. We identify Tweety homolog 1 (TTYH1) as an astrocyte-enriched endolysosomal protein that facilitates autophagic flux and LD degradation. Astrocyte-specific deletion of mouse Ttyh1 and loss of its Drosophila ortholog lead to brain accumulation of neutral lipids. Computational and experimental evidence suggests that TTYH1 mediates endolysosomal clearance of ceramide 1-phosphate (C1P), a sphingolipid that dampens autophagic flux and LD breakdown in mouse and human astrocytes. Furthermore, neuronal C1P secretion induced by inflammatory cytokine interleukin-1β causes TTYH1-dependent autophagic flux and LD adaptations in astrocytes. These findings reveal a neuron-initiated signaling paradigm that culminates in the regulation of catabolic activities in astrocytes.
Similar content being viewed by others
Introduction
Metabolism in the central nervous system (CNS) is highly compartmentalized among different brain cell types to efficiently support various energy-demanding processes. For instance, synaptic transmission in neurons generates a steep demand for energy substrates, which are produced either locally or by neighboring glial cells1. Microglia exhibit high catabolic activity, ensuring efficient clearance of extracellular neuron-derived components2. Mature oligodendrocytes modulate axonal processes by providing specific energy substrates3. Efficient distribution of metabolic precursors supports anabolism during neural development and regeneration4. These examples highlight the importance of cell type-specific metabolic signatures in supporting brain functions. Such compartmentalized metabolic processes necessitate concerted transcellular mechanisms to ensure proper processing and exchange of metabolites between different brain cell types.
Astrocytes are regarded as the metabolic workhorses of the brain. They regulate neurotransmitter homeostasis at the synaptic cleft by neurotransmitter uptake and dynamically adjusting their metabolism and bioenergetics5. Astrocytes supply metabolites such as lactate, serine, and glutamine to support neuronal activity and bioenergetics1. In addition to small metabolites, astrocytes also process neurotoxic protein species, mediating both disease protection and pathogenesis6. With high expression of genes related to lipid metabolism, astrocytes specialize in handling, processing, storing, and allocating lipid metabolites7. They facilitate transcellular lipid mobilization through lipoprotein secretion, a process crucial for maintaining brain lipid homeostasis. Dysregulated lipid secretion can induce neurotoxicity, underlining the importance of proper lipid handling by astrocytes8. On the other hand, excessive lipids generated by neurons are received and degraded by the astrocyte endolysosomal system9. Neuronal stress triggers the transfer of lipids from neurons to astrocytes, where these lipids are stored in lipid droplets (LDs)10,11,12,13. Although these processes involve other brain cells, the precise mechanisms by which astrocytes coordinate transcellular signaling and metabolism remain poorly understood.
Endolysosomes serve as a critical interface between the extracellular environment and the intracellular machinery of astrocytes. These organelles are equipped with a diverse array of hydrolases, enabling the processing and degradation of endocytosed substances and autophagy substrates. Beyond their degradative functions, endolysosomes play significant roles in cellular signaling by transducing metabolite signals into metabolic responses14,15. Indeed, endolysosomal transmembrane proteins, such as ion channels and transporters, function as signal transducers16. Furthermore, endolysosomes interact with other organelles, such as autophagosomes, endoplasmic reticulum, and lipid droplets, to modulate their metabolic activities. These properties confer endolysosomes a critical function in the integration of various metabolic signals14,15.
In this study, we uncover the role of an endolysosomal transmembrane protein in mediating catabolic processes in astrocytes. Tweety homolog 1 (TTYH1) is a member of the Tweety homolog (TTYH) protein family, which consists of multi-pass transmembrane proteins17. Orthologs of TTYH1 have been identified across animal species, including worm (Caenorhabditis elegans) and fruit fly (Drosophila melanogaster)17,18. Designated as an understudied protein by the Illuminating the Druggable Genome Program of the United States National Institutes of Health19, the functional implications of TTYH1 have been limited to neural stem cell signaling and glioma invasion20,21,22. TTYH1 expression is enriched in the human brain where reduced expression levels were observed in Alzheimer’s disease and Parkinson’s disease patients23,24,25. However, the molecular function of TTYH1 remained undefined, especially after recent cryo-electron microscopy (cryo-EM) studies refuted its previously proposed ion channel property26,27. Here, we leveraged Drosophila brain glia, conditional Ttyh1 knockout mouse astrocytes, and human induced pluripotent cell (iPSC)-derived cell models to elucidate the function of TTYH1 in astrocytes. Our investigations reveal a neuron-initiated signaling paradigm that culminates in the regulation of catabolic activities in astrocytes.
Results
Ttyh1 mediates astrocyte autophagic flux at endolysosomes
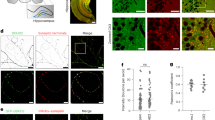
The mouse TTYH1 ortholog was identified as an astrocyte-enriched gene in adult brains28,29,30. Single-cell RNA sequencing datasets reveal that TTYH1 is predominantly expressed by both human and mouse brain astrocytes (Supplementary Fig. 1a)31,32. In adult mouse brains, Ttyh1 immunofluorescence signals were found in cells that expressed astrocytic marker Gfap (Fig. 1a), confirming its astrocyte-enriched expression pattern. While rat Ttyh1 exhibited vesicular localization in astrocytes33, the primary subcellular site of function for TTYH1 orthologs remained undefined. In astrocytes isolated from neonatal mouse brain cortices (Supplementary Fig. 1b), we found that Ttyh1 is primarily present in Lamp1-positive endolysosomes and colocalizes with autophagosome marker LC3 (Fig. 1b, c). In contrast, low colocalization with Eea1 and Rab11 suggests that Ttyh1 is not a resident in early and recycling endosomes (Supplementary Fig. 1c−e). We also observed endolysosomal localization of TTYH1 in human iPSC-derived astrocytes (iAstrocytes) (Supplementary Fig. 2). These data are consistent with our recent findings that both Drosophila and human TTYH1 orthologs are localized to endolysosomes and autolysosomes in Drosophila brain glia34. Interestingly, Ttyh1 was present on the outer membrane of brain-derived autophagic vesicles35, indicating that it does not constitute the proteome of phagophore or nascent autophagosome. Collectively, these findings suggest that TTYH1 might participate in the interaction between autophagosomes and endolysosomes.
a Ttyh1 is primarily expressed in mouse astrocytes. Representative (n = 2 mouse brains; 16-weeks-old female) immunofluorescence image of a mouse hippocampal sagittal section shows nucleus (lavender), Ttyh1 (magenta), and Gfap (green). Insets show magnification of boxed region. b, c Ttyh1 localizes to endolysosomes in astrocytes. Confocal images of primary mouse cortical astrocytes show nucleus (blue), Ttyh1 (magenta), Lamp1 (green) (c) (n = 4 astrocytes), and LC3 (green) (d) (n = 7 astrocytes). d Schematic representations of mouse Ttyh1flox allele and strategy to delete Ttyh1 in cortical astrocytes isolated from Aldh1l1-cre/ERT2; Ttyh1fl/fl conditional knockout (cKO) mice. e Loss of Ttyh1 leads to accumulation of autophagosome markers upon autophagy induction. Shown are representative immunoblot images and quantifications of autophagosome markers LC3 (three independent experiments) and p62 (four independent experiment) in primary cortical astrocytes of the cKO mice. Treatment with culture medium was used as control. Nutrient deprivation for 2 h in artificial cerebrospinal fluid (ACSF) was used to induce autophagy. Bafilomycin A1 (BafA1; 100 nM) inhibits lysosomal acidification and blocks autophagic flux. High LC3-II/I ratio and normalized p62 levels indicate autophagic flux blockage. Values are normalized to those of the Ttyh1 WT, medium condition, on the same blot. Data were presented as mean ± SEM. Number of biological replicates (n) is shown in brackets at the bottom of each bar. Unpaired two-tailed t-test (LC3-II/I: t = 5.464, df = 4; p62/αTub: t = 2.877, df = 6). f Ttyh1 mediates autophagic flux. Primary cortical astrocytes from the cKO mice expressing autophagic flux probe GFP-LC3-RFP-LC3ΔG were treated with ACSF for 0, 30, and 90 min. The ratio between GFP and RFP fluorescence inversely correlates with autophagic flux. Shown are representative fluorescence images and quantification of the GFP/RFP ratio of individual astrocytes. All GFP/RFP ratios are normalized to the mean of those at 0 min. Each datapoint represents one astrocyte. Data were presented as mean ± SEM. Number of astrocytes (n) is shown in brackets at the bottom of each bar. Unpaired two-tailed t-test (Ttyh1 WT: t = 4.135, df = 44; Ttyh1 KO: t = 1.200, df = 43; n.s not significant, P = 0.2366). Source data are provided as a Source Data file.
To elucidate the biological function of TTYH1, we created a conditional Ttyh1 knockout allele (Ttyh1flox) in mouse by flanking the fourth exon of Ttyh1 with a pair of loxP sites (Fig. 1d). By crossing in the astrocyte-specific tamoxifen-inducible Cre recombinase allele, tamoxifen treatment should abolish the expression of all Ttyh1 transcripts in astrocytes (Fig. 1d). We isolated cortical astrocytes from these mice and cultured the cells with 4-hydroxy tamoxifen (4OH-TAM) or vehicle for one week. As expected, Ttyh1 expression was significantly diminished in astrocytes treated with 4OH-TAM (Fig. 1e and Supplementary Fig. 1f). We refer to these tamoxifen-treated astrocytes as Ttyh1 KO hereafter. To test if loss of Ttyh1 impacts autophagy, we measured the protein levels of autophagy markers, LC3 and p62. While we did not detect significant changes in LC3 or p62 levels when astrocytes were cultured with supplemented culture media, nutrient deprivation induced by artificial cerebrospinal fluid (ACSF) resulted in markedly increased p62 and LC3-II levels in Ttyh1 KO astrocytes (Fig. 1e). Addition of lysosomal inhibitor bafilomycin A1 (BafA1) did not further increase autophagic markers in Ttyh1 KO but normalized the difference between WT and KO (Fig. 1e), indicating that the ACSF-induced increase was caused by impaired lysosomal turnover of autophagosome upon Ttyh1 deficiency. These data were consistent with findings that the Drosophila TTYH1 ortholog is required for autophagic flux maintenance in glia34. To validate the autophagy phenotype in Ttyh1 KO astrocytes, we utilized the fluorescent autophagic flux probe GFP-LC3-RFP-LC3ΔG36. Upon expressing the probe in target cells, the RFP fluorescence serves as an internal control for probe expression and autophagosome levels. The autophagosome-localized GFP-LC3 is subject to degradation upon fusion with endolysosomes. Thus, the signal ratio of GFP/RFP quantitatively reflects autophagic flux. We found that upon switching culture medium to ACSF for 30 min, the GFP/RFP ratio was significantly reduced in control (Ttyh1 WT) astrocytes (Fig. 1f), indicative of elevated autophagic flux. In contrast, Ttyh1 KO astrocytes did not exhibit significant changes in the GFP/RFP ratio throughout the 90-minute course of nutrient deprivation (Fig. 1f). Thus, these data demonstrate that Ttyh1 is required for induced autophagic flux in astrocytes.
Ttyh1 mediates the clearance of ceramide 1-phosphate
TTYH1 orthologs were characterized as transmembrane proteins that form anion channels37. However, recent studies leveraging high-resolution cryo-EM and electrophysiology did not find any structural or functional evidence to support anion conductance26,27. The undefined molecular function of TTYH1 posed a challenge to elucidating its mechanistic role in endolysosomes. Interestingly, both cryo-EM studies identified an evolutionarily conserved hydrophobic cavity that spans across the aqueous and the membrane phases26,27. Such a solvent-exposed cavity could potentially support the transport of membrane lipids between the two phases, thereby modulating the membrane abundance of certain lipid species. Since membrane lipid composition and dynamics regulate inter-organellar interactions38, lipids that modulate autophagic flux might be recognized by the hydrophobic cavity of TTYH1 and be extracted from the endolysosomal membrane. Indeed, 18 lipid species were co-purified with TTYH2, a paralog of TTYH126. Among them, a sphingolipid with a 702.57 mass-to-charge (m/z) ratio matches a ceramide 1-phosphate (CerP(d18:1/22:0) or a glucosylceramide (GlcCer(d18:0/16:0)) according to the LIPID MAPS structure database39. Notably, ceramide 1-phosphate (C1P) was found to inhibit autophagic flux in rat neurons40. Thus, TTYH1 might facilitate membrane extraction of C1P, in addition to other lipid substrates, to create a membrane lipid composition permissible to fusion with autophagosomes.
Consistent with the proposed lipid-extraction role of the conserved hydrophobic cavity, in silico protein-ligand docking simulations revealed energetically favorable binding between multiple C1P species and the noted cavity on TTYH1 (Fig. 2a, b and Supplementary Fig. 3a). Other sphingolipids, such as ceramides and glucosylceramides, were also docked to the hydrophobic cavity (Supplementary Fig. 3a−c). As expected, the acyl chains of docking lipids form hydrophobic interactions with multiple amino acid residues of the cavity (shown in green in Fig. 2a). Intriguingly, we noticed that the head groups of multiple lipids were docked in proximity to three neighboring amino acids, E210, R213, and Y217, located on transmembrane helix 3 of TTYH1 (Fig. 2b and Supplementary Fig. 3a). Hydrogen bond interactions were also predicted between these amino acids and the docking lipids (Fig. 2b and Supplementary Fig. 3a). Among these amino acid residues, E210 and R213 are conserved in TTYH1 orthologs from worms to humans (Fig. 2c). Electrostatic interactions between these residues and the polar or charged lipid head group might anchor the lipid to TTYH1 when the acyl chains are displaced from the membrane. We speculate that such lipid-TTYH1 interaction is critical for shuttling lipid substrates into the luminal space of the endolysosome, where degradation by lipid hydrolases occurs. If so, endocytosed C1P will require TTYH1 for its removal from the endolysosomal membrane.
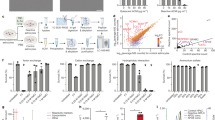
a, b Simulated 3-dimensional docking between a human TTYH1 monomer and ceramide 1-phosphate (CerP(d18:1/22:0)). TMD transmembrane ___domain. Constituent residues of the docking cavity are shown in surface representation. Residues in green were predicted to form hydrophobic interactions with the docked C1P. E210, R213, and Y217 are highlighted in orange. Magnification of the docking region is shown in (b). c Peptide sequence alignment between TTYH1 orthologs of the indicated species. Shown are the partial sequences of the third transmembrane helix. Positions of E210, R213, and Y217 of human TTYH1 are indicated by orange dots. The evolutionarily conserved E210 and R213 are colored in red. d Quantifications of NBD-C1P pulse-chase assay. Ttyh1 WT and Ttyh1 KO astrocytes were pulsed with 2 µM NBD-C1P for 15 min, before chasing in ACSF for 0, 15, 30, and 60 min. NBD fluorescence intensities for each time-point were normalized to the mean of Ttyh1 WT values. Box plots show the median, the bounds of the box (25th and 75th percentiles) and whiskers representing minimum and maximum values. Number of astrocytes (n) from two independent experiments is shown in bracket underneath each box plot. Mann–Whitney test. n.s. not significant, P = 0.379. e E210 and R213 confer C1P clearance function. The two residues in human TTYH1 were substituted with nonpolar alanine to create the TTYH1ER/AA mutant. Astrocytes expressing either wild-type TTYH1 or TTYH1ER/AA mutant were subject to the NBD-C1P pulse-chase assay. Astrocytes were pulsed with 2 µM NBD-C1P for 15 min, followed by chasing for 60 min. Shown are quantifications of NBD fluorescence before and after the chase. Fluorescence intensities were normalized to the mean of the TTYH1-expressing Ttyh1 WT astrocytes values at 0-min chase. Box plots show the median, the bounds of the box (25th and 75th percentiles) and whiskers representing minimum and maximum values. Number of astrocytes (n) from two independent experiments is shown in brackets underneath each box plot. One-way ANOVA (F = 19.3, df = 188) followed by Bonferroni’s multiple comparisons post hoc test. n.s. not significant, P > 0.9999. Source data are provided as a Source Data file.
After 1-h incubation with exogenous C1P, we found that Ttyh1 KO astrocytes contained significantly more C1P than WT control astrocytes (Supplementary Fig. 3d). Lysosomal inhibition by BafA1 further increased intracellular C1P levels in both WT and KO (Supplementary Fig. 3d), indicating that internalized C1P was subject to TTYH1-dependent lysosomal degradation. On the contrary, degradation of internalized bovine serum albumin (BSA) was not affected by the loss of Ttyh1 (Supplementary Fig. 3e), suggesting that TTYH1 specifically mediates lysosomal degradation of C1P. To distinguish between C1P uptake and degradation, we performed an imaging-based pulse-chase assay. Astrocytes were pulsed with fluorescent NBD-C1P added to the bath solution. Upon removal of extracellular NBD-C1P, we chased the intracellular NBD fluorescence using wide-field fluorescence microscopy. Whereas the fluorescence intensities were comparable between control and Ttyh1 KO astrocytes at the onset of the chase, Ttyh1 KO astrocytes exhibited significantly higher intensity over the entire 60-minute course of the chase (Fig. 2d and Supplementary Fig. 3f), indicating that clearance of NBD-C1P requires Ttyh1. We then tested whether the evolutionarily conserved E210 and R213 residues are required for the NBD-C1P clearance function of TTYH1. We created a mutant TTYH1 construct, TTYH1ER/AA, by substituting these two charged residues with nonpolar alanine (Supplementary Fig. 4). The nonpolar alanine residues should weaken the interaction between C1P and TTYH1 and thus are expected to impair the extraction of C1P from the endolysosomal membrane. After 60-minute chasing, NBD-C1P fluorescence was significantly reduced in Ttyh1 KO astrocytes expressing TTYH1 (Fig. 2e). In contrast, Ttyh1 KO astrocytes expressing mutant TTYH1ER/AA failed to clear the NBD-C1P fluorescence (Fig. 2e). Expression of mutant TTYH1ER/AA did not impact the clearance of NBD-C1P fluorescence in WT control astrocytes (Fig. 2e), suggesting loss of function in TTYH1ER/AA. These results demonstrate that TTYH1 is required for intracellular clearance of exogenous C1P, a function that is conferred by the E210 and R213 residues. Thus, loss of TTYH1 will likely result in the accumulation of internalized C1P at the endolysosomal compartment.
Ttyh1 prevents C1P from inhibiting autophagic flux
Next, we tested how the absence of TTYH1 impacts autophagic flux when astrocytes are challenged by C1P (Fig. 3a). We found that exogenous C1P led to a dose-dependent accumulation of autophagosome markers p62 and LC3-II in mouse astrocytes (Fig. 3b), indicative of impeded autophagic flux. Additionally, while TTYH1 expression in human iAstrocytes was not altered by C1P (Supplementary Fig. 5a), p62 and LC3-II accumulated in response to increasing C1P concentrations (Supplementary Fig. 5b, c). Notably, Ttyh1/TTYH1 deficiency exacerbated the effects of C1P, resulting in significantly higher levels of p62 and LC3-II accumulations (Fig. 3b and Supplementary Fig. 5b, c). These data are consistent with the notion that TTYH1 downregulates C1P levels in endolysosomes, the failure of which causes autophagy impairment. To further validate that TTYH1 is required for counteracting C1P’s inhibitory effect on autophagic flux, we performed a rescue experiment by expressing human TTYH1 in Ttyh1 KO astrocytes. Whereas 0.5 µM C1P caused markedly increased p62 and LC3-II levels in mock-transfected Ttyh1 KO astrocytes, ectopic expression of human TTYH1 nullified the effects of C1P (Fig. 3c). Consistent with the findings that E210 and R213 are required for removing internalized C1P (Fig. 2e), TTYH1ER/AA was unable to rescue autophagic flux inhibition caused by C1P, resulting in p62 and LC3-II accumulations comparable to mock-transfected Ttyh1 KO astrocytes (Fig. 3c). Thus, TTYH1 relies on E210 and R213 to mitigate C1P’s effect on autophagic flux. We also tested if TTYH1 is sufficient to restore autophagic flux in wild-type astrocytes treated with excessive C1P. In primary astrocytes isolated from postmortem adult human brain (Supplementary Fig. 6), we found that C1P drastically elevated the levels of autophagic markers p62 and LC3-II (Fig. 3d). Overexpression of TTYH1, but not TTYH1ER/AA, reversed p62 and LC3-II levels to those in vehicle control (Fig. 3d). Together, these data indicate that astrocytes require TTYH1 to mitigate the inhibitory effect of C1P on autophagic flux.
a Schematic representation of TTYH1-mediated endolysosomal clearance of C1P. b Ttyh1 deficiency exacerbates autophagic flux inhibition by exogenous C1P. Levels of p62 and LC3 in cKO astrocytes in response to exogenous C1P were analyzed. Astrocytes were treated with 0 (0.1% DMSO), 0.1, or 0.5 µM of C1P in 0.1% BSA-supplemented culture media for 2 h. Shown are representative immunoblot images and quantifications of three independent experiments. Values were normalized to those of Ttyh1 WT, 0 µM C1P condition on the same blot. Data were presented as mean ± SEM. Number of biological replicates (n) is shown in brackets at the bottom of each bar. ***P < 0.001, **P < 0.01, One-way ANOVA (p62/αTub: F = 56.91, df = 17; LC3-II/I: F = 42.95, df = 23) followed by Bonferroni’s multiple comparisons post hoc test. c Human TTYH1 rescues C1P-induced autophagic flux blockage in Ttyh1 KO astrocytes. Levels of LC3 and p62 in Ttyh1 KO astrocytes in response to C1P (0.5 µM) were analyzed. Shown are representative immunoblot images and quantifications of four independent experiments. Values were normalized to those of the untreated mock-transfected control on the same blot. Data are presented as mean ± SEM. Number of biological replicates (n) is shown in brackets at the bottom of each bar. One-way ANOVA (p62/αTub: F = 15.43, df = 23; LC3-II/I: F = 40.88, df = 23) followed by Bonferroni’s multiple comparisons post hoc test. n.s. not significant, P > 0.9999. d Overexpression of TTYH1 in astrocytes restores autophagic flux upon C1P. Levels of LC3 and p62 in primary human brain astrocytes in response to C1P (50 µM) were analyzed. Shown are representative immunoblot images and quantifications of four independent experiments. Values were normalized to those of the untreated mock-transfected control on the same blot. Data were presented as mean ± SEM. Number of biological replicates (n) is shown in brackets at the bottom of each bar. One-way ANOVA (p62/αTub: F = 17.98, df = 23; LC3-II/I: F = 117.5, df = 23) followed by Bonferroni’s multiple comparisons post hoc test. n.s. not significant, P > 0.9999. Source data are provided as a Source Data file.
Ttyh1 deficiency impairs lipid droplet degradation
Astrocytes play a crucial role in supporting lipid metabolism within the nervous system. Their unique feature of possessing cytoplasmic lipid droplets (LDs) necessitates concerted pathways to mobilize lipids from these lipid depots for biosynthesis and bioenergetics. Endolysosomes are involved in four major LD catabolic pathways: lysosomal lipolysis, chaperone-mediated autophagy, microlipophagy, and macrolipophagy. Consequently, defective endolysosomal and autophagic machinery leads to the buildup of LDs and neutral lipids such as triacylglycerols (TAG)41. The astrocyte-enriched expression and the endolysosomal function of TTYH1 insinuate its potential role in LD metabolism. We therefore examined cellular TAG levels as a readout of LDs in Ttyh1 KO astrocytes. After feeding astrocytes with palmitic acid to promote LD synthesis, we found that Ttyh1 KO astrocytes contained comparable TAG levels to those of control astrocytes (Fig. 4a), suggesting that TTYH1 does not directly participate in LD biogenesis. Upon nutrient deprivation to stimulate LD breakdown, TAG in control astrocytes decreased to significantly lower levels compared to those in Ttyh1 KO astrocytes (Fig. 4a), indicating that Ttyh1 loss impedes LD degradation. TAG levels in both control and KO astrocytes remained high when endolysosomal acidification was inhibited by bafilomycin A1 (BafA1) (Fig. 4a), validating that TAG reduction upon LD degradation requires endolysosomal degradation. We then tested if the LD breakdown function is conserved in Drosophila TTYH1 ortholog, tweety (tty)34. Drosophila larval brain cells containing both neurons and glia were isolated and cultured34. Cells were then stained with fluorescent LD dye BODIPY493/503. Glial cells were identified by genetically encoded nuclear RFP whose expression was driven by a pan-glial driver (repo-GAL4) (Fig. 4b). We found that tty−/− mutant glia possessed more LDs compared to wild-type glia (Fig. 4c). Like in mammalian astrocytes, nutrient deprivation caused significant reduction in LD levels in wild-type but not tty−/− glia (Fig. 4c). To examine the impact of tty deficiency in vivo, we quantified the neutral lipid species extracted from fly heads using liquid chromatography-coupled mass spectrometry (LC-MS). Supporting the notion that TTYH1 orthologs regulate LD homeostasis, multiple species of TAG and diacylglycerides (DAG) were at significantly higher levels in tty−/− mutants compared to wild-type (Fig. 4d). Given that tty functions in glia and that glia are the major cell type that makes LDs in fly brain34,42, the neutral lipid buildup is likely attributed to glial LD accumulation. We further examined the impact of Ttyh1 loss in mouse astrocytes on brain neutral lipid homeostasis. After inducing astrocyte-specific deletion of Ttyh1 in the conditional knockout (cKO) mice (Supplementary Fig. 7a, b), we found that the mouse cortices contained significantly higher TAG levels (Fig. 4e). Taken together, these data demonstrate that TTYH1 facilitates LD degradation in astrocytes.
a Ttyh1 mediates lipid droplet (LD) breakdown in astrocytes. Triglycerides (TAG) levels in astrocytes were measured at 0 and 120 min after nutrient deprivation. Bafilomycin A1 (BafA1) inhibits lysosome degradation. Relative absorbance values were reported by the colorimetric TAG assay, and are normalized to the mean of Ttyh1 WT values at 0 min. Data were presented as mean ± SEM. Number of independent lipid extracts (n) is shown at the bottom of each bar. Unpaired two-tailed t-test (t = 105.2, df = 4). b, c Drosophila TTYH1 ortholog mediates LD breakdown in glia. Representative confocal images of primary glial cells isolated from wildtype (WT) and tty−/− Drosophila brains in (b) show glial nuclei (nucRFP; magenta) and BODIPY493/503 (BODIPY; green) stained LDs. Quantifications of BODIPY signals in c reveal LD contents in the glial cells. Each datapoint represents one glial cell. Data are presented as mean ± SEM. Number of glial cells (n) from ≥3 primary cell culture is shown in bracket underneath each bar. Mann–Whitney test. d Lipidomic profiles show increased amounts of TAG and DAG species in tty−/− Drosophila heads. Relative fold-changes are shown. Box plots show the median, the bounds of the box (25th and 75th percentiles) and whiskers representing minimum and maximum values. Data were from seven sample cohorts for each genotype; each cohort contained lipid extracts from ≥40 fly heads. Mann–Whitney test. e Astrocyte-specific knockout of Ttyh1 leads to TAG accumulation in mouse cortices. Two-month-old Aldh1l1-cre/ERT2; Ttyh1fl/fl mice were injected with either vehicle or tamoxifen (TAM) for 5 consecutive days. Protein and TAG contents in the dissected cortices were measured. TAG contents were quantitated by enzyme-coupled assay. TAG contents in the cortical tissues of female (n = 3) and male (n = 6) mice were shown separately. [TAG] quantity in each cortical tissue sample was normalized to the respective protein quantity. Box plots show the median, the bounds of the box (25th and 75th percentiles) and whiskers representing minimum and maximum values. Unpaired two-tailed t-test (Female: t = 41.04, df = 4; male: t = 33.33, df = 10). Source data are provided as a Source Data file.
Ttyh1 prevents LD accumulation induced by C1P
Our findings thus far indicate that TTYH1 mitigates C1P levels at endolysosomal membrane to facilitate degradative activity in astrocytes (Figs. 2, 3). We speculated that excessive C1P might overwhelm the capacity of TTYH1 and therefore dampen LD degradation. Indeed, C1P dose-dependently increased TAG levels in astrocytes (Fig. 5a), suggesting that C1P antagonizes LD breakdown. Notably, loss of Ttyh1 exacerbated TAG accumulation in response to increasing C1P concentrations (Fig. 5a). On the contrary, TTYH1 overexpression prevented the TAG accumulation in response to C1P (Supplementary Fig. 8). These results support the notion that TTYH1 mitigates endolysosomal levels of C1P to facilitate LD degradation. We further validated these findings by quantifying LD abundance in single astrocytes by LD staining (Fig. 5b). Loss of Ttyh1 led to significantly more LDs in astrocytes upon C1P exposure (Fig. 5b, c). Thus, Ttyh1 deficiency renders astrocytes more sensitive to C1P, thus shifting LD homeostasis further away from catabolism. These results reveal the function of C1P in astrocytic LD homeostasis and underscore the importance of TTYH1 in regulating such process.
a Loss of Ttyh1 sensitizes C1P-induced TAG accumulation in astrocytes. Cellular TAG contents in cKO astrocytes were measured after treatment with 0 (0.1% DMSO), 0.5, or 5 µM of C1P in 0.1% BSA-supplemented culture media for 2 h. Data were presented as mean ± SEM. Number of independent lipid extracts (n) is shown at the bottom of each bar. One-way ANOVA (F = 581.7; df = 20) followed by Bonferroni’s multiple comparisons post hoc test. b, c Loss of Ttyh1 exacerbates C1P-induced LD buildup in astrocytes. Representative confocal images in (b) show cKO astrocytes stained with BODIPY (green) and DAPI (blue) after indicated treatments. Quantifications of BODIPY signals in (c) reveal LD contents in Ttyh1 WT and KO astrocytes. BODIPY-positive area per cell values were normalized to the mean values of Ttyh1 WT control. Violin plots show median (solid line), interquartile range (dotted line), and probability density of data (smoothed shape). Number of astrocytes (n) from ≥3 independent experiments is shown underneath each violin plot. Mann–Whitney test. Source data are provided as a Source Data file.
Neuronal C1P signals autophagic flux inhibition in astrocytes
Neurons transfer lipids to astrocytes for metabolic processing and storage42. Conceivably, some of these lipids might signal astrocytes to initiate metabolic adaptation. C1P produced by neurons might be one such signal to alter endolysosomal function in astrocytes. C1P can be found in extracellular vesicles, including rat synaptic vesicles, where it was first identified43,44,45. Ceramide kinase (CERK), localized to the trans-Golgi, is the major biosynthetic enzyme of C1P46. CERK activation requires cytosolic Ca2+ rise, a condition that can be fulfilled in neurons by the inflammatory cytokine IL-1β47,48. In both mouse cortical neurons and human iPSC-derived neurons (iNeurons), we found that intracellular C1P levels were significantly increased upon IL-1β stimulation (Fig. 6a and Supplementary Fig. 9a). CERK activation likely accounts for the increase since cotreatment with CERK inhibitor NVP-231 restored C1P levels (Fig. 6a and Supplementary Fig. 9a). Importantly, C1P concentration in the neuronal conditioned medium (NCM) followed intracellular C1P levels in these conditions (Fig. 6a), demonstrating that IL-1β promotes neuronal secretion of C1P. Next, we tested whether lipids secreted by IL-1β-stimulated neurons inhibit astrocyte autophagy like C1P does (Fig. 3). To avoid collecting IL-1β or CERK inhibitor in the lipids secreted by neurons, we conditioned NCM after withdrawing IL-1β from the neuronal culture. Extracellular lipids were then extracted from the NCM conditioned by naïve or IL-1β-stimulated neurons (Fig. 6b). The lipid extracts, referred to as naïve-lipids and IL-1β-lipids hereafter, were then used to treat astrocytes before assessing autophagic flux by Western blot (Fig. 6b). Both control and Ttyh1 KO astrocytes exhibited accumulations of p62 and LC3-II upon treatment with IL-1β-lipids derived from mouse cortical neurons (Fig. 6c, d). Importantly, neuronal CERK inhibition (NVP-231) abolished the effect of IL-1β-lipids (Fig. 6c, d). Validating the role of CERK, neuronal Cerk knockdown reproduced the effect of NVP-231, in which autophagic flux was not altered despite IL-1β stimulation (Supplementary Fig. 9b−e). Furthermore, adding exogenous C1P to the IL-1β-lipids derived from Cerk-inhibited neurons reinstated p62 and LC3-II accumulations (Fig. 6c, d and Supplementary Fig. 9d, e). These data demonstrate that IL-1β elicits neuronal signaling lipid secretion to impede autophagy in astrocytes and that such lipids are associated with C1P. In addition, Ttyh1 KO astrocytes accumulated more LC3-II when treated with IL-1β-lipids (Fig. 6c, d). Thus, astrocytes require TTYH1 to maintain autophagic flux in response to neuron-to-astrocyte lipid signaling induced by IL-1β. We also tested if such paradigm applies to human neurons and astrocytes. IL-1β-lipids derived from human iNeurons also caused LC3-II accumulation in human iAstrocytes (Supplementary Fig. 10). Like in mouse cells, the LC3-II accumulation was exacerbated by TTYH1 knockdown in iAstrocytes and was completely restored by CERK inhibition in iNeurons (Supplementary Fig. 10). On the other hand, overexpression of TTYH1, but not TTYH1ER/AA, protected primary human brain astrocytes from autophagic flux inhibition caused by IL-1β-lipids (Fig. 6e, f). Taken together, our findings reveal a C1P-dependent neuron-to-astrocyte lipid signaling paradigm and highlight the importance of TTYH1 in mitigating the effect of neuron-derived lipids on astrocyte autophagy.
a IL-1β induces C1P production and secretion in mouse cortical neurons. Data were presented as mean ± SEM. Number of independent lipid samples (n) is shown in bracket at the bottom of each bar. One-way ANOVA (Intracellular: F = 33.81, df = 8; Extracellular: F = 3214, df = 11) followed by Tukey’s multiple comparisons post hoc test. b Schematic representation of extracting lipids from neuronal conditioned medium (NCM). NCM lipids derived from IL-1β-treated and untreated neurons are referred to as IL-1β-lipids and naïve-lipids, respectively. c, d Autophagic flux inhibition by IL-1β-lipids is exacerbated by Ttyh1 deficiency and is neuronal Cerk-dependent. cKO astrocytes were treated with NCM lipids for 30 min. For the C1P (+) groups, 0.5 µM C1P was added with NCM lipids. Shown are representative immunoblot images (c) and quantifications of p62 and LC3 levels (d) from n = 3 independent experiments. Same samples were also loaded on a separate gel for probing Ttyh1 expression. Ratios were normalized to that of Ttyh1 WT-naïve-lipids control on the same blot. Data were presented as mean ± SEM. One-way ANOVA (p62/αTub: F = 40.74, df = 23; LC3-II/LC3-I: F = 118.1, df = 23) followed by Bonferroni’s multiple comparisons post hoc test. e, f TTYH1 overexpression in human astrocytes rescues IL-1β-lipids-induced autophagic flux inhibition. NCM lipids were derived from human iNeurons. Shown are representative immunoblot images (e) and quantifications of p62 and LC3 levels (f). Ratios were normalized to that of mock-naïve-lipids control on the same blot. Data were presented as mean ± SEM. Number of biological replicates (n) is shown at the bottom of each bar. One-way ANOVA (p62/αTUB: F = 13.5, df = 26; LC3-II/LC3-I: F = 35.09, df = 50) followed by Bonferroni’s multiple comparisons post hoc test. n.s. not significant, P > 0.999. Source data are provided as a Source Data file.
Ttyh1 prevents LD accumulation induced by neuronal lipids
Does C1P mediate the neuron-astrocyte signaling to regulate LD homeostasis? Given that exogenous C1P increases LD abundance in astrocytes (Fig. 5), we speculated that neuronal CERK activity might initiate LD alterations in astrocytes. To examine such a possibility, we first perturbed neuronal CERK expression in Drosophila brain by either overexpressing or knocking down Cerk in neurons. Drosophila Cerk is the only CERK ortholog in flies and is associated with phototransduction and photoreceptor degeneration49. Induction of ectopic Cerk expression in neurons significantly increased TAG levels in fly heads (Fig. 7a), indicative of LD accumulation in the brain. Conversely, neuronal knockdown of Cerk drastically reduced TAG levels (Fig. 7a), demonstrating that neuronal C1P production correlates with brain LD abundance. Since glia are the primary cell type that synthesize LDs in the fly brain50,51, these findings support the model that C1P is a neuron-derived signaling lipid that regulates LD homeostasis in glia. Since IL-1β promotes C1P production in neurons (Fig. 6a), we next tested whether the IL-1β-induced neuron-to-astrocyte lipid signaling might also regulate astrocyte LDs. Indeed, BODIPY-positive LDs in both control and TTYH1-deficient human iAstrocytes were significantly increased in response to IL-1β-lipids (Fig. 7b, c). Whereas the BODIPY-positive LDs in mouse Ttyh1 WT astrocytes were not altered by IL-1β-lipids, loss of Ttyh1 sensitized astrocytes to IL-1β-lipids, resulting in the buildup of LDs and TAG (Fig. 7d and Supplementary Fig. 11). Since Ttyh1 deficiency impairs LD degradation (Fig. 4a), we reason that the LD accumulation is likely due to perturbed LD catabolism when endolysosomes fail to process IL-1β-lipids. Importantly, neuronal CERK inhibition abolished the effect of IL-1β-lipids in Ttyh1 KO astrocytes (Fig. 7d and Supplementary Fig. 11), indicating that C1P underlies the LD modulating function of IL-1β-lipids. Collectively, these data suggest that astrocytes require TTYH1 to coordinate between neuron-astrocyte lipid signaling and LD homeostasis.
a Neuronal C1P production regulates TAG abundance in Drosophila heads. Each datapoint represents one cohort of ≥20 fly heads. Absorbance values from the colorimetric TAG assay were normalized to the mean of controls (Switch OFF). Number of independent lipid extracts (n) from fly heads is shown in brackets at the bottom of each bar. Unpaired two-tailed t-test (Cerk: t = 6.881, df = 12; Cerk-IR: t = 28.87, df = 4). b, c Loss of TTYH1 exacerbates IL-1β-lipids-induced lipid droplet (LD) accumulation in astrocytes. Shown are representative confocal images (b) and quantifications of BODIPY signals (c). Values were normalized to the mean of the ctrl-shR-naïve-lipids controls. One-way ANOVA (F = 11.79, df = 190) followed by Bonferroni’s multiple comparisons post hoc test. d Astrocytic Ttyh1 and neuronal C1P production are associated with the IL-1β-lipids-induced LD accumulation in astrocytes. Quantifications of BODIPY signals are shown. Values were normalized to the mean of Ttyh1 WT-naïve-lipids controls. One-way ANOVA (F = 11.79, df = 190) followed by Bonferroni’s multiple comparisons post hoc test. e Schematic representation of the neuron-astrocyte coculture system. f Ttyh1 KO astrocytes cocultured with IL-1β-stimulated neurons accumulate LDs. Shown are quantifications of BODIPY signals in astrocytes after coculture. Values were normalized to the mean of Ttyh1 WT-beads controls. One-way ANOVA (F = 4.395, df = 108) followed by Bonferroni’s multiple comparisons post hoc test. n.s. not significant, P > 0.9999. g Loss of Ttyh1 exacerbated TAG accumulation in astrocytes cocultured with IL-1β-stimulated neurons. TAG contents in astrocytes were measured after coculture. In C1P (+) groups, 0.5 µM C1P was added to the coculture medium. Absorbance values from the colorimetric TAG assay were normalized to the mean of Ttyh1 WT-naïve-ctrl-shR controls. Number of independent lipid extracts (n) from ≥3 independent experiments is shown in brackets at the bottom of each bar. One-way ANOVA (F = 937.6, df = 77) followed by Bonferroni’s multiple comparisons post hoc test. ***P < 0.001. Violin plots in (c–f) represent median (solid line), interquartile range (dotted line), and probability density of data (smoothed shape). Number of astrocytes (n) from ≥3 independent experiments is shown in brackets underneath each violin plot. Bar charts in (a, g) represent mean ± SEM. Source data are provided as a Source Data file.
IL-1β-stimulated neurons signal astrocytic LD accumulation
To avoid the presence of IL-1β or CERK inhibitor in the lipid extracts, we conditioned NCM after withdrawing IL-1β from the neuronal culture. Whereas the robust effects on autophagy and LD homeostasis can be attributed to the extracted lipids, they might be an outcome of neuronal resolution to prior inflammatory cytokine stimulation. It was unclear whether neurons produce similar signaling lipids during persistent cytokine challenge in contexts such as neuroinflammation. Of relevance to our findings, emerging evidence indicates that glial LD accumulation is associated with neuroinflammation52,53. To model the neuron-astrocyte lipid signaling during chronic cytokine challenge, we adapted a conventional neuron-astrocyte coculture system for our experiments12. Neurons were grown in transwell inserts that were placed inside astrocyte-seeded wells (Fig. 7e). While both neuronal and astrocytic compartments shared the same medium, the 0.4-µm pores on the transwell membrane permitted the exchange of materials such as lipoproteins and exosomes between the two cell types. By conjugating IL-1β to 0.8-µm beads, we restricted IL-1β exposure in the neuronal compartment during coculture (Supplementary Fig. 12). After 24 h of coculture, LD levels in the astrocytes were quantified. BODIPY staining revealed that both control and Ttyh1 KO astrocytes possessed comparable LD contents when cocultured with unstimulated (beads only) neurons. Interestingly, when neurons were exposed to IL-1β-conjugated beads, the LD contents in control astrocytes appeared to decrease, while those in Ttyh1 KO appeared to increase (Fig. 7f). However, neither of these changes were statistically significant. Nevertheless, Ttyh1 KO astrocytes exhibited significantly more LDs compared to control astrocytes when cocultured with IL-1β-stimulated neurons (Fig. 7f), consistent with the notion that TTYH1 mediates the astrocytic response to neuron-derived lipids. Furthermore, we assessed the role of C1P by knocking down Cerk in the cocultured neurons. In line with earlier findings made by using NCM, TAG levels in astrocytes were significantly elevated by coculturing with IL-1β-stimulated neurons with unperturbed Cerk (Fig. 7g). The increase in TAG was substantially higher in Ttyh1 KO astrocytes compared to control astrocytes (Fig. 7g). Furthermore, whereas neuronal Cerk knockdown nullified the IL-1β-induced TAG increase in both control and Ttyh1 KO astrocytes, addition of exogenous C1P reinstated TAG accumulation despite Cerk knockdown (Fig. 7g). Thus, our coculture model demonstrates that neuroinflammatory cytokine triggers a transcellular lipid signaling paradigm that stems from neuronal C1P production and is modulated by TTYH1 in astrocytes. This signaling mechanism culminates in regulating autophagic flux and lipid droplet degradation in astrocytes (Fig. 8).
Inflammatory cytokine stimulates C1P biosynthesis in neurons. C1P and other lipid metabolites secreted by neurons are taken up by the astrocyte endolysosomes, where they are processed by resident enzymes. TTYH1 facilitates the extraction of C1P from endolysosomal membrane, which is necessary for maintaining autophagic flux and lipid droplet degradation. Excessive C1P internalized by astrocytes slows down autophagic flux and reduces lipid droplet degradation. These catabolic activities are further impaired upon TTYH1 deficiency, resulting in autophagic flux blockage and lipid droplet accumulation.
Discussion
Our investigations on TTYH1 uncover its endolysosomal functions in regulating catabolic processes in astrocytes and mediating a neuron-astrocyte lipid signaling paradigm. TTYH1 orthologs in Drosophila glia, mouse astrocytes, and human iPSC-derived astrocytes are essential for two endolysosome-dependent processes: autophagic flux and lipid droplet (LD) degradation. Computational and experimental evidence demonstrate that TTYH1 facilitates the endolysosomal removal of ceramide 1-phosphate (C1P), a sphingolipid metabolite that impairs both autophagic flux and LD breakdown in astrocytes. Additionally, TTYH1 mitigates the effects of neuron-derived C1P, produced in response to inflammatory cytokine stimulation, on astrocyte autophagy and LD breakdown. Our findings highlight the importance of TTYH1 in the endolysosomal processing of signaling lipids that regulate metabolism in astrocytes, potentially impacting neuroinflammation and related neurological conditions.
Recent cryo-EM studies have shed light on the structural features of TTYH proteins that are likely critical to their molecular functions. Our in silico docking results support the hypothesis of a lipid-interacting function for TTYH1, suggesting it could interact with multiple sphingolipid species via a hydrophobic cavity, thereby facilitating their extraction from the membrane. Notably, we identified two charged amino acid residues, E210 and R213, as crucial for interaction with the sphingolipid C1P. E210A/R213A substitutions render TTYH1 non-functional in rescuing the inhibitory effect of C1P on autophagic flux. Since endolysosomes are the primary site of sphingolipid degradation, it remains to be tested whether E210 and R213 also contribute to the clearance of other sphingolipids associated with autophagic impairment [30651381]. Beyond the sphingolipid-binding function of saposins54, little is known about how sphingolipids enter the endolysosomal lumen, where acid hydrolases are located. We hypothesize that the charged residues anchor the sphingolipid headgroup to TTYH1 through strong charge-charge or hydrogen bond interactions as the acyl chains leave the membrane, thereby promoting their displacement to the lumen. Our findings thus suggest that TTYH1, along with TTYH2 and TTYH3, may play a broader role in sphingolipid metabolism.
We demonstrate that endolysosomal processing of signaling lipid C1P modulates organellar interactions. While our data reveal a robust inhibitory effect of C1P on autophagic flux in astrocytes, the mechanism by which exogenous C1P influences interactions between endolysosomes and other organelles remains poorly understood. Thus far, such inhibition of autophagy has only been observed in neurons, where it is mediated by the activation of cytosolic phospholipase A240. C1P may induce ectopic vesicular fusion through its fusogenic properties55,56, thereby disrupting autophagosome-endolysosome fusion. Conversely, the site of C1P accumulation may determine its effect on autophagy. Loss of C1P transfer protein (CPTP) induces autophagy and inflammasome formation, presumably due to the accumulation of C1P produced in the trans-Golgi57. Further study is required to determine whether endolysosomal C1P can be transferred to the trans-Golgi when TTYH1-mediated C1P clearance is impaired.
Our findings reveal that neurons respond to the inflammatory cytokine IL-1β by upregulating C1P synthesis. Besides its potential role in inflammasome induction57, neuronal C1P synthesis may modulate inflammatory signaling by inhibiting tumor necrosis factor production58. Furthermore, increased C1P levels have recently been associated with a disrupted lipid profile in the secretory pathway59. Thus, elevated C1P synthesis in response to IL-1β may initiate a reprogramming of the neuronal secretory profile—one that signals to neighboring cells for metabolic adaptations. Other neuronal processes might also initiate a similar CERK-dependent transcellular lipid signaling paradigm. Since Ca2+ activates CERK, sustained cytosolic Ca2+ levels during neuronal hyperexcitability or excessive neuronal firing will likely promote neuronal C1P production. The significance of neuron-to-astrocyte C1P signaling during these pathophysiological conditions warrants further examination. Since astrocytic LD accumulation was recently associated with epilepsy60, we speculate that TTYH1 and transcellular lipid signaling might be involved in the perturbed LD homeostasis. Nevertheless, since hyperexcitability and neuroinflammation cooccur in conditions such as neurodegeneration and epilepsy61,62, neuronal CERK activation may represent a common mediator to trigger a neuron-astrocyte lipid signaling paradigm in neurologic disorders. Although neuronal CERK regulates neutral lipid storage in Drosophila brain (Fig. 7a), our findings on this lipid signaling paradigm are primarily derived from controlled ex vivo experimental conditions. Delineating how the transcellular lipid signaling functions under complex cytokine dynamics will require in vivo examinations in neuroinflammation models.
What could be the potential benefit of slowing down catabolic processes in astrocytes? Glial LD expansion observed upon neuronal stress appears to be a protective response11,13,63,64. Peroxidated lipids generated by stressed neurons are transferred to glial LDs10,11,12,13. Likewise, during neuroinflammation or hyperexcitation, neurons might release peroxidated lipids to neighboring glial cells. In this context, a transcellular signaling paradigm that slows down LD catabolism in glia could facilitate the sequestration of these toxic lipids in LDs and prevent their mobilization to other subcellular compartments and neighboring cells. Additionally, autophagy promotes lipid mobilization via recycling of membrane components. Impeded autophagic flux may work in tandem with reduced LD catabolism to limit the mobilization of toxic lipid species. Furthermore, inhibition of autophagy or TTYH1 ortholog was recently found to increase glycolytic energy production in glia34. The neuron-astrocyte lipid signaling identified here might also reprogram bioenergetics in astrocytes. Further research is needed to determine whether such bioenergetic adaptation occurs in astrocytes during neuroinflammation.
Methods
Research in this study complies with all relevant ethical regulations and has been approved by the Institutional Animal Care & Use Committee and the Institutional Biosafety Committee of Rutgers University.
Antibodies and chemicals
Antibodies: Anti-TTYH1 (Western blot (WB): 1:2500; immunofluorescence (IF): 1:500) (26973-1-AP; Proteintech); anti-ALDH1L1 (WB: 1:1000; IF: 1:500) (NBP2-50045; Novus Biologics); anti-LAMP1 (IF: 1:100) (H4A3-c; Developmental Studies Hybridoma Bank); anti-SQSTM1 (p62) (WB: 1:5000) (AB109012-1001; Abcam); Anti-LC3 (WB: 1:1000; IF: 1:200) (83506S; Cell Signaling); anti-alpha-tubulin (WB: 1:2500) (12G10; Developmental Studies Hybridoma Bank); anti-RFP (WB: 1:3000) (600-401-379; Rockland); anti-GFAP (WB: 1:2000; IF: 1:500) (3670S; Cell Signaling); anti-β-III-TUBULIN (WB: 1:2000; IF: 1:400) (MAB1195; Bio-techne); anti-Synapsin-1 (IF: 1:200) (5297S; Cell Signaling); anti-NESTIN (WB: 1:1000) (33475S; Cell Signaling); anti-S100B (WB: 1:1000) (66616-1-Ig; Proteintech); anti-Eea1 (IF: 1:200) (48453S; Cell Signaling); anti-Rab11 (IF: 1:500) (610656; BD Biosciences); anti-serum albumin (WB: 1:5000) (A11133; Invitrogen); goat anti-mouse IgG IRDye 800CW (WB: 1:10000) (926-32210; Li-COR); goat anti-rabbit IgG IRDye 680LT (WB: 1:15000) (926-68021; Li-COR); donkey anti-mouse IgG Alexa Fluor 488 (IF: 1:500) (A-21202; Invitrogen); donkey anti-rabbit IgG Alexa Fluor 488 (IF: 1:500) (A-21206; Invitrogen); donkey anti-mouse IgG Alexa Fluor 568 (IF: 1:500) (A10037; Invitrogen); donkey anti-rabbit IgG Alexa Fluor 568 (IF: 1:500) (A10042; Invitrogen).
Chemicals: Ceramide 1-phosphate (d18:1/16:0) (C1P; 22542; Cayman); bafilomycin A1 (BafA1; B1793; Sigma-Aldrich); NVP-231 (HY-13945; MedChem Express); human IL-1β (SRP3083; Sigma-Aldrich); biotinylated human IL-1β (ILB-H82E9; Acro Biosystem); bicuculine (Bic; HY-N0219; MedChem Express); 4-aminopyridine (4AP; 275875; Sigma-Aldrich); methyl tert-butyl ether (MTBE; 34875; Sigma-Aldrich).
Generation of Ttyh1 flox conditional knockout mouse
Research activities involving mice in this study have been approved by the Institutional Animal Care & Use Committee of Rutgers University. All mice were housed in cages with free access to water and diet, in a temperature (20 to 22 °C) and humidity (45 to 55%) controlled room with 12/12-hour light/dark cycle, at the Rutgers Animal Care Facility. The Ttyh1flox conditional knockout mouse strain was generated by the Rutgers University Genome Editing Shared Resource core. LoxP (ATAACTTCGTATAATGTATGCTATACGAAGTTAT) sites were inserted into the introns immediately flanking exon 4 of Ttyh1 in C57BL/6 strain (The Jackson Laboratory) using CRISPR/Cas9. Founder mice were cross-bred to generate homozygous Ttyh1fl/fl. Genotyping was performed by Sanger sequencing on the PCR amplicons of the loxP-inserted intron sequences using the following primers:
5’-loxP: CTGGAGACCAAAAGCAGCCAAATC and AGTTCCTGAACTTGCATCTGAGCC
3’-loxP: TGTCCCACTGGTTGAATGAGAAGC and AGACGAGTTGCACGATGAACACAG
Homozygous Ttyh1fl/fl mice were crossed with Aldh1l1-Cre/ERT2 BAC transgenic mice (031008; The Jackson Laboratory) to generate the Aldh1l1-Cre/ERT2; Ttyh1fl/fl mice. Genotyping for the Aldh1l1-Cre/ERT2 allele was performed by PCR amplification using the following primers:
CTTCAACAGGTGCCTTCCA and GGCAAACGGACAGAAGCA
Isolation and primary culture of mouse cortical neurons and astrocytes
Mouse brains of P0 newborn mice of both sexes were dissected, and the isolated brains were placed in ice-cold phosphate-buffered saline (PBS; 10010023; Gibco). Meninges were removed above the cortex region, and the cortex was separated, and the cortical tissues were dissected into small pieces. After washing three times with PBS, the cortical tissues were incubated with collagenase (1 mg/mL; C2674; Sigma-Aldrich) for 30 min at 37 °C. Collagenase was then removed, and the residual collagenase was neutralized with fetal bovine serum (FBS; F4135; Sigma-Aldrich) by incubating for 5 min at room temperature. After removing FBS, the tissues were triturated through pipette tips to make a single-cell suspension. For cortical neurons, cells were suspended in neurobasal medium A supplemented with B-27 (10888022; Gibco), Glutamax (35050061; Gibco), and 1% penicillin and streptomycin. Neurons were plated on the surface coated with poly-d-lysine. Medium was refreshed after 30 min to remove unattached cells. Neurons were maintained with 50% medium change every 2 days for 7 days before experiments. For cortical astrocytes, cells were suspended in primary astrocyte medium (A1261301; Gibco) supplemented with 1% penicillin and streptomycin, and were plated on the surface coated with Geltrex (A1413201; Gibco). The culture dish was then shaken for 30 min on an orbital shaker before replacing the medium with fresh primary astrocyte medium. Medium was refreshed every 3 days until reaching confluency for passage.
Astrocyte culture
Both primary mouse cortical astrocytes and primary human brain astrocytes (HMP202; Neuromics; adult female postmortem) were expanded and maintained in primary astrocyte medium (A1261301; Gibco) supplemented with 1% penicillin and streptomycin. For cortical astrocytes derived from Aldh1l1-Cre/ERT2; Ttyh1fl/fl mice, cells were treated with either 1 µg/mL 4-hydroxy tamoxifen (4-OHT; 14854; Cayman) or vehicle (DMSO) for 1 week before experiments. Astrocytes seeded for experiments were cultured in low-serum astrocyte medium (1801; ScienCell) for at least 2 days, followed by overnight serum-free medium synchronization before experiments.
Neuronal conditioned medium (NCM)
Mouse cortical neurons were seeded on a six-well plate at a density of ~50k cells per well. Mouse neurons were cultured in B-27 supplemented neural basal medium (10888022; Gibco) for 7 days before treatment and medium conditioning. For Cerk knockdown, mouse neurons were transduced with a lentiviral vector and cultured for an additional 3 days. For iNeurons, cells were seeded on a 24-well plate at a density of ~57k cells per well. iNeurons were cultured in STEMdiff forebrain maturation medium (08605; Stemcell Tech) for 14 days before treatment and medium conditioning. Neurons were treated with 10 ng/mL of IL-1β together with NVP-231 (12 nM; HY-13945; MedChem Express) or vehicle DMSO (0.1%) for 2 h. IL-1β was omitted in the naïve treatment condition. After treatment, the culture medium was replaced with plain neural basal medium (10888022; Gibco) for 2 h to condition the medium. The neuronal conditioned medium (NCM) was first centrifuged for 5 min at 1500 × g to remove cell debris. The supernatant was collected for solvent extraction of secreted lipids. After solvent extraction, dried lipids were reconstituted in 800 µL DMEM (10569-010; Gibco). The lipid extract was diluted in half to treat one well of 25k astrocytes seeded in a 24-well plate for 2 h, before lysate collection for Western blotting or immunofluorescence for lipid droplets. NCM lipids derived from mouse neurons were used to treat primary mouse and human brain astrocytes. NCM lipids derived from iNeurons were used to treat iAstrocytes. For lipid droplet experiments, the treatment time was 30 min, before BODIPY staining.
Neuron-astrocyte coculture
Primary mouse cortical neurons were seeded in poly-D-lysine-coated transwell inserts (230635; CELLTREAT) and cultured in B-27 supplemented neurobasal medium for 7 days. For shRNA-mediated Cerk knockdown, neurons were transduced with lentiviral vectors and cultured for an additional 3 days. One day before coculture, mouse cortical astrocytes were seeded in 24-well plates (P24-1.5P; Cellvis) and cultured in neurobasal medium. The neuron-containing transwells were then inserted into the astrocyte-seeded wells. Neurons and astrocytes were cocultured in neurobasal medium for 1 day before the addition of beads to the transwell. Approximately 1.9 nmol of biotinylated human IL-1β (ILB-H82E9; Acro Biosystem) was incubated with 0.5 mg of streptavidin magnetic beads (CCT-1497; Vector Laboratories) in DMEM/F-12 medium (12660012; Gibco) for 30 min before washing with DMEM/F-12 for three times on a magnetic rack. The IL-1β-conjugated beads were resuspended at 1 µg IL-1β (12.5 mg beads) per milliliter of DMEM/F-12 (100x stock). Control unconjugated beads were prepared the same way, omitting IL-1β. Beads were added at 1/100 dilution to the neuron-containing transwells for an additional 24 h of coculture. Astrocytes in the bottom wells were then either stained for lipid droplets or lysed for lipid extraction.
Differentiation of human neurons (iNeurons) and astrocytes (iAstrocytes) from iPSC-derived neural progenitor cells (NPCs)
Human iPSC-derived NPCs were differentiated from the iPSC line SCTi003-A (derived from female blood mononuclear cells) and were obtained directly from the manufacturer, Stemcell Technologies (200-0620; Stemcell Tech). The NPCs were cultured in STEMdiff Neural Progenitor Medium (05833; Stemcell Tech) for fewer than four passages before differentiation into iNeurons and iAstrocytes, respectively. Neuronal differentiation was performed using the STEMdiff Forebrain Neuron Differentiation kit (08600; Stemcell Tech). NPCs were seeded on a culture plate coated with poly-L-ornithine and laminin (PLO + Lam), and were cultured in STEMdiff Forebrain Neuron Differentiation Medium (08600; Stemcell Tech) for one week. Cells were then passaged and seeded on a 24-well plate coated with PLO + Lam and cultured in STEMdiff Forebrain Neuron Maturation Medium (08605; Stemcell Tech) for 2 to 3 weeks. The resulting iNeurons were validated by the expression of MAP2 and β-III-tubulin detected by Western blot of cell lysate and immunofluorescence of fixed cells. For iAstrocytes, differentiation was performed by adapting the protocol developed by ref. 65 NPCs were seeded on a culture plate coated with Geltrex (A1413202; Gibco) and cultured in astrocyte medium (1801, ScienCell). Cells were passaged when confluency reached 80% and maintained for at least 3 weeks, before assessing for astrocytic markers and transduction with shRNA-containing lentiviral vectors. Expression of astrocytic markers, GFAP and ALDH1L1, was validated by Western blot of cell lysate. After transduction with lentiviral vectors, iAstrocytes were cultured in astrocyte medium containing 10 µg/mL hygromycin for at least two weeks. The resulting shRNA-expressing iAstrocytes were then cultured in serum-free astrocyte medium for at least 3 days before passage and seeding for experimentation.
shRNA-mediated knockdown
Lentiviral vectors carrying shRNAs targeting human TTYH1 and mouse Cerk were synthesized by GeneCopoeia. For human TTYH1 knockdown in iAstrocytes, the psi-LVRU6MH vector containing U6 promoter, mCherry expression construct, and hygromycin resistance cassette was used. The target sequences for TTYH1 and control shRNAs are as follows.
TTYH1: GGCATTGGCATCGGTTTCTAT
Control: GCTTCGCGCCGTAGTCTTA
For Cerk knockdown in mouse cortical neurons, the psi-LVRU6GP vector containing U6 promoter, GFP expression construct, and puromycin resistance cassette was used. The target sequences for Cerk and control shRNAs are as follows.
Cerk: AATCCGTTCGCATTCACAGTC
Control: GCTTCGCGCCGTAGTCTTA
Lentiviral vectors were produced using the 3rd generation packaging system and further purified by centrifugation in a lentivirus concentrator (Viral-PEG LVG100; OZBioscience). Upon transduction, iAstrocytes were cultured with 10 µg/mL hygromycin for at least two passages. Hygromycin was withdrawn when cells were seeded for experiments. For mouse cortical neurons, cells were not subject to selection and were used for experiments 3 days after transduction. Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to assess Cerk knockdown efficiency in mouse neurons using the following primers:
Cerk: CAATCAAGGCAACCTCTCACAGACTTGGAT (forward), TGGCACAGGTCTTTCACCAGGCTTTGGACA (reverse)
Actb: CTGTCGAGTCGCGTCCA (forward), ACGATGGAGGGGAATCAGC (reverse)
Generation of expression constructs
Complementary DNA sequence of C-terminally RFP-tagged human TTYH1 (NM_001005367.2) was synthesized as described previously34. Site-directed mutagenesis to generate the TTYH1ER/AA mutant was performed using NEBuilder HiFi (New England Biolabs) and the following primers (mutated codons are underlined): TGGCGGAGTACGCGTGGCTGGCCTA and CACGCGTACTCCGCCACAAAGGACACA. The cDNA sequences of TTYH1 and TTYH1ER/AA were respectively subcloned into the expression vector pQCXI-Puro, which drives transgene expression by a CMV promoter.
Transfection of astrocytes with expression constructs
Astrocytes were transfected with expression constructs of GFP-LC3-RFP-LC3ΔG, TTYH1, or TTYH1ER/AA using Lipofectamine 3000 (L3000001; Invitrogen). Plasmid DNA of pMRX-IP-GFP-LC3-RFP-LC3ΔG was a kind gift from Dr. Noboru Mizushima and was obtained from Addgene. Four hours after transfection, the transfection complex in Opti-MEM reduced serum medium (31985070; Invitrogen) was removed and replaced with fresh astrocyte medium. One day after transfection, cells were synchronized in serum-free medium overnight, before recovery in astrocyte medium for 4 h. Cells were then treated before imaging or lysate collection for Western blotting.
Western blotting
Cells were lysed in RIPA buffer (9806S; Cell Signaling) supplemented with protease and phosphatase inhibitor cocktail (78447; Thermo Scientific). Laemmli buffer (161-0747; Bio-Rad) was added to lysate before loading onto 4–20% gradient gel (4561096; Bio-Rad) for electrophoresis. Separated proteins were transferred onto a nitrocellulose membrane (10600004; Amersham), followed by blocking (927-80001; Li-COR). The membrane was cut at the 25 kDa ladder mark into two parts. The lower part was used for LC3 detection, while the upper part was used for other protein targets. After incubation with primary antibodies, IRDye 680LT- and IRDye 800CW-conjugated secondary antibodies were used to detect the protein targets on the membrane. The membrane image captured by a ChemiDoc MP imager (Bio-Rad) was exported to a TIFF image file. The images were processed and analysed using ImageJ (National Institutes of Health).
Immunofluorescence
Cultured cells: Astrocytes seeded on glass-bottom dishes were cultured in astrocyte medium (1801; ScienCell) for 48 h before overnight synchronization in serum-free medium. Cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. After washing three times with PBS, cells were permeabilized by incubating with 0.1% Triton X-100-containing PBS (PBST) for 15 min. Samples were then blocked with 5% normal donkey serum (017-000-121; Jackson Immuno). Samples were incubated overnight at 4 °C in primary antibodies diluted in PBST with 1% BSA (PBST-BSA). After primary antibody incubation, samples were washed three times with PBS and incubated with the fluorophore-conjugated secondary antibodies in PBST-BSA for 1 h at room temperature in the dark. After washing with PBST three times, samples were immersed in mounting medium with DAPI (P36985; Invitrogen). Images were taken using confocal microscopy (Zeiss LSM 980 with Airyscan; 100x oil objective) with appropriate laser power and filter selection. Representative images were processed and generated using Zen 3.3 (Zeiss) and ImageJ (National Institutes of Health).
Mouse brain tissues: Mice were fully anesthetized with a cocktail of ketamine (K; 100 mg/kg), xylazine (X; 10 mg/kg), and acepromazine (A; 10 mg/kg) (KXA; 0.1 ml/10 g body weight; intraperitoneal injections). They were then perfused transcardially through the left ventricle with cold PBS, followed by 4% paraformaldehyde (PFA) in PBS. The brains were placed in 30% sucrose after 24-h fixation in PFA. Brains were sectioned and stored in an antifreeze solution at −20 °C. Coronal sections (40-μm thick) spanning the entire brain from anterior to posterior were prepared in serial order. For histological analysis, an antigen retrieval procedure was performed as previously described66. Briefly, the sections were placed in hot citrate buffer (1.8 mM citric acid, 8.2 mM tri-sodium citrate) and incubated for 7 min. The sections were then cooled to room temperature in the citrate buffer for 1 h. Immunostaining was conducted with primary antibodies, followed by secondary antibodies. DAPI (D1306; Molecular Probes) was used for counterstaining. Images were acquired using a Leica THUNDER Imaging System with a multi-track configuration.
Lipid droplet staining
Astrocytes were synchronized with serum-free medium supplemented with palmitic acid (PA) conjugated BSA (50 µM PA and 142 µM BSA) for overnight before experiments. After respective treatments, cells were stained with 10 µM BODIPY493/503 (D3922; Invitrogen) for 30 min at room temperature. After washing with PBS, cells were fixed with 4% paraformaldehyde for 30 min. Fixed cells were washed with PBS three times before mounting with mounting medium containing DAPI. Images taken by confocal microscopy were processed by CellProfiler (Broad Institute) for analyses and by ImageJ for representative images.
NBD-C1P pulse-chase assay
Ttyh1 WT and Ttyh1 KO astrocytes were seeded on Geltrex-coated glass-bottom culture dish and cultured in astrocyte medium (1801; ScienCell) for 1 to 2 days. Cells were then incubated with 2 µM NBD-C1P (S-500N6, Echelon Biosciences) in 0.005% fatty acid-free bovine serum albumin (A7030; Sigma-Aldrich) supplemented artificial cerebrospinal fluid (ACSF; 126 mM NaCl, 24 mM NaHCO3, 0.375 mM KCl, 1.25 mM KH2PO4, 2 mM MgCl2, 1 mM CaCl2, 1 mM glucose; pH 7.4) for 15 min at 37 °C. After washing with ACSF twice to get rid of extracellular NBD-C1P, cells were either fixed with 4% paraformaldehyde (0-min chase) or chased in ACSF with 0.5 µM LysoTracker (L7528; Invitrogen) at 37 °C for 15, 30, or 60 min. Chase was terminated by fixation in 4% paraformaldehyde for 15 min. Fixed cells were immersed in DAPI Floromount-G mounting medium (Southern Biotech) before imaging. NBD-C1P fluorescence intensity was measured by imaging single cells using a Zeiss Axio Observer microscope equipped with a Colibri 5 LED light source and a 90 HE LED multi-bandpass filter with excitation wavelength/bandwidth of 469/38 nm. Ttyh1 WT and KO samples corresponding to the same chasing time-point were imaged successively within 1 h. Images were captured using the same exposure ang image export settings for each WT and KO pair. Some of the samples were imaged again using a Zeiss LSM 980 with Airyscan2 confocal microscope to generate high-resolution subcellular localization images.
BSA internalization and lysosomal degradation assay
Ttyh1 WT and Ttyh1 KO astrocytes were fed with 0.1% fatty acid-free BSA for 30 min at 37 °C, followed by chasing for 60 min in the presence of serum-free DMEM. After the chase, cell lysates were collected and analyzed for BSA levels. To inhibit lysosomal degradation, cells were treated with Bafilomycin A1 (100 nM) or chloroquine (50 µM) during the chase period.
Image analyses
Lipid droplets
A CellProfiler (Broad Institute) pipeline was built for lipid droplet analysis. Confocal images were loaded into the pipeline as merged channel images (405 nm laser channel and 488 nm laser channel). The 405 nm laser channel image was used to detect cell nuclei, while the 488 nm laser channel image was optimized to highlight primary objects (lipid droplets) after being enhanced. The nuclei signal was utilized to generate a mask on the 488 nm laser channel image to exclude lipid droplets within nuclei. The 488 nm laser channel image was also used to delineate the cell area by thresholding the signals. Cytoplasmic lipid droplet signals within the cell boundary were counted by the pipeline. Both lipid droplet and cell areas were computed for quantification.
GFP-LC3-RFP-LC3ΔG: ImageJ (National Institutes of Health) was employed to measure the fluorescence intensity of GFP-LC3-RFP-LC3ΔG. Images of GFP and RFP fluorescence emissions were captured by a Zeiss Axio Observer epifluorescence microscope with a 40x oil objective. A cell boundary mask was generated by thresholding the GFP- and RFP-channel images. The mask was used for cell area measurement. GFP and RFP emission intensities within the respective masks were then measured. Mean intensity was generated by standardizing total intensity within the mask to cell area. The ratio between the mean GFP intensity and the mean RFP intensity for each cell was then calculated.
NBD-C1P pulse-chase analysis
A CellProfiler (Broad Institute) pipeline was built for evaluating the NBD fluorescence intensity across different time points (0, 15, 30, 60 min). Images were captured by a Zeiss Axio Observer epifluorescence microscope with a 100x oil objective. A cell boundary mask was generated by thresholding. The mask was used for cell area measurement. NBD emission intensity within the mask was then measured. Mean intensity was generated by standardizing total intensity within the mask to cell area.
Fly strains and husbandry
Flies were grown with standard fly food (3 L of food contained 25.6 g agar, 80.6 g brewer’s yeast, 190.6 g of cornmeal, 40.3 g of sugar, 16.6 g of propionic acid, and 13.3 g of tegosept) in a 25 °C-incubator with 12-h light-dark cycle. The wild-type (WT; w1118) and tty−/− (ttyMI12269) strains were described previously34. The following fly strains were obtained from the Bloomington Drosophila Stock Center: repo-GAL4 7415, UAS-nucRFP 30556, nSyb-GS 80699 (neuron-switch), UAS-Cerk 38410, UAS-Cerk-RNAi 67256 (Cerk-IR; TRiP.HMC06359). In 1-week-old flies with inducible neuronal overexpression (nSyb-GS>Cerk) or knockdown of Cerk (nSyb-GS>Cerk-IR), neuron-switch diver (nSyb-GS) activity was induced by feeding flies with 500 µM RU486 (mifepristone; 459980010, Acros Organics) for 2 weeks. After feeding with either regular or RU486-containing fly food, fly heads of 3-week-old flies were dissected and collected for solvent extraction of brain lipids.
Primary culture and lipid droplet staining of fly brain cells
The procedures for primary fly brain cell culture were described previously34,67. Briefly, wandering third-instar larvae were washed with 70% ethanol and sterile water successively. Brains were dissected from pinned larvae submersed in Schneider’s medium (Sigma-Aldrich) supplemented with 10% heat-inactivated FBS and 1% antibiotic/antimycotic solution (Sigma-Aldrich). Dissected brains were transferred to filtered HL-3 solution (70 mM NaCl, 5 mM KCl, 1 mM CaCl2, 20 mM MgCl2, 10 mM NaHCO3, 115 mM sucrose, 5 mM trehalose, and 5 mM HEPES) containing 0.423 mM l-cysteine (Sigma-Aldrich) and 5 U/mL papain (Worthington) and incubated at ~29 for 20 min The brains were washed twice with Schneider’s medium before triturating the tissues through the pore of a 300 µL pipette tip in supplemented Schneider’s medium. Dissociated cells were seeded in 35 mm glass-bottom dishes (Cellvis) precoated with concanavalin A (Sigma-Aldrich) and cultured in supplemented Schneider’s medium at 25 °C for 4 days.
For lipid droplet fluorescence imaging, primary glia expressing nuclear RFP driven by repo-GAL4 were incubated with 10 µM BODIPY493/503 (D3922; Invitrogen) in either HL-3 or sugar-free HL-3 (sucrose and trehalose replaced by 120 mM mannitol) for ~15 min before imaging using a Zeiss Axio Observer microscope equipped with a Colibri 5 LED light source and a 90 HE LED multi-bandpass filter with excitation wavelength/bandwidth of 469/38 nm and 555/30 nm. The recorded signals of sequential fluorescence emission from Em469ex and Em555ex were analyzed by CellProfiler (Broad Institute), where Em555ex signals were selected to identify locations of glia and Em469ex signals were selected to identify punctate structures representing lipid droplets. The total area of lipid droplets per cell was then measured by a CellProfiler pipeline. For each genotype and treatment group, the average lipid droplet area of a total of 30 individual cells randomly selected were compared, using the average value of wild-type glia in HL-3 as the baseline.
Lipid extraction
Lipids were extracted from cells, fly heads and conditioned medium using organic methyl tert-butyl ether (MTBE) solvent phase separation. Cells or fly heads were homogenized in standard diluent assay buffer (700732; Cayman) at 100 µL per 1 million cells or 2 µL per fly head, respectively. MTBE (34875; Sigma-Aldrich) was added to the lysate/conditioned medium at 2:1 volume ratio. The mixture was vortexed for 30 s. Upon 10-min incubation at room temperature, the mixture was centrifuged at 1000 × g for 10 min. The upper (organic) phase was collected, which contains the extracted lipids. The separated organic phase was dried in a vacuum centrifuge. Dry lipid extracts were stored at −80 °C.
Lipidomics
Total lipids were extracted from adult Drosophila heads using the MTBE solvent extraction method. For each biological replicate, a total of 40 fly heads collected from snap freezing was homogenized in MTBE at 4 °C. After the addition of methanol spiked with SPLASH LIPIDOMIX (330707; Avanti) as the internal standard, the mixture was mixed briefly and incubated for 5 min. At the endpoint, the upper organic phase was transferred into a collection tube and dried. Seven biological replicates for each of the two genotypes (WT and tty−/−) were prepared for LC-MS. Two independent LC-MS analyses were performed on the samples: the first analysis included four biological replicates for each of the two genotypes; the second analysis included the remaining three biological replicates for each of the two genotypes. LC-MS lipid profiling was performed by the Rutgers University Metabolomics Shared Resource core. Reverse-phase separation of the lipid extract was conducted on a Vanquish Horizon UHPLC system (Thermo Fisher Scientific) using a Poroshell 120 EC-C18 column (2.7-μm particle size, Agilent InfinityLab). The gradient elution employed solvent A (90% H2O:10% MeOH with 34.2 mM acetic acid, 1 mM ammonium acetate, pH 9.4) and solvent B (75% IPA:25% methanol with 34.2 mM acetic acid, 1 mM ammonium acetate, pH 9.4). The gradient program was set as follows: 0 min, 25% B; 2 min, 25% B; 5.5 min, 65% B; 12.5 min, 100% B; 19.5 min, 100% B; 20.0 min, 25% B; 30 min, 25% B. The flow rate was set to 200 μL/min, with an injection volume of 5 μL, and the column temperature was maintained at 55 °C. The autosampler was kept at 4 °C. Full-scan mass spectrometry analysis was performed using a Thermo Q Exactive PLUS equipped with a HESI source. The spray voltage was −2.7 kV in negative mode and 3.5 kV in positive mode. Sheath gas, auxiliary gas, and sweep gas flow rates were set at 40, 10, and 2 (arbitrary units), respectively. The capillary temperature was 300 °C, and the auxiliary gas heater was set to 360 °C. The S-lens RF level was 45, with an m/z scan range of 100 to 1200 m/z for both positive and negative ionization modes. Metabolite data were processed using the MAVEN software package68 with a mass accuracy window of 5 ppm. For the lipidomics data analyses, the normalized mean values of diacylglycerides (DAG) and triglycerides (TAG) for fly heads from wildtype and tty−/− mutants from seven biological replicates each were used. Normalization was achieved by comparing the response signal of target lipids and their respective internal standards, i.e., 5:0-18:1(d7) DAG for diacylglycerides and 5:0-18:1(d7)-15:0 TAG for triglycerides. Individual species from the lipid family were reported only when they were detected above the limit of quantitation (signal/noise = 10:1).
C1P quantitation by enzyme-linked immunosorbent assay (ELISA)
C1P was measured using a colorimetric C1P ELISA kit (AMS.E4332hu; Amsbio), which consists of an anti-C1P antibody precoated well plate, biotinylated anti-C1P antibody, and streptavidin-HRP. Lipids were solvent-extracted from the cell lysates and vacuum-dried. The dried lipids were dissolved in the standard diluent provided by the ELISA kit. After completing the colorimetric reaction, the optical density (OD; 450 nm) of each sample was measured using a microplate reader. Individual OD values were normalized to the mean values of respective control samples. To determine C1P concentration in conditioned medium, OD values were calibrated using a standard curve derived from C1P(d18:1/16:0) at defined concentrations. In experiments shown in Fig. 6a, neurons were treated with IL-1β (10 ng/mL) and ceramide kinase (CERK) inhibitor NVP-231 (12 nM) for 2 h. Vehicle control was 0.1% DMSO without IL-1β. Intracellular and extracellular C1P levels were then measured by ELISA. Intracellular levels were normalized to naïve vehicle control. Extracellular C1P concentrations were calculated using C1P (d18:1/16:0) as standards.
IL-1β quantitation by ELISA
IL-1β in the transwell coculture medium was measured using the Human IL-1 beta Sandwich ELISA kit (KE00021; Proteintech).
Triglyceride quantitation assay
Triglyceride (TAG) contents were measured using an enzyme-coupled colorimetric kit (10010303; Cayman). For astrocytes, cells were seeded in the six-well plate at around 1 million cells/well or 24-well plate at around 25k cells/well. After respective treatments, cells in each well were washed with PBS before being scraped into 100 µL (six-well) or 50 µL (24-well) of standard diluent assay buffer provided by the kit. For fly head lipid extracts, each extract (from 30 fly heads) was reconstituted in 60 µL of standard diluent assay buffer. After completing the colorimetric reaction, the optical density (OD; 540 nm) of each sample was measured using a microplate reader. Individual OD values were normalized to the mean values of respective control samples.
Quantitation of protein and TAG contents in mouse cortical tissue
Two-month-old Aldh1l1-Cre/ERT2; Ttyh1fl/fl mice received intraperitoneal injections of either corn oil (vehicle) or tamoxifen-containing (75 mg/kg-mouse) corn oil everyday for 5 consecutive days. Three days after the last injection, the brain cortices were dissected out from euthanized mice. Each cortical tissue sample was homogenized in 1 mL of standard diluent assay buffer (700732; Cayman) using a Dounce homogenizer. Protein concentration in the tissue lysate was measured using Bradford reagent (5000205; Bio-Rad). Lipids in the tissue lysate were extracted using MTBE solvent phase separation. Extracted lipids were vacuum-dried and reconstituted in standard diluent assay buffer (700732; Cayman). TAG contents were measured by the triglyceride quantitation assay.
Protein-lipid in silico docking
The protein structure of human TTYH1 (accession number 7P5J) was obtained from the Protein Data Bank (PDB). The lipid structures were collected from the LIPID MAPS Structure Database39. The structure of CerP(d18:0/22:0) was manually constructed in ChemDraw software. The ligands were prepared using the LigPrep tool by protonating molecules at a pH of 7 ± 1 using the Epik tool, minimizing the energy with the OPLS3e force field, and generating the tautomeric conformers. CB-Dock2 was used to identify the coordinates of the TTYH1 surface cavity and the constituent amino acid residues by matching to the hydrophobic cavity reported in recent cryo-EM studies26,27. Isomeric chain A (7P5J-A) was prepared using the ProPrep tool of Schrödinger, where high-confidence protein structures were generated through adding missed hydrogen atoms, filling the missed side-chain atoms, generating the het states at pH: 7 ± 2, deleting the het & water molecules (within 5.0 Å), assigning Hydrogen bonds using PROPKA pH:7 and converging the heavy atoms to root mean square deviation (RMSD) at greater than 0.30 Å. The receptor-bound inhibitor molecule was chosen to generate a GRID for the ligand binding using the GRID-GLIDE module. The constituent residues of the cavity, including Phe 33, Pro 35, Tyr 40, Leu 44, Thr 115, Val 119, Leu 122, Leu 126, Val 202, Asn 205, Val 206, Val 209, Glu 210, Arg 213, Tyr 217, Ser 260, Leu 263, Glu 264, Ala 265, Ala 266, Thr 267, Gly 270, leu 271, Phe 274, Tyr 281, Val 282, Leu 285, Thr 286, Tyr 377, Ala 380, Leu 381, Leu 384, Ala 388, and Leu 392 were picked to define the grid centroid to the active site. To minimize the penalties, the van der Waals radii were scaled at 0.80, and the default partial cut-off charge was set at 0.15. Using the GLIDE module, each ligand was docked with its corresponding protein binding site. The extra precision (XP) module and OPLS3e force field were used for GLIDE-docking. The Epik state penalties were added to the XP GLIDE score function. The Glide score was used to align the compounds based on their specific interactions with the proteins, e.g., hydrogen bonding. The top-scored docking poses were visualized with the PyMol graphics system v3.0, Schrödinger. The lipid ligands were represented in a stick model. The constituent residues of the cavity were displayed in Fig. 2 with surface representations: hydrophobic interacting residues were shown in green; residues E210, R213, and Y217 were shown in orange; and the rest were shown in gray.
Data presentation and statistical analyses
Statistical comparisons were performed using two-tailed Student’s t-test, Mann–Whitney test, ANOVA, and Bonferroni’s post hoc test. Prism 8 (GraphPad) were used for statistical analyses. t values, F values, and the degree of freedom (df) of relevant tests are shown in the figure legends. Statistical significance was defined as P < 0.05. P values are shown on the graphs or in the figure legends. Graphs were generated in Prism 8 (GraphPad). Box plots show median, interquartile range (box), and data range (whiskers). Violin plots show median (solid line), interquartile range (dotted line), and probability density of data (smoothed shape). Bar charts show mean average (bar) and standard error mean (whiskers). Data points on bar charts represent values from individual cells, individual Western blots, individual lipid extracts, or biological replicates. Distinct biological samples were used for Western blot and lipid quantitation assays. The sample sizes for the data shown in the figures are indicated in brackets on each graph.
Statistics and reproducibility
No statistical methods were used to pre-determine the sample size. The experiments were not randomized. Investigators were not blinded to allocation during experiments and outcome assessment. In lipid droplet staining experiments, outliers were excluded from statistical analyses after the BODIPY area per cell values were determined due to the concern of staining artifact. Outlier test was performed using the ROUT method with coefficient Q set to 1. No data were excluded from the analyses in all other experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data reported as graphs or figures are available in the Source Data file provided with this paper and are also available in the public repository Figshare via the https://doi.org/10.6084/m9.figshare.28914281. Raw lipidomics data are available in the Figshare repository via the DOI 10.6084/m9.figshare.28914281. Previously published RNA-seq datasets used in this study were retrieved from the Gene Expression Omnibus database under the accession codes GSE52564 and GSE73721. Additional raw data will be shared by the corresponding author upon request. Source data are provided with this paper.
References
Bonvento, G. & Bolanos, J. P. Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33, 1546–1564 (2021).
Quick, J. D. et al. Lysosomal acidification dysfunction in microglia: an emerging pathogenic mechanism of neuroinflammation and neurodegeneration. J. Neuroinflammation 20, 185 (2023).
Mishra, S. K. & Tiwari, S. P. Bioenergetics of axon integrity and its regulation by oligodendrocytes and schwann cells. Mol. Neurobiol. 61, 5928–5934 (2024).
Mahmoud, A. I. Metabolic switches during development and regeneration. Development 150, dev202008 (2023).
Pellerin, L. et al. Activity-dependent regulation of energy metabolism by astrocytes: an update. Glia 55, 1251–1262 (2007).
Wong, C. O. Endosomal-lysosomal processing of neurodegeneration-associated proteins in astrocytes. Int. J. Mol. Sci. 21, 5149 (2020).
Lee, J. A., Hall, B., Allsop, J., Alqarni, R. & Allen, S. P. Lipid metabolism in astrocytic structure and function. Semin. Cell Dev. Biol. 112, 123–136 (2021).
Guttenplan, K. A. et al. Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature 599, 102–107 (2021).
Wang, L. et al. Neuronal activity induces glucosylceramide that is secreted via exosomes for lysosomal degradation in glia. Sci. Adv. 8, eabn3326 (2022).
Liu, L., MacKenzie, K. R., Putluri, N., Maletic-Savatic, M. & Bellen, H. J. The glia-neuron lactate shuttle and elevated ROS promote lipid synthesis in neurons and lipid droplet accumulation in glia via APOE/D. Cell Metab. 26, 719–737.e716 (2017).
Ioannou, M. S. et al. Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell 177, 1522–1535 e1514 (2019).
Qi, G. et al. ApoE4 impairs neuron-astrocyte coupling of fatty acid metabolism. Cell Rep. 34, 108572 (2021).
Moulton, M. J. et al. Neuronal ROS-induced glial lipid droplet formation is altered by loss of Alzheimer’s disease-associated genes. Proc. Natl Acad. Sci. USA 118, e2112095118 (2021).
Savini, M., Zhao, Q. & Wang, M. C. Lysosomes: signaling hubs for metabolic sensing and longevity. Trends Cell Biol. 29, 876–887 (2019).
Afghah, Z., Chen, X. & Geiger, J. D. Role of endolysosomes and inter-organellar signaling in brain disease. Neurobiol. Dis. 134, 104670 (2020).
Li, P., Gu, M. & Xu, H. Lysosomal ion channels as decoders of cellular signals. Trends Biochem. Sci. 44, 110–124 (2019).
Nalamalapu, R. R., Yue, M., Stone, A. R., Murphy, S. & Saha, M. S. The tweety gene family: from embryo to disease. Front. Mol. Neurosci. 14, 672511 (2021).
Matthews, C. A. et al. Expression and evolution of the mammalian brain gene Ttyh1. J. Neurochem. 100, 693–707 (2007).
Sharma, K. R., Colvis, C. M., Rodgers, G. P. & Sheeley, D. M. Illuminating the druggable genome: Pathways to progress. Drug Discov. Today 29, 103805 (2024).
Jung, E. et al. Tweety-homolog 1 drives brain colonization of gliomas. J. Neurosci. 37, 6837–6850 (2017).
Kim, J. et al. Ttyh1 regulates embryonic neural stem cell properties by enhancing the Notch signaling pathway. EMBO Rep. 19, e45472 (2018).
Wu, H. N. et al. Deficiency of Ttyh1 downstream to Notch signaling results in precocious differentiation of neural stem cells. Biochem. Biophys. Res. Commun. 514, 842–847 (2019).
Xu, P. T. et al. Differences in apolipoprotein E3/3 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Neurobiol. Dis. 21, 256–275 (2006).
van Dijk, K. D. et al. The proteome of the locus ceruleus in Parkinson’s disease: relevance to pathogenesis. Brain Pathol. 22, 485–498 (2012).
Uhlen, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Sukalskaia, A., Straub, M. S., Deneka, D., Sawicka, M. & Dutzler, R. Cryo-EM structures of the TTYH family reveal a novel architecture for lipid interactions. Nat. Commun. 12, 4893 (2021).
Li, B., Hoel, C. M. & Brohawn, S. G. Structures of tweety homolog proteins TTYH2 and TTYH3 reveal a Ca(2+)-dependent switch from intra- to intermembrane dimerization. Nat. Commun. 12, 6913 (2021).
Lovatt, D. et al. The transcriptome and metabolic gene signature of protoplasmic astrocytes in the adult murine cortex. J. Neurosci. 27, 12255–12266 (2007).
Cahoy, J. D. et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 28, 264–278 (2008).
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Zhang, Y. et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014).
Zhang, Y. et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron 89, 37–53 (2016).
Wiernasz, E. et al. Ttyh1 protein is expressed in glia in vitro and shows elevated expression in activated astrocytes following status epilepticus. Neurochem. Res. 39, 2516–2526 (2014).
Leung, H. H. et al. Drosophila tweety facilitates autophagy to regulate mitochondrial homeostasis and bioenergetics in Glia. Glia 72, 433–451 (2024).
Goldsmith, J., Ordureau, A., Harper, J. W. & Holzbaur, E. L. F. Brain-derived autophagosome profiling reveals the engulfment of nucleoid-enriched mitochondrial fragments by basal autophagy in neurons. Neuron 110, 967–976 e968 (2022).
Kaizuka, T. et al. An autophagic flux probe that releases an internal control. Mol. Cell 64, 835–849 (2016).
Han, Y. E. et al. Tweety-homolog (Ttyh) family encodes the pore-forming subunits of the swelling-dependent volume-regulated anion channel (VRAC(swell)) in the brain. Exp. Neurobiol. 28, 183–215 (2019).
Voeltz, G. K., Sawyer, E. M., Hajnoczky, G. & Prinz, W. A. Making the connection: how membrane contact sites have changed our view of organelle biology. Cell 187, 257–270 (2024).
Conroy, M. J. et al. LIPID MAPS: update to databases and tools for the lipidomics community. Nucleic Acids Res. 52, D1677–D1682 (2024).
Sarkar, C. et al. PLA2G4A/cPLA2-mediated lysosomal membrane damage leads to inhibition of autophagy and neurodegeneration after brain trauma. Autophagy 16, 466–485 (2020).
Lan, Z. Q., Ge, Z. Y., Lv, S. K., Zhao, B. & Li, C. X. The regulatory role of lipophagy in central nervous system diseases. Cell Death Discov. 9, 229 (2023).
Ralhan, I., Chang, C. L., Lippincott-Schwartz, J. & Ioannou, M. S. Lipid droplets in the nervous system. J. Cell Biol. 220, e202102136 (2021).
Bajjalieh, S. M., Martin, T. F. & Floor, E. Synaptic vesicle ceramide kinase. A calcium-stimulated lipid kinase that co-purifies with brain synaptic vesicles. J. Biol. Chem. 264, 14354–14360 (1989).
Boath, A. et al. Regulation and traffic of ceramide 1-phosphate produced by ceramide kinase: comparative analysis to glucosylceramide and sphingomyelin. J. Biol. Chem. 283, 8517–8526 (2008).
Kuc, N. et al. Pancreatic ductal adenocarcinoma cell secreted extracellular vesicles containing ceramide-1-phosphate promote pancreatic cancer stem cell motility. Biochem. Pharm. 156, 458–466 (2018).
Graf, C., Niwa, S., Muller, M., Kinzel, B. & Bornancin, F. Wild-type levels of ceramide and ceramide-1-phosphate in the retina of ceramide kinase-like-deficient mice. Biochem. Biophys. Res. Commun. 373, 159–163 (2008).
Pettus, B. J. et al. Ceramide kinase mediates cytokine- and calcium ionophore-induced arachidonic acid release. J. Biol. Chem. 278, 38206–38213 (2003).
Clarkson, B. D. S., Kahoud, R. J., McCarthy, C. B. & Howe, C. L. Inflammatory cytokine-induced changes in neural network activity measured by waveform analysis of high-content calcium imaging in murine cortical neurons. Sci. Rep. 7, 9037 (2017).
Dasgupta, U. et al. Ceramide kinase regulates phospholipase C and phosphatidylinositol 4, 5, bisphosphate in phototransduction. Proc. Natl Acad. Sci. USA 106, 20063–20068 (2009).
Kremer, M. C., Jung, C., Batelli, S., Rubin, G. M. & Gaul, U. The glia of the adult Drosophila nervous system. Glia 65, 606–638 (2017).
Cabirol-Pol, M. J., Khalil, B., Rival, T., Faivre-Sarrailh, C. & Besson, M. T. Glial lipid droplets and neurodegeneration in a Drosophila model of complex I deficiency. Glia 66, 874–888 (2018).
Marschallinger, J. et al. Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208 (2020).
Lee, J. et al. Microglial REV-ERBalpha regulates inflammation and lipid droplet formation to drive tauopathy in male mice. Nat. Commun. 14, 5197 (2023).
Breiden, B. & Sandhoff, K. Lysosomal glycosphingolipid storage diseases. Annu. Rev. Biochem. 88, 461–485 (2019).
Hinkovska-Galcheva, V. T. et al. The formation of ceramide-1-phosphate during neutrophil phagocytosis and its role in liposome fusion. J. Biol. Chem. 273, 33203–33209 (1998).
Hinkovska-Galcheva, V. et al. Ceramide 1-phosphate, a mediator of phagocytosis. J. Biol. Chem. 280, 26612–26621 (2005).
Mishra, S. K. et al. CPTP: A sphingolipid transfer protein that regulates autophagy and inflammasome activation. Autophagy 14, 862–879 (2018).
Lamour, N. F. et al. Ceramide kinase regulates the production of tumor necrosis factor alpha (TNFalpha) via inhibition of TNFalpha-converting enzyme. J. Biol. Chem. 286, 42808–42817 (2011).
Sokoya, T. et al. Pathogenic variants of sphingomyelin synthase SMS2 disrupt lipid landscapes in the secretory pathway. Elife 11, e79278 (2022).
Chen, Z. P. et al. Lipid-accumulated reactive astrocytes promote disease progression in epilepsy. Nat. Neurosci. 26, 542–554 (2023).
Targa Dias Anastacio, H., Matosin, N. & Ooi, L. Neuronal hyperexcitability in Alzheimer’s disease: what are the drivers behind this aberrant phenotype?. Transl. Psychiatry 12, 257 (2022).
Kurki, S. N. et al. Acute neuroinflammation leads to disruption of neuronal chloride regulation and consequent hyperexcitability in the dentate gyrus. Cell Rep. 42, 113379 (2023).
Van Den Brink, D. M. et al. Physiological and pathological roles of FATP-mediated lipid droplets in Drosophila and mice retina. PLoS Genet. 14, e1007627 (2018).
Smolic, T. et al. Astrocytes in stress accumulate lipid droplets. Glia 69, 1540–1562 (2021).
Tcw, J. et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 9, 600–614 (2017).
Jang, M. H. et al. Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell 12, 215–223 (2013).
Karagas, N. E. et al. Loss of activity-induced mitochondrial ATP production underlies the synaptic defects in a Drosophila model of ALS. J. Neurosci. 42, 8019–8037 (2022).
Melamud, E., Vastag, L. & Rabinowitz, J. D. Metabolomic analysis and visualization engine for LC-MS data. Anal. Chem. 82, 9818–9826 (2010).
Acknowledgements
We thank Drs. Peter Romanienko and Ghassan Yehia of the Rutgers University Genome Editing Shared Resource Core for their expertise in generating the conditional Ttyh1 knockout mouse strain. We also thank Dr. Xiaoyang Su of the Rutgers University Metabolomic Shared Resource for his expertise in lipidomics analyses. We thank Dr. Nan Gao of the New Jersey Medical School for the Rab11 antibody. Fly strains were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). Confocal microscopy was performed with the equipment partly supported by the National Science Foundation MRI grant 2117484. This work was supported by the NIH grants R03AG063251 (C.-O.W.), R03TR004191 (C.-O.W.) and R01AG081379 (C.-O.W.).
Author information
Authors and Affiliations
Contributions
C.-O.W. conceived and supervised the project; J.N.B., A.P., H.H.L., M.A.R., and C.-O.W. designed the experiments; J.N.B. and A.P. performed most of the experiments; H.H.L. performed fly-related experiments, lipid extractions and quantifications, and lipidomics analyses; M.A.R. performed in silico docking experiments; Y.T. performed iPSC differentiation; S.H.K. and M.-H.J. performed mouse brain immunofluorescence analyses; J.N.B., A.P., H.H.L., and C.-O.W. analyzed experiments and prepared images and graphs. J.N.B. and C.-O.W. wrote the manuscript with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bhupana, J.N., Pabon, A., Leung, H.H. et al. Endolysosomal processing of neuron-derived signaling lipids regulates autophagy and lipid droplet degradation in astrocytes. Nat Commun 16, 5073 (2025). https://doi.org/10.1038/s41467-025-60402-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-025-60402-3