Abstract
The tetraploid genome and clonal propagation of the cultivated potato (Solanum tuberosum L.)1,2 dictate a slow, non-accumulative breeding mode of the most important tuber crop. Transitioning potato breeding to a seed-propagated hybrid system based on diploid inbred lines has the potential to greatly accelerate its improvement3. Crucially, the development of inbred lines is impeded by manifold deleterious variants; explaining their nature and finding ways to eliminate them is the current focus of hybrid potato research4,5,6,7,8,9,10. However, most published diploid potato genomes are unphased, concealing crucial information on haplotype diversity and heterozygosity11,12,13. Here we develop a phased potato pangenome graph of 60 haplotypes from cultivated diploids and the ancestral wild species, and find evidence for the prevalence of transposable elements in generating structural variants. Compared with the linear reference, the graph pangenome represents a broader diversity (3,076 Mb versus 742 Mb). Notably, we observe enhanced heterozygosity in cultivated diploids compared with wild ones (14.0% versus 9.5%), indicating extensive hybridization during potato domestication. Using conservative criteria, we identify 19,625 putatively deleterious structural variants (dSVs) and reveal a biased accumulation of deleterious single nucleotide polymorphisms (dSNPs) around dSVs in coupling phase. Based on the graph pangenome, we computationally design ideal potato haplotypes with minimal dSNPs and dSVs. These advances provide critical insights into the genomic basis of clonal propagation and will guide breeders to develop a suite of promising inbred lines.
Similar content being viewed by others
Main
Potato (S. tuberosum L.) is the most important tuber crop, feeding around 1.3 billion people annually in more than 120 countries1. Recent studies have decoded the genomes of monoploid, diploid and tetraploid potatoes, resolved self-incompatibility and dissected the genetic basis of inbreeding depression4,5,6,7,8,9,10,11,12,13,14,15. These efforts have led to the development of a first generation of highly homozygous inbred lines and subsequently, the first uniform hybrid as a proof-of-concept3, demonstrating the potential of genome design for revolutionizing potato breeding towards a fast, iterative mode.
A central focus of hybrid potato breeding programmes is development of a better understanding of deleterious variants4,5,6,7,8,9,10, which compromise growth and overall fitness9. dSNPs have been widely studied in many species such as humans, dogs and rice16,17,18. Structural variants (SVs), however, are expected to affect more genomic regions and are more likely to have stronger effects on fitness than SNPs; nonetheless, dSVs have rarely been studied. Phenotype-based selection is neither efficient nor accurate in purging deleterious variants closely linked in repulsion phase in diploid potatoes5. Haplotype-based selection based on in-depth understanding of deleterious variants and knowledge-guided recombination strategies are essential for integrating superior genomic fragments from different donors, paving the way for the eventual development of ideal potato haplotypes (IPHs).
Thus far, our knowledge of haplotype diversity in potatoes is limited. Only a few genomes have been published and most diploid potatoes are assembled into a single chimeric haploid genome, with only a few examples of haplotype-resolved assemblies11,12,13. Collapsed assemblies of two haplotypes from diploid genomes lose valuable information on haplotype diversity and the phase state, introducing multiple types of errors, including switch errors, false duplications and nucleotide consensus errors19,20. Haplotype-resolved assemblies provide a more accurate representation of genetic diversity and promise to become essential for future potato breeding5,7,14. Concurrently, there is a need to shift from single reference genomes to phased pangenome references that better capture diversity across multiple populations21.
In this study, we carried out de novo assembly for 60 haplotypes from 31 diploid potatoes, including 20 haplotypes from 10 wild accessions, 38 haplotypes from 19 domesticated diploid accessions and two haplotypes from two inbred lines. From these 60 potato haplotypes, we constructed a potato pangenome graph, which enabled us to uncover extensive non-reference sequences and document multiallelic SVs. We further reveal the origin and fate of SVs in evolution and domestication and design IPHs for future breeding. Our study provides in-depth understanding of the haplotype diversity of a clonal crop, establishing the theoretical foundation for the reinvention of potato.
The potato pangenome graph
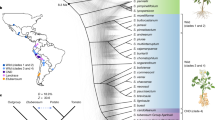
To capture and characterize haplotype diversity in potato populations, we selected 31 accessions based on previous studies of genetic diversity and principal component analysis (PCA) of 193 accessions3,5,9,13 (Fig. 1a and Supplementary Figs. 1 and 2). The selected accessions comprise 19 landraces covering three indigenous diploid cultivated groups (S. tuberosum Group Phureja (PHU), S. tuberosum Group Stenotomum (STN) and S. tuberosum Group Goniocalyx (GON)), two inbred lines (A6-26 and E4-63, the 5th-generation selfed offspring of diploid PG6359 and E86-69, respectively)3 and 10 wild accessions from Solanum candolleanum (CND), which is believed to be the progenitor of domesticated potatoes22 (Supplementary Table 1). We generated an average of 28.15 Gb (approximately 38-fold) PacBio high-fidelity (HiFi) reads and 110.43 Gb (approximately 149-fold) high‐throughput chromosome conformation capture technique (Hi-C) reads (Supplementary Tables 2 and 3). Genome surveys using the HiFi reads revealed an average of around 1.55% genomic heterozygosity across cultivated diploid potato genomes (excluding inbred lines; Supplementary Table 4). Our initial de novo assembled haplotypes have an average genome size of 811 Mb, with contig N50 of 12.25 Mb (Fig. 1b and Supplementary Table 4). Paired haplotypes of each heterozygous diploid were further scaffolded on the basis of Hi-C contact maps (Extended Data Fig. 1a and Supplementary Fig. 3), resulting in pseudo-chromosome assemblies with an average anchor rate of 95.17% (Supplementary Table 5). Assessment using BUSCO23 demonstrated a high level of completeness with an average of 99.2% (Supplementary Table 5).
a, PCA from 193 potato accessions; the 31 selected accessions are highlighted with solid dots. b, Assessment of genome contiguity by contig N50 for haplotype 1 (solid lines) and haplotype 2 (dashed lines). The Nx value represents x% of the total contigs length that is covered by the shortest contig length. c, Assessment of correctness by switch errors and Hamming errors for each diploid assembly. d, Reliability of the 29 diploid assemblies (excluding 2 inbred lines). For each haplotype-resolved assembly, the left bar represents haplotype H1 and the right bar represents H2. Regions identified as haploid are considered trustworthy (green). The y axis is broken to illustrate both the prominence of the reliable haploid component and the stratification of the unreliable segments. e, Pangenome growth curves for wild, cultivated, and wild plus cultivated groups. Pie charts represent conserved sequence (present in at least 90% of the haplotypes) and variable sequence in the PGGB pangenome graph. Numbers in the pie charts represent sequence length (Mb). f, Non-reference nodes (>50 bp) in the potato PGGB pangenome were annotated as four primary types, including TEs, satellite repeats, low-complexity (simple repeats) and non-repetitive sequences. Six different TE families were identified.
Three additional independent analyses confirm the quality of our assemblies: (1) the low switch error rates (0.21% on average) and Hamming errors (1.05% on average) indicate the high correctness of our assemblies (Fig. 1c); (2) the high read mapping rate (mean 99.99%) and consistent k-mer spectrum (Extended Data Fig. 1b and Supplementary Fig. 4) confirm the completeness of the assemblies; and (3) to further estimate the reliability of our assemblies, we also used Flagger24 to estimate the potentially erroneous regions. This confirmed that 98.9% of our assemblies are not collapsed or duplicated (Fig. 1d and Supplementary Table 6). We annotated an average of 449.0 Mb (59.69%) as being composed of transposable elements (TEs) (Supplementary Table 7) and predict 36,421–40,781 gene structure models for each haplotype (Supplementary Table 8).
To better represent non-reference sequences in potato genomes, we built two pangenome graphs for 60 haplotype assemblies plus the reference DMv6.125 using the PanGenome Graph Builder (PGGB)26 and Minigraph-Cactus27, respectively. We found that the Minigraph-Cactus pipeline removed highly diverged and large inverted regions, resulting in discarding of an average of 14.5% of sequence per chromosome (Supplementary Fig. 5). Therefore, we utilized PGGB to build a potato pangenome graph, which reduces reference bias28 and is more sensitive for variation calling than methods based on mapping to a linear reference genome (Supplementary Fig. 6 and Supplementary Tables 9 and 10). This unbiased potato pangenome graph (PPG-v.1.0) comprises 248.64 million nodes and 345.61 million edges with total sequence length of 3,076 Mb. Wild potato accessions contribute a larger portion to the pangenome graph (2,286 Mb) than cultivated potato assemblies (1,807 Mb), indicating a higher pangenome graph growth rate in wild potatoes (Supplementary Table 11). We determined that PPG-v.1.0 consists of 365 Mb conserved sequences (present in at least 90% of the haplotypes) and 2,711 Mb variable sequences (Fig. 1e). Using the DMv6.1 reference as a baseline, PPG-v.1.0 also reveals prevalent rearrangements (Extended Data Fig. 2). Furthermore, we annotated non-reference nodes in each haplotype path: 52.6% of them overlap with TEs, 22.7% overlap with satellite repeats, and 23.8% are classified as non-repetitive sequences (Fig. 1f). Notably, 13.3% of repetitive sequences exhibit characteristics of both transposons and satellites (Supplementary Table 12) potentially originating from atypical centromere or subtelomere repeats29 in the potato genome.
Finally, we deconstructed genome variation from the graph structure for small variants (SNPs and insertion–deletion mutations (indels) smaller than 50 bp in size) and structural variants (SVs ≥ 50 bp) based on DMv6.1 coordinates. In total, the constructed potato haplotype variation map includes 46,012,502 SNPs, 3,587,850 indels and 133,264 SVs. Notably, 16.9% of indels and 87.0% of SVs are multiallelic, with three or more distinct alleles. The graph generated by the Minigraph-Cactus pipeline shows a similar trend (Extended Data Fig. 1c,d and Supplementary Table 13). Collectively, the PPG-v.1.0 generated from phased diploid assemblies provides a fundamental resource for in-depth exploration of haplotype diversity and its consequences for potato breeding.
TEs drive SV formation
To reveal the mechanistic origins and dynamics of SVs in potato genomes, we analysed the pangenome SV sequences and their residual flanking sequences (±100 bp), which we annotated as one of three types: TEs, tandem repeats (TRs) and segmental duplications. We found that TEs account for 90.6% of repetitive elements associated with SVs (Fig. 2a and Supplementary Table 14). Additionally, 28.2% of SVs are covered by a single TE sequence (Supplementary Fig. 7), suggesting that these features are likely to reflect TE insertion events30,31. Recent reports suggest that TEs could serve as substrates for ectopic DNA repair leading to SV formation32,33. Therefore, we examined SV breakpoints and found that 33.8% could be classified as being compatible with ectopic recombination events from TE-mediated rearrangement (TEMR), characterized by both breakpoints (100 bp) having the same TE class (Supplementary Table 15, Supplementary Fig. 8 and Methods). This observed TEMR rate is significantly higher than the expected rate of 7.7% (P < 2.2 × 10–16, two-tailed Student’s t-test), supporting a significant contribution of homologous TEs in mediating SV formation. TEMR events were further classified on the basis of TE families, revealing that Gypsy long terminal repeat retrotransposon (LTR/Gypsy)-mediated TEMR (42.4%) is seven times more prevalent than that involving Copia long terminal repeat retrotransposons (LTR/Copia (7.0%)) (Fig. 2b). Moreover, SVs mediated by LTR/Gypsy TEMR are significantly longer than those mediated by LTR/Copia TEMR (average of 7,005 bp versus 1,899 bp; P < 2.2 × 10–16, two-sided Student’s t-test), and exhibit more recent and frequent activity (Fig. 2c and Extended Data Fig. 3).
a, Proportion diagram of structural variation formation driven by different repetitive sequences, including TEs, TRs and segmental duplications (SDs). b, Left, percentage of TEMR through ectopic recombination by different families. Right, percentage of different TE families in TEMR among the three SV types. Del, deletion; Inv, inversion; Dup, duplication. Inserted sequences can be formed by TE movement and do not fall under our definition of TEMR. c, Comparison of SV length of TEMR for different TE families. A two-sided Student’s t-test was used to calculate the P values. NS, not significant; ***P < 2.2 × 10−16, n = 2,768 (LTR/Gypsy), 361 (LTR/Copia), 596 (DNA/Helitron) and 1,044 (DNA/TIR). d, Validation for the 4.0-Mb de novo inversion using Hi-C data from potato E86-69; the haplotype of inbred line E4-63 was used as the reference genome. e, Sequence analysis in the de novo inversion. Top, synteny plot of the de novo inversion. Middle, distribution of TEs in the de novo inversion. Bottom, the 466-bp homologous sequence stretch of the LTR mediating the de novo inversion.
To investigate the potential recent TE activity in potato genomes, we also compared two homozygous inbred lines with their heterozygous founders. Notably, we identified an approximately 4.0 Mb (chromosome (chr.) 6: 41.7–45.7 Mb) long terminal repeat retrotransposon (LTR)-mediated de novo paracentric inversion (Fig. 2d,e and Supplementary Fig. 9) in the fifth-generation inbred line E4-63, but not in the two founder haplotypes of the parental E86-69. Therefore, it is advisable to approach this de novo inversion with caution when implementing recurrent selection in hybrid potato breeding34,35.
Domestication boosts heterozygosity
Previous studies suggested that there was increased heterozygosity in cultivated potatoes compared to wild ones22, consistent with heterozygosity evaluated in our study on the basis of k-mer statistics using HiFi reads. To better describe heterozygosity and the loss of collinearity between the two haplotypes within the same diploid individual, we introduce the concept of genome heterozygosity by sequence length (GHSL). This approach complements existing k-mer-based methods by quantifying the total sequence length of non-redundant variants (SNPs, indels and SVs) between two haplotypes in phased diploid assemblies (Supplementary Fig. 10). We found the GHSL to be approximately 93.8 Mb in our phased assemblies, constituting 12.5% of the average haplotype length (Fig. 3a and Supplementary Table 16). This estimate is similar to estimates by read mapping in clonally propagated grapevine36. Moreover, our analysis shows that inversions account for a large proportion of haplotype-specific SVs (59.4% on average) (Supplementary Table 16).
a, Genomic length affected by haplotype-specific (heterozygous) variants by comparing haplotype H1 to H2 across 29 diploid potato accessions. b, GHSL in wild and cultivated potatoes. The boxes represent 75% and 25% quartiles, the central line indicates the median and the whiskers extend to 1.5 times the interquartile range; P = 2.6 × 10−4, two-tailed Student’s t-test, n = 19 (cultivated) and 10 (wild). c, PCA of chromosome 1 haplotypes, with dotted lines connecting paired haplotypes in heterozygous diploid potatoes. d, The phylogeny of 63 haplotypes for chromosome 1 was constructed using the maximum-likelihood method based on 1,074 single-copy genes. Dashed lines (right) connect the paired haplotypes in each diploid genome. RH, RH89-039-16. S. lycopersicum, Solanum lycopersicum; S. etuberosum, Solanum etuberosum; S. wrightii, Solanum wrightii. e, Unfolded site frequency spectrum of four types of SVs and non-synonymous single nucleotide polymorphisms (nSNPs) compared to putatively neutral synonymous SNPs (sSNPs) across cultivated potatoes.
We found that haplotype divergence in cultivated potatoes, as quantified by GHSL, is 1.46 times greater than that observed across wild potatoes (approximately 105.2 Mb versus 72.1 Mb; Fig. 3b, P = 2.6 × 10−4, two-sided Student’s t-test). Notably, two haplotype-specific inversions are quite prevalent in cultivated potatoes (Extended Data Fig. 4). One inversion has previously been reported and is associated with a locus that regulates yellow tuber flesh13 (DMv6.1, chr. 3: 42.9–48.7 Mb). The other inversion (DMv6.1, chr. 10: 52.7–59.1 Mb) contains 661 genes, including StPIN7 (Soltu.DM.10G026500.1) and StPTB6 (Soltu.DM.10G026670.1), which may be associated with auxin transport37 and tuberization38, respectively (Extended Data Fig. 5).
To explore the extent of genome heterozygosity and haplotype divergence during potato domestication, we conducted PCA separately for each chromosome and found that the two haplotypes of each wild potato form ‘nearest neighbours’, whereas the two haplotypes of each cultivated potato exhibit a more dispersed pattern (Fig. 3c and Supplementary Fig. 11). We constructed haplotype trees for each chromosome pair based on single-copy genes, using three additional Solanum species as outgroups (Fig. 3d and Supplementary Fig. 12). Analogous to the PCA-based patterns, the two haplotypes of each cultivated potato typically reveal more pronounced genetic divergence than those of each wild potato. We conducted additional validation through pairwise Jaccard similarity analysis and alignment for split windows, both of which indicate that multiple hybridization events have affected the haplotype landscape in cultivated potatoes (Extended Data Fig. 6 and Supplementary Fig. 13). Molecular evidence in maize hybrids suggests that the length of nonsyntenic regions between parental lines is strongly correlated with levels of heterosis39. Our findings of increased genome heterozygosity and haplotype divergence in cultivated potatoes are similar to those in grapevine, another clonal crop40. The enhanced genome heterozygosity in cultivated potatoes compared with wild potatoes results in reduced homozygous deleterious burden (Extended Data Fig. 7a,b), indicating that potato domestication included an exploration of heterosis.
Fate of dSVs under breeding
To characterize the fitness effects of SVs in potato cultivars, we computed the unfolded site frequency spectrum on the basis of SVs and SNPs from domesticated potatoes (Fig. 3e). We found that SVs are over-represented for singletons and other minor-allele frequency classes, compared to both synonymous SNPs and even non-synonymous SNPs at minor frequencies, consistent with equivalent observations in grapevine genomes36. Cultivated potatoes have around 2.7 times more heterozygous SVs (average of 20,613 per individual) than homozygous SVs (average of 7,561) (Extended Data Fig. 7a). Additionally, 39.4% of the heterozygous SVs are located either in the gene bodies or in putative promoter regions (Supplementary Table 17). These findings suggest that SVs are more likely to be strongly deleterious than SNPs and are typically present in heterozygous state with relatively low frequencies across diploid potato cultivars.
The presence of dSVs in heterozygous state could lead to strong inbreeding depression upon forced inbreeding, thus hindering the development of inbred lines, with important implications for hybrid potato breeding3,9,41. Inferred purifying selection against SVs is stronger than against SNPs, implying that SV frequencies can be useful for an initial assessment of putative selection pressure42,43. Recently, deep phylogenomic analyses of 92 species in the family Solanaceae assessed genome-wide evolutionary constraints (with highly constrained regions amounting to 31.7 Mb under the threshold genomic evolutionary rate profiling (GERP) score ≥ 2.0, including non-synonymous, synonymous and non-coding sites), forming the basis for identifying and quantifying dSNPs at genome-wide scales in potato10. Analogous to dSNP identification, we developed an unweighted approach to infer putative dSVs in the potato genome on the basis of the joint criteria of evolutionarily derived state, low allele frequency and being within gene-coding or evolutionarily constrained regions (Fig. 4a and Methods). In total, we characterized 19,625 dSVs, 50.4% of which are located within non-coding regions (Extended Data Fig. 7c,d and Supplementary Table 18). For example, we identified a 51.1-kb deleterious deletion occurring in only one haplotype of cultivar PG6090. This deletion leads to truncated proteins or gene loss and is associated with three genes (Supplementary Fig. 14).
a, The pipeline for identifying dSVs includes the joint criteria of derived state of low-frequency SVs (≤0.05) in gene-coding and/or evolutionarily constrained regions. b, The percentage of homozygous and heterozygous dSVs in wild and cultivated potato populations. c, The number of dSVs per chromosome in the founder PG6359 (average of the two haplotypes) and its derivative inbred line A6-26. d, Schematic diagram of recombination events in chromosome 2 (left) and haplotype selection in chromosome 4 (right). The potential recombination events are marked with dashed lines. Deleterious variants from the H1 and H2 haplotypes are shown in different colours. e, Pearson correlation coefficient (r = 0.78) and P value (2.6 × 10−16) between numbers of dSNPs and dSVs across all chromosomes (n = 696). The shaded area represents the bootstrapped 95% confidence interval. f, Distribution of the number of dSNPs flanking focal dSVs for 10-kb non-overlapping windows in the dSV coupling phase (orange) and the dSV repulsion phase (grey). The points represent means of 58 haplotypes and error bars represent 95% confidence intervals. g, Average distance of a focal dSV to the nearest dSV in coupling phase (same haplotype; dis-coupling) and to the nearest dSV in syntenic regions of the other haplotype (dis-repulsion). Only distances <1 Mb are included. The boxes represent 75% and 25% quartiles, the central line indicates the median and the whiskers extend to 1.5 times the interquartile range; P < 2.2 × 10−16, two-tailed Student’s t-test, n = 13,717 (dis-coupling) and 11,679 (dis-repulsion). h, A zoomed-in view of biased dSNP accumulation (blue bars) around a heterozygous dSV (red triangle) within a 20-kb segment of chromosome 4 in E86-69 H2.
Excluding the inbred lines, we found that each diploid potato individual has an average of 843 dSVs, comprising 23.1 Mb of genomic sequence. Our analyses revealed that 73% of dSVs are in heterozygous state across wild potatoes, and this proportion increases to 97% in cultivated lines (Fig. 4b and Supplementary Fig. 15). These patterns indicate that dSVs tend to be sheltered in heterozygous state, potentially largely avoiding negative selection on them36,44. Thus these heterozygous dSVs require attention from breeders when choosing suitable materials for breeding.
Purging dSVs is crucial for developing elite inbred lines in hybrid potato breeding. We established that the available inbred lines, A6-26 (88 dSVs) and E4-63 (303 dSVs), carry fewer dSVs compared to the average haplotypes from the two parents, PG6359 (135 dSVs) and E86-69 (351 dSVs), respectively (Fig. 4c and Supplementary Fig. 16). To investigate the process of purging dSVs at the haplotype level, we constructed a genome-wide recombination map based on inbred lines and their respective parental haplotypes (Extended Data Fig. 8). Our results track at least 16 recombination events in one of the inbred lines across 5 generations of self-fertilization. For example, the 2 haplotypes from accession PG6359 carry 17 and 11 dSVs on chromosome 2; recombination events decreased the number of dSVs to seven in the derivative inbred line A6-26. For chromosome 4 of PG6359 H2, which carries fewer dSVs than PG6359 H1 (14 versus 30), 85.2% of dSVs are retained in the inbred line A6-26 (chr. 4: 7.5–59.5 Mb; 14 dSVs) (Fig. 4d and Supplementary Table 19).
The ‘broken-window’ effect of dSVs
On the basis of the genome-wide map of deleterious variants, we found a significant positive correlation between the numbers of dSVs and dSNPs per haplotype (r = 0.78, P < 2.2 × 10–16, Pearson correlation coefficient; Fig. 4e and Supplementary Tables 19 and 20). In diploid genomes, we define the coupling phase as the haplotype that contains a focal dSV, whereas the repulsion phase refers to the other haplotype. To assess the potential effect of dSVs on dSNPs, we compared the number of dSNPs surrounding dSVs in the coupling phase to the number of dSNPs in the syntenic region of the repulsion phase. We partitioned the genome into intervals on the basis of dSV distribution across both haplotypes and calculated statistical significance to identify affected genomic regions (Extended Data Fig. 9a and Methods). Our results suggest that dSNPs are significantly more frequently present in dSV coupling phase than in the dSV repulsion phase (Fig. 4f, Extended Data Fig. 9b and Supplementary Table 21). This accumulation signal of dSNPs tends to be more pronounced in close proximity to dSVs.
Furthermore, we investigated whether dSVs form clusters by calculating two types of distances: the distance between the focal dSV to the nearest dSV in coupling phase (same haplotype; dis-coupling), and its distance to the nearest dSV in repulsion phase, (dis-repulsion) (Extended Data Fig. 9c). Dis-repulsion is significantly larger than dis-coupling (Fig. 4g, 264.1 kb versus 202.1 kb, P < 2.2 × 10–16, two-sided Student’s t-test), suggesting that dSVs tend to form clusters within the same haplotype (Extended Data Fig. 9d). We provide an illustrative example of this phenomenon using the diploid potato line E86-69 (Supplementary Fig. 17).
The presence of dSVs, enhancing the occurrence of dSVs and dSNPs in the same phase, is analogous to a sociological hypothesis called the broken-window effect45. Therefore, we refer to this pattern as the ‘broken-window’ effect of dSVs. We validated this pattern by examining local assemblies, which cover the 10-kb regions flanking dSVs with an average coverage of 19 reads, as well as 6 long reads spanning the flanking regions. Theoretical considerations as well as empirical evidence suggest that SVs affect recombination rates of closely linked genomic regions46,47,48. Those associations may result from insufficient purging around the dSV owing to reduced recombination, whereas the dSV repulsion phase tends to remain functional, thus preventing strong selection on deleterious recessive variants9,49, leading to gradual differences in deleterious burden between the two haplotypes (Fig. 4h). The broken-window effect reveals that dSVs and dSNPs are not randomly distributed across potato genomes, as they tend to form clusters in coupling phase. Such deleterious clusters are important features of potato genomes, and they are easy to spot in our phased pangenome. We found that many deleterious variants were not efficiently purged during our previous development of two inbred lines3 (A6-26 and E4-63). Line A6-26 carries 88 dSVs and 61,017 dSNPs, whereas E4-63 has 303 dSVs and 71,200 dSNPs (Supplementary Tables 19 and 20). These deleterious variants in inbred lines substantially reduce their fitness3 and thus need to be purged as a foundation for future potato breeding.
Genome design of ideal haplotypes
Plant breeders adopted the concept of ideal plant architecture (IPA) to guide breeding of superior varieties by combining multiple desirable traits50. Similar to the IPA, we propose the IPHs strategy to guide breeders to develop a suite of inbred lines that are as close as possible to the ideal genotype (see Supplementary Methods).
Based on PPG-v.1.0, the starting point of computationally designed IPHs-v.1.0 are two heterotic groups; nine cultivars from the STN and GON groups with the inbred line A6-26 (heterotic group A), and eight cultivars from the PHU group with the inbred line E4-63 (heterotic group E) (Extended Data Fig. 10a,b). In principle, recombination events can reduce genetic burden in simulated haplotype combinations (Extended Data Fig. 10c,d). On the basis of the recombination map of two inbred lines (Extended Data Fig. 8), we assumed no more than four recombination events per chromosome in our IPHs. The consideration of recombination breakpoints takes into account the recombination coldspots in certain regions, such as heterochromatic centromeric regions and inversions. We estimated the distribution of dSVs and dSNPs under various haplotype donors and recombination events to identify the optimal combinations of IPHs, IPHs-A and IPHs-E (Supplementary Fig. 18).
We found that ideally, all dSVs could be removed from IPHs-A and IPHs-E and the number of dSNPs could be reduced to 41,262 and 35,360, implying a reduction of 32.4% (A6-26) and 50.3% (E4-63) compared with the previous inbred lines (Fig. 5a,b and Supplementary Table 22). Haplotype distances predict that hypothetical F1 hybrid genomes between IPHs-A and -E would carry only 10,830 homozygous dSNPs (Fig. 5c), implying a reduction of 54.5% compared with the previous F1 hybrid potato3. By integrating the genetic linkage map51, we found that achieving a IPHs-A chromosome 1 would require at least five generations of recurrent selection from hybrid progenies, and that one of the designed breakpoints is located in a low-recombination region (near chr. 1: 44.3 Mb), requiring a population size of at least 1,321 individuals to obtain at least 1 copy of IPHs-A chromosome 1 (Supplementary Fig. 19). It is worth noting that relying solely on natural recombination is unlikely to be sufficient (Supplementary Fig. 20). New technologies such as targeted recombination, gene editing and synthetic biology to precisely eliminate deleterious variants may provide future pathways for approaching IPHs52,53,54.
a, Chimeric map of the IPHs-A genome and the most ideal graph path of 12 chromosomes from heterotic A group (left). The numbers of dSVs and dSNPs for each chromosome are indicated. Haplotypes for chromosome 1 are shown in detail (right), and numbers of dSVs and dSNPs for each haplotype are represented by five parts derived from the ideal designed recombination events. The chimeric graph path of IPHs-A ‘ideal’ chromosome 1 is highlighted with red frames. Inversions and centromeres are coloured in genomic collinear blocks and chromosomes, respectively. b, Similar to a, the chimeric map of the IPHs-E genome and the ideal chimeric graph paths from the heterotic E group. c, The number of dSNPs of IPHs-A and IPHs-E, the overlap indicating the number of homozygous dSNPs in the inferred ideal F1 hybrid generated from IPHs-A and IPHs-E.
Discussion
In this study, we developed a graph-based phased potato pangenome reference comprising 60 haplotype sequences, reducing reference bias and identifying more variants that remain unaccounted by reads-based methods55,56. Our phased pangenome provides an example of a typical clonally propagated crop with a highly heterozygous genome. The integration of pedigree information and high-accuracy ultra-long reads can further improve phase accuracy in highly repetitive regions.
Consistent with observations in grapevine36, SVs are over-represented among singletons and other minor-allele frequency classes, suggesting that SVs are under stronger purifying selection. SVs can be associated with agronomic traits57,58,59, and by disrupting gene structure and cis-regulatory elements, dSVs can exert strong effects. We classified 14% of all SVs to be putative dSVs. However, inference of dSVs shows lower efficacy in certain cases owing to overlapping SVs and ambiguous ancestral state. Therefore, quantifying their fitness penalties will be crucial to guide breeding practices in the future.
Asexually reproducing crops tend to accumulate more heterozygous deleterious mutations36,49. The broken-window effect of dSVs may be reinforced owing to very limited recombination under long-term clonal propagation. After one haplotype is hit by a deleterious mutation with major effects, it seems less likely that deleterious mutations accumulate in the dSV repulsion phase, otherwise the affected individual might not survive to reproduce. This inference underscores the importance of preserving at least one functional copy of genes to maintain genetic robustness60,61. Eradicating dSVs is therefore a new focus in the development of inbred lines to generate diverse hybrid potato combinations, a principle that should also be relevant for other clonally propagated crops.
To develop inbred lines, it is more favourable to start from haplotypes of diploid landraces with a lower deleterious burden10. The relatively high deleterious burden per haplotype in a dihaploid generated from tetraploid varieties poses formidable obstacles for this step (Supplementary Fig. 21). Therefore, we plan to develop the first generation of inbred lines from diploid landraces concurrently with inducing dihaploids from elite tetraploid varieties. The IPHs strategy provides a blueprint for the combination of haplotypes to reduce the negative impact of deleterious variants. Although achieving IPHs at the chromosome level through crossing is possible, it is challenging to combine the ideal chromosomes into one genome without additional recombination (Supplementary Figs. 22 and 23). Currently, the primary value of IPHs lies in their ability to outline an optimal goal for haplotype recombination, allowing breeders to compare real haplotypes, including dSVs and dSNPs, against an idealized standard, thus facilitating refinement of breeding strategies.
Methods
Plant materials and sequencing
We selected 31 potato accessions based on phylogenetic trees generated by previous studies. Of these accessions, 10 are from S. candolleanum and 21 are from 3 groups of diploid potato cultivars. Among these potato cultivars, nine accessions represent S. tuberosum Group Phureja (including inbred line E4-63), eight accessions represent S. tuberosum Group Stenotomum (including inbred line A6-26), and four are from S. tuberosum Group Goniocalyx. Previous HiFi data are available for 21 accessions and previous Hi-C reads are available for five accessions (PRJNA754534). Additionally, we newly sequenced 10 accessions using HiFi and 25 accessions using Hi-C in this study. HiFi reads for 10 accessions were generated using the ccs program version 6.4.0 (https://github.com/PacificBiosciences/ccs) and subreads obtained from the Pacific Biosciences Sequel II platform, which were then converted to FASTQ format by SAMtools (v.1.17)62. In total, we generated 318.49 Gb of HiFi data, ranging from 27.72 to 35.32 Gb per sample. For Hi-C libraries, DNA was extracted from in vitro seedlings and digested with the restriction enzyme MboI using previously described Hi-C library preparation protocols63,64. A total of 2.70 Tb of Hi-C data were generated based on the Illumina HiSeq platform. To facilitate genome annotation, we used RNA-sequencing (RNA-seq) data from six plant tissues (including roots, stems, leaves, stolon, tubers and flowers) from a previous study13.
De novo genome assembly of 60 haplotypes
For genome size and heterozygosity estimation, we used Jellyfish (v.2.3.0)65 to obtain a frequency distribution of the k-mers and estimated the histograms by GenomeScope (v.2.0)66. We then assembled haplotype-resolved assemblies with the HiFi reads and Hi-C reads using hifiasm (https://github.com/chhylp123/hifiasm) (v.0.16)67 with default parameters. Subsequently, we aligned haplotypes using the Juicer pipeline and then generated Hi-C maps for the 3D-DNA pipeline (v.201008)68 with parameter “-q 0”. The assembly from two haplotypes was scaffolded and ordered using the Hi-C data-based rough scaffold and compared with the reference (accession DM1-3 516 R44)25. Two sets of pseudo-chromosomes were constructed for each accession, and this workflow was applied to all 29 diploid potato accessions, resulting in 58 haplotypes. For the two inbred lines E4-63 and A6-26, we generated haploid assemblies. The Hi-C format file was visualized using the Juicebox program (v.2.16.00)69 and misassembled contigs were manually curated. Finally, each haplotype was integrated into the 3D-DNA pipeline based on the run-asm-pipeline-post-review.sh script. To ensure the accuracy of the assemblies, we filtered out contigs smaller than 50 kb and used chromosome-based haplotypes for subsequent analyses.
Evaluation of the genome assemblies
The contig N50 statistic was calculated using assembly-stats (https://github.com/sanger-pathogens/assembly-stats). BUSCO (v.5.4.4)23 was used to evaluate assembly quality with the embryophyte_odb10 protein database. Both switch and Hamming errors were calculated using two variant call format (VCF) files produced by the pipeline calc_switchErr (https://github.com/tangerzhang/calc_switchErr), based on the ‘compare’ function of WhatsHap (v.1.1)70. Genome completeness was estimated via the KAT (v.2.4.2)71 ‘comp’ command. Reliability of our assemblies was estimated by Flagger, a reads-mapping-based pipeline optimized for diploid assemblies24. A detailed workflow and commands can be found in the Supplementary Information and GitHub (https://github.com/Chenglin20170390/Haplotype-diversity).
Annotation of repetitive elements
Different repeat elements are identified based on 60 haplotypes, including TEs, segmental duplications, TRs and satellite repeats. To identify TEs, we employed the Extensive de novo TE Annotator (EDTA) (v.2.1.0)72, including LTRs, DNA transposons with terminal inverted repeat sequences and helitron transposons. For the remaining transposon elements, RepeatModeler73 was used to search for a second round. To search for large segmental duplications (>1 kb), we employed Asgart (v.2.4.0)74 using a k-mer-based method with the parameter “-CRSV”. TRs were identified using Tandem Repeats Finder (v.4.09.1)75 with default parameters. Satellite repeats were identified using Satellite Repeat Finder (SRF, commit e54ca8c; https://github.com/lh3/srf) with HiFi reads.
Prediction of protein-coding genes
A comprehensive strategy consisting of transcript evidence, ab initio prediction, and homology alignment was applied for gene prediction. First, we aligned RNA-seq reads to assembled haplotypes using HISAT2 (v.2.2.1)76 with the “--dta” parameter and then assembled by StringTie (v.2.2.1)77 with the “--rf” parameter. Subsequently, we used the BRAKER2 (v.2.1.5)78 program to train the ab initio prediction model from AUGUSTUS (v.3.4.0)79 (https://github.com/Gaius-Augustus/Augustus) and collected high-quality RNA-seq hints using the Hidden Markov Model (HMM) from GeneMark-ET (v.3.67)80 with the parameter “--nocleanup --softmasking”. To improve gene structure prediction, we performed a homology search using a curated plant protein dataset downloaded from the UniProt Swiss-Prot database (https://www.uniprot.org/downloads). We merged these homologous proteins with previously published peptides from tomato81 and potato4 and eliminated potential redundancy using the CD-HIT-est (v.4.6.8)82 program with default parameters. The MAKER2 (v.3.01.03)83 program was used to combine the homology search, expression evidence and ab initio prediction through two rounds. Finally, we used the Mikado (v.2.3.4)84 program to identify the representative set of transcripts from transcript assemblies, before those were fed to the PASA pipeline (v.2.5.1)85 to update gene structures.
To perform functional gene annotation, we utilized the InterProScan (v.5.34-73.0)86 program, which identifies potential protein domains and Gene Ontology terms based on sequence signatures. We applied the following parameters to the program: “-cli -iprlookup -tsv -gotermd -appl Pfam”. In particular, we extracted protein ___domain information from Pfam by enabling the “-appl Pfam” parameter. For each of the genes, we assigned the functional description of the best hit.
Construction of the potato pangenome
The Minigraph-Cactus pipeline27 and PGGB26 were used to construct a pseudo-phased pangenome with all 61 haplotypes based on the whole-genome alignment (including the DMv6.1 reference genome). For the PGGB, we estimated the divergence of each chromosome with mash distances and confirmed chromosome community with wfmash87 mapping. Then, we used “pggb -s 10000 -n 61 -p 90 -k 47 -P asm20 -O 0.001” to build each chromosome graph. We visualized the 1D layout of the graph and estimated presence and absence ratios to the DMv6.1 reference in 100-kb sliding windows using ODGI88. The small variants and SVs were detected by vg deconstruct from snarls, and we only kept top-level and <1-Mb variants with vcfbub. For the Minigraph-Cactus pipeline, we assigned DMv6.1 as the guide for the paths, and progressively aligned the 60 haplotypes to it. We used the cactus-pangenome script with parameters “--gfa full --gbz full --vcfReference DMv6.1” to generate complete workflows and execute commands. The generated graph fragment assembly (GFA) format graph was used for edge, node and coverage statistics and subgraph generation from a BED input. The VCF output file comprises all graph variations based on the DMv6.1 reference, enabling the calculation of polymorphisms.
Pangenome size and growth
To fully capture the genome diversity of our potato populations, we used Panacus89 to assess pangenome size and growth ratio, which estimates pangenome openness directly by applying the binomial formula. We calculated cumulative bases based on quorum (minimum fraction of haplotypes sharing a graph feature after haplotypes are sequentially added to the growth histogram), and proportion of conserved (≥90% of haplotypes) and variable sequences (<90% of haplotypes) in the pangenome graphs. These statistics were obtained using Panacus hist and growth scripts with parameters “-c bp -l 1,1,1 -q 0,0.1,0.5,0.9 -S”, and we selected different haplotype paths for group-specific statistics.
Phylogeny and synteny analysis
To build haplotype phylogenetic trees at the chromosome level, annotated protein sequences from the 60 newly assembled haplotypes and three published genomes (outgroups: S. wrightii10; S. etuberosum13 and S. lycopersicum90) were aligned to produce all-versus-all alignments using Diamond (v.0.9.21)91. The gene families were inferred using the OrthoFinder (v.2.5.4)92 program, which utilizes the Markov cluster algorithm. Separately for each chromosome pair, all single-copy orthologous protein sequences were merged into a single FASTA file, which was then fed into IQ-TREE (v.2.0.6)93 using the maximum-likelihood method. Each chromosome tree was visualized with the function ggtree (v.3.0.2)94 in R (v.4.2.0). Synteny plots and pangenome annotations were generated by the R package Genespace (v.0.9)95.
Identification of SNPs, indels and SVs
Assembly-based calling was used to detect SVs with a minimum length of 50 bp. Genome sequences were aligned to the DMv6.1 reference genome to produce alignment BAM files using Minimap2 (v.2.17)96 with “-ax asm5”. SyRI (v.1.5)97 was then utilized to call genome-wide variants and generate assemblies-based VCFs. We kept variants of Ins, Del, Inv and Dup from the output of SyRI as the individual SV dataset. The accuracy of the SV dataset was validated by randomly selecting and manually verifying 100 SVs longer than 100 kb using Hi-C maps. Finally, we merged the individual SVs based on 80% overlap using SURVIVOR with the parameters “0.8 1 1 -1 -1 -1” to produce the final population SVs file. For SNPs and indels, we merged the output of assembly-based variation calling from SyRI using BCFtools (v.1.13)98.
Haplotype diversity and GHSL statistic
The length of the haplotype-specific (heterozygous) variants for each diploid accession was calculated by summing all variants present in only one haplotype. In detail, we aligned the two haplotypes of one accession to each other using Minimap2 (v 2.17), identified variants by SyRI (v.1.5), and then calculated the number of haplotype-specific SNPs and the length of haplotype-specific variants (indels and SVs). The GHSL approach considers reference bias and regions potentially affected by haplotype-specific variants (https://github.com/Chenglin20170390/Haplotype-diversity). PCA based on haplotypes (12 chromosomes) was performed using Plink (v.1.90)99 on the VCF file with the parameter “--pca 5”; the results were visualized using the R package ggplot2100. Pairwise Jaccard similarities between haplotypes were estimated using BEDTools (v.2.30.0) with the ‘jaccard’ command.
SV dynamics in potato domestication
To calculate the unfolded site frequency spectrum, we analysed 19 cultivated potato accessions (excluding the two inbred lines) from VCFs. The genome of S. lycopersicum (SL 5.090) was used as an outgroup to infer the ancestral states of genotypes. We distinguish different types of SNPs (synonymous and non-synonymous) and SVs (deletions, insertions, inversions and duplications) and counted the number of haplotypes carrying the derived variants. For each accession, we also calculated the incidences (numbers) of heterozygous and homozygous SVs and the sums of all SVs (twice the number of homozygous SVs plus the number of heterozygous SVs).
Identification of TEMRs
SV breakpoints with additional ±100-bp flanking sequences were used to identify repeat elements (TEs, segmental duplications and TRs) associated with SVs. To detect the potentially causal relationship between TE movement and SV formation, the documented genomic position of a focal SV and overlapping TEs determine the following categories: ‘No TE SV’ for SVs without any TE overlap, ‘Incomplete TE SVs’ for SVs overlapping with a single TE with <95% coverage, ‘Single TE SVs’ for SVs overlapping with a single TE with coverage ≥95%, and ‘Multi TE SVs’ for SVs overlapping with multiple TEs with coverage >95%. Since no single TE SV events were considered, insertions were excluded. To quantify the formation of SVs (deletions, inversions and duplications) by ectopic recombination via TEMRs, SV breakpoints (±100 bp) overlapping with the identical class of TEs were calculated. To assess the distribution of repeats in SV breakpoints, we randomly simulated SVs using the shuffle function of BEDTools with the number and length of SVs set according to the potato SV variation map. Two-sided Student’s t-tests were used to test the significance of the proportions between observed and simulated TE-related SVs. To infer the insertion times of TEMRs, we extracted insertion times from the pass.list of EDTA and kept intact TE sequences overlapping with the breakpoints of SVs. Sequence homology of TEMRs was calculated using 200-bp flanking sequences based on global pairwise sequence alignment from Needle (https://github.com/nanjiangshu/my_needle).
Haplotype recombination in inbred lines
The fifth-generation inbred lines A6-26 and E4-63 were generated from the diploid cultivars E86-69 and PG6359, respectively. To evaluate haplotype recombination events in each inbred line, we utilized two different methods: heterozygosity analysis and haplotype-specific k-mer analysis. Genome-wide heterozygous peaks were identified by aligning the sequences of the haplotypes. The different heterozygous peaks were compared between haplotypes from inbred lines and their founders. The regions inherited from the founders’ haplotypes in the alignment would be in homozygous state. These switch signals from homozygosity to heterozygosity were used to characterize putative recombination events. Haplotype-specific k-mers were calculated by the number of specific k-mers from the inbred line based on non-overlapping 500-kb regions and compared with parental haplotypes using the Meryl program (available at https://github.com/marbl/meryl).
Identification of dSVs and dSNPs
Putative dSVs were identified based on three criteria: (1) ancestral versus derived state of SVs. S. lycopersicum (SL 5.0)90 was used as the outgroup for ancestral state inference, as phylogenetic analyses have placed this species in a clade relatively close to Solanum section Petota13. (2) SVs with frequencies below 0.05 in our 60 haplotypes were considered, as most deleterious variants occur at low population frequencies16,42,43. (3) SVs in coding regions that may damage proteins’ function or SVs in the 92 Solanaceae evolutionarily constrained regions are considered to be deleterious10. We thus identified putative dSVs as being derived-state SVs with low-frequency (<0.05) that overlap with coding regions or evolutionarily constrained regions; we implemented this with the BEDTools ‘overlap’ command to infer putative dSVs. Putative dSNPs were identified by showing GERP values >2.75 and overlapping with evolutionarily constrained regions10.
Estimation of the broken-window effect
The Pearson correlation coefficient was used to examine the correlation between the number of dSNPs and dSVs per chromosome. Focal dSVs located in haplotype 1 (H1) indicates H1 was considered to be the dSV coupling phase and corresponding syntenic regions of H2 were considered to be the dSV repulsion phase. We calculated the distance of the focal dSV to the nearest dSV in coupling phase and to the nearest dSV in repulsion phase (Extended Data Fig. 9). To exclude the potential concurrent influence of dSVs on both coupling and repulsion phases, we segmented chromosomes into fragments based on the midpoint between adjacent dSVs (irrespective whether in coupling or in repulsion phase). Regions that may be affected by the broken-window effect were estimated based on 10-kb non-overlapping sliding windows. For each sliding window, we calculated the number of dSNPs flanking each dSV in the coupling and repulsion phases. To assess dSNP enrichment, for each sliding window we performed a two-tailed Student’s t-test for the numbers of associated dSNPs in coupling and repulsion phase, respectively (regions significantly affected by the broken-window effect of dSVs). The depth of reads near the dSV is calculated using the SAMtools (v.1.17) ‘depth’ command. Finally, we employed SafFire (https://github.com/mrvollger/SafFire) to visualize schematic representations of dSVs and dSNPs within haplotypes.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the findings of this work are available within the paper and its supplementary information files. Whole-genome sequencing data are accessible through NCBI under the BioProject accession number PRJNA1020967 and from the National Genomics Data Center (NGDC) Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) with the BioProject accession number PRJCA020375. Genome sequences, gene annotations and variation maps were uploaded to the Potato Repositories Website (http://solomics.agis.org.cn/potato/) and Figshare (https://figshare.com/articles/dataset/IPHs/25846003 (ref. 101)). Publicly available sequencing data were downloaded from the NCBI with BioProject accession numbers PRJNA754534, PRJNA641265, PRJNA573826 and PRJNA766763.
Code availability
Scripts used in this Article are available at https://github.com/Chenglin20170390/Haplotype-diversity.
References
Stokstad, E. The new potato. Science 363, 574–577 (2019).
Spooner, D. M., Ghislain, M., Simon, R., Jansky, S. H. & Gavrilenko, T. Systematics, diversity, genetics, and evolution of wild and cultivated potatoes. Bot. Rev. 80, 283–383 (2014).
Zhang, C. et al. Genome design of hybrid potato. Cell 184, 3873–3883 (2021).
The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011).
Zhou, Q. et al. Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat. Genet. 52, 1018–1023 (2020).
Lian, Q. et al. Acquisition of deleterious mutations during potato polyploidization. J. Integr. Plant Biol. 61, 7–11 (2019).
Bao, Z. et al. Genome architecture and tetrasomic inheritance of autotetraploid potato. Mol. Plant 15, 1211–1226 (2022).
Jansky, S. H. et al. Reinventing potato as a diploid inbred line-based crop. Crop Sci. 56, 1412–1422 (2016).
Zhang, C. et al. The genetic basis of inbreeding depression in potato. Nat. Genet. 51, 374–378 (2019).
Wu, Y. et al. Phylogenomic discovery of deleterious mutations facilitates hybrid potato breeding. Cell 186, 2313–2328.e15 (2023).
van Lieshout, N. et al. Solyntus, the new highly contiguous reference genome for potato (Solanum tuberosum). G3 10, 3489–3495 (2020).
Freire, R. et al. Chromosome-scale reference genome assembly of a diploid potato clone derived from an elite variety. G3 11, jkab330 (2021).
Tang, D. et al. Genome evolution and diversity of wild and cultivated potatoes. Nature 606, 535–541 (2022).
Sun, H. et al. Chromosome-scale and haplotype-resolved genome assembly of a tetraploid potato cultivar. Nat. Genet. 54, 342–348 (2022).
Ye, M. et al. Generation of self-compatible diploid potato by knockout of S-RNase. Nat. Plants 4, 651–654 (2018).
Chun, S. & Fay, J. C. Identification of deleterious mutations within three human genomes. Genome Res. 19, 1553–1561 (2009).
Marsden, C. D. et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc. Natl Acad. Sci. USA 113, 152–157 (2016).
Liu, Q., Zhou, Y., Morrell, P. L. & Gaut, B. S. Deleterious variants in Asian rice and the potential cost of domestication. Mol. Biol. Evol. 34, 908–924 (2017).
Zhang, X. et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 53, 1250–1259 (2021).
Jarvis, E. D. et al. Semi-automated assembly of high-quality diploid human reference genomes. Nature 611, 519–531 (2022).
Schreiber, M., Jayakodi, M., Stein, N. & Mascher, M. Plant pangenomes for crop improvement, biodiversity and evolution. Nat. Rev. Genet. 25, 563–577 (2024).
Hardigan, M. A. et al. Genome diversity of tuber-bearing Solanum uncovers complex evolutionary history and targets of domestication in the cultivated potato. Proc. Natl Acad. Sci. USA 114, E9999–E10008 (2017).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Liao, W.-W. et al. A draft human pangenome reference. Nature 617, 312–324 (2023).
Pham, G. M. et al. Construction of a chromosome-scale long-read reference genome assembly for potato. GigaScience 9, giaa100 (2020).
Garrison, E. et al. Building pangenome graphs. Nat. Methods 21, 2008–2012 (2024).
Hickey, G. et al. Pangenome graph construction from genome alignments with Minigraph-Cactus. Nat. Biotechnol. 42, 663–673 (2024).
Garrison, E. & Guarracino, A. Unbiased pangenome graphs. Bioinformatics 39, btac743 (2023).
Gong, Z. et al. Repeatless and repeat-based centromeres in potato: implications for centromere evolution. Plant Cell 24, 3559–3574 (2012).
Bozan, I. et al. Pangenome analyses reveal impact of transposable elements and ploidy on the evolution of potato species. Proc. Natl Acad. Sci. USA 120, e2211117120 (2023).
Domínguez, M. et al. The impact of transposable elements on tomato diversity. Nat. Commun. 11, 4058 (2020).
Wicker, T. et al. A detailed look at 7 million years of genome evolution in a 439 kb contiguous sequence at the barley Hv‐eIF4E locus: recombination, rearrangements and repeats. Plant J. 41, 184–194 (2005).
Balachandran, P. et al. Transposable element-mediated rearrangements are prevalent in human genomes. Nat. Commun. 13, 7115 (2022).
Fang, Z. et al. Megabase-scale inversion polymorphism in the wild ancestor of maize. Genetics 191, 883–894 (2012).
Berdan, E. L., Blanckaert, A., Butlin, R. K. & Bank, C. Deleterious mutation accumulation and the long-term fate of chromosomal inversions. PLoS Genet. 17, e1009411 (2021).
Zhou, Y. et al. The population genetics of structural variants in grapevine domestication. Nat. Plants 5, 965–979 (2019).
Roumeliotis, E., Kloosterman, B., Oortwijn, M., Visser, R. G. & Bachem, C. W. The PIN family of proteins in potato and their putative role in tuberization. Front. Plant Sci. 4, 524 (2013).
Cho, S. K. et al. Polypyrimidine tract-binding proteins of potato mediate tuberization through an interaction with StBEL5 RNA. J. Exp. Bot. 66, 6835–6847 (2015).
Wang, B. et al. De novo genome assembly and analyses of 12 founder inbred lines provide insights into maize heterosis. Nat. Genet. 55, 312–323 (2023).
Xiao, H. et al. Adaptive and maladaptive introgression in grapevine domestication. Proc. Natl Acad. Sci. USA 120, e2222041120 (2023).
Bethke, P. C. et al. Diploid potatoes as a catalyst for change in the potato industry. Am. J. Potato Res. 99, 337–357 (2022).
Mezmouk, S. & Ross-Ibarra, J. The pattern and distribution of deleterious mutations in maize. G3 4, 163–171 (2014).
Li, Y. et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat. Genet. 42, 969–972 (2010).
Wang, N. et al. Structural variation and parallel evolution of apomixis in citrus during domestication and diversification. Natl Sci. Rev. 9, nwac114 (2022).
Harcourt, B. E. Reflecting on the subject: a critique of the social influence conception of deterrence, the broken windows theory, and order-maintenance policing New York style. Mich. Law Rev. 97, 291 (1998).
Morgan, A. P. et al. Structural variation shapes the landscape of recombination in mouse. Genetics 206, 603–619 (2017).
Porubsky, D. et al. Recurrent inversion polymorphisms in humans associate with genetic instability and genomic disorders. Cell 185, 1986–2005 (2022).
Rowan, B. A. et al. An ultra high-density Arabidopsis thaliana crossover map that refines the influences of structural variation and epigenetic features. Genetics 213, 771–787 (2019).
Ramu, P. et al. Cassava haplotype map highlights fixation of deleterious mutations during clonal propagation. Nat. Genet. 49, 959–963 (2017).
Jiao, Y. et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544 (2010).
Jiang, X. et al. Genomic features of meiotic crossovers in diploid potato. Hortic. Res. 10, uhad079 (2023).
Hayes, B. J. et al. Potential approaches to create ultimate genotypes in crops and livestock. Nat. Genet. 56, 2310–2317 (2024).
Filler-Hayut, S., Kniazev, K., Melamed-Bessudo, C. & Levy, A. A. Targeted inter-homologs recombination in Arabidopsis euchromatin and heterochromatin. Int. J. Mol. Sci. 22, 12096 (2021).
Schmidt, C., Schindele, P. & Puchta, H. From gene editing to genome engineering: restructuring plant chromosomes via CRISPR/Cas. Abiotech 1, 21–31 (2020).
Eizenga, J. M. et al. Pangenome graphs. Annu. Rev. Genomics Hum. Genet. 21, 139–162 (2020).
Rakocevic, G. et al. Fast and accurate genomic analyses using genome graphs. Nat. Genet. 51, 354–362 (2019).
Alonge, M. et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145–161. e23 (2020).
Leffler, E. M. et al. Resistance to malaria through structural variation of red blood cell invasion receptors. Science 356, eaam6393 (2017).
Yan, H. et al. Pangenomic analysis identifies structural variation associated with heat tolerance in pearl millet. Nat. Genet. 55, 507–518 (2023).
Conrad, D. F. et al. Origins and functional impact of copy number variation in the human genome. Nature 464, 704–712 (2010).
Xue, J. R. et al. The functional and evolutionary impacts of human-specific deletions in conserved elements. Science 380, eabn2253 (2023).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Burton, J. N. et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat. Biotechnol. 31, 1119–1125 (2013).
Kaplan, N. & Dekker, J. High-throughput genome scaffolding from in vivo DNA interaction frequency. Nat. Biotechnol. 31, 1143–1147 (2013).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Ranallo-Benavidez, T. R., Jaron, K. S. & Schatz, M. C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020).
Cheng, H., Concepcion, G. T., Feng, X., Zhang, H. & Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 18, 170–175 (2021).
Dudchenko, O. et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95 (2017).
Durand, N. C. et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016).
Martin, M. et al. WhatsHap: fast and accurate read-based phasing. Preprint at BioRxiv https://doi.org/10.1101/085050 (2016).
Mapleson, D., Garcia Accinelli, G., Kettleborough, G., Wright, J. & Clavijo, B. J. KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33, 574–576 (2017).
Ou, S. et al. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20, 275 (2019).
Flynn, J. M. et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl Acad. Sci. USA 117, 9451–9457 (2020).
Delehelle, F., Cussat-Blanc, S., Alliot, J.-M., Luga, H. & Balaresque, P. ASGART: fast and parallel genome scale segmental duplications mapping. Bioinformatics 34, 2708–2714 (2018).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Brůna, T., Hoff, K. J., Lomsadze, A., Stanke, M. & Borodovsky, M. BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genomics Bioinformatics 3, lqaa108 (2021).
Stanke, M. et al. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 34, W435–W439 (2006).
Lukashin, A. V. & Borodovsky, M. GeneMark. hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107–1115 (1998).
The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
Li, W. & Godzik, A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Holt, C. & Yandell, M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011).
Venturini, L., Caim, S., Kaithakottil, G. G., Mapleson, D. L. & Swarbreck, D. Leveraging multiple transcriptome assembly methods for improved gene structure annotation. GigaScience 7, giy093 (2018).
Haas, B. J. et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003).
Jones, P. et al. InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014).
Marco-Sola, S., Moure, J. C., Moreto, M. & Espinosa, A. Fast gap-affine pairwise alignment using the wavefront algorithm. Bioinformatics 37, 456–463 (2021).
Guarracino, A., Heumos, S., Nahnsen, S., Prins, P. & Garrison, E. ODGI: understanding pangenome graphs. Bioinformatics 38, 3319–3326 (2022).
Parmigiani, L., Garrison, E., Stoye, J., Marschall, T. & Doerr, D. Panacus: fast and exact pangenome growth and core size estimation. Bioinformatics https://doi.org/10.1093/bioinformatics/btae720 (2024).
Zhou, Y. et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 606, 527–534 (2022).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Emms, D. M. & Kelly, S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019).
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Yu, G., Smith, D. K., Zhu, H., Guan, Y. & Lam, T. T. Y. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 8, 28–36 (2017).
Lovell, J. T. et al. GENESPACE tracks regions of interest and gene copy number variation across multiple genomes. eLife 11, e78526 (2022).
Li, H. New strategies to improve minimap2 alignment accuracy. Bioinformatics 37, 4572–4574 (2021).
Goel, M., Sun, H., Jiao, W.-B. & Schneeberger, K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 20, 277 (2019).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. GigaScience 10, giab008 (2021).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 3, 180–185 (2011).
Cheng, L. IPHs. Figshare https://doi.org/10.6084/m9.figshare.25846003.v8 (2024).
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32488302), Guangdong Major Project of Basic and Applied Basic Research (2021B03C1030004), the National Key Research and Development Program of China (2019FA0906200), Agricultural Science and Technology Innovation Program (CAAS-ZDRW202404), the National Natural Science Foundation of China (32422079) and Shenzhen Outstanding Talents Training Fund.
Author information
Authors and Affiliations
Contributions
S.H. designed the project. S.H. and Yongfeng Zhou supervised the project. L.C. performed analyses of genome assembly, annotation, TE-mediated SV formation and GHSL of SVs. L.C. and N.W. analysed deleterious variants and the construction of IPHs. N.W. simulated breeding routines to achieve IPHs. Z.B., A.G. and E.G. performed graph construction and evaluation. P.W. contributed to the greenhouse work. Y.Y., Z.Z., D.T., Y.H., Z.M. and Q.L. assisted in bioinformatics analyses. P.Z., Y.W., Yi Zheng, Yao Zhou and L.L. provided essential technology support. L.C., N.W. and Z.B. wrote the draft. Q.Z., A.G., C.Z., W.J.L., E.G., N.S., T.S., Yongfeng Zhou and S.H. revised the manuscript. All authors contributed to manuscript preparation and read, commented on and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Todd Michael, Peter Morrell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Assembly strategy and validation of the potato pangenome.
a, Schematic diagram of the assembly process of the haplotype-resolved potato genome. b, Assessment of genome completeness by the read-mapping rate. c, Pangenome growth curves for 61 haplotypes (plus DMv6.1 reference) based on the Minigraph-Cactus (MC) graph, including conserved (present in at least 90% haplotypes) and variable sequence (<90% of haplotypes). d, Percentage of multiallelic SVs and indels for the PGGB and MC potato pangenomes.
Extended Data Fig. 2 Visualization of the potato chromosome 10 pangenome graph.
The visualization utilizes a 1D representation of the pangenome graph, with the DMv6.1 reference genome serving as the baseline for orientation. Graph nodes are arranged from left to right corresponding to their positions within the reference genome. The black lines under the paths are the links, which represent the graph topology. Black and red bars indicate regions oriented in the forward and reverse direction relative to the pangenome sequence (the complete sequence formed by concatenating all graph nodes). Access to visualizations of the 1D PGGB pangenome graphs for all chromosomes can be found in Figshare (https://figshare.com/articles/dataset/IPHs/25846003).
Extended Data Fig. 3 The statistics of TEMR mediated by LTR/Gypsy and LTR/Copia.
a, The density of estimated insertion time of TEMRs for LTR/Copia and LTR/Gypsy. b, The intact length of long terminal repeats in TEMRs for LTR/Copia and LTR/Gypsy. The boxes represent 75% and 25% quartiles, the central line denotes the median and the whiskers extend to 1.5 times the interquartile range. P < 2.2 × 10−16, two-tailed Student’s t-test, n = 70,000 for both LTR/Copia and LTR/Gypsy. Recently active TE events exhibit a higher degree of intact sequences and are associated with more recent TEMR events. c, The number and length of flanking homologous sequence of TEMRs for LTR/Copia and LTR/Gypsy. d, An example of a LTR/Gypsy-mediated deletion on chromosome 2 of the potato accession PG5003 genome.
Extended Data Fig. 4 The gene synteny ideogram constructed by protein sequences of all haplotypes.
Different chromosomes are distinguished by alternating light grey and dark grey. Two heterozygous inversions prevalent in cultivated potatoes are marked with dashed-line red rectangles.
Extended Data Fig. 5 An example of a heterozygous inversion in the PG6244 genome assembly.
a, Synteny plot for two haplotypes aligned to the DM genome. The haplotype H1 of PG6244 exhibits a 6.4-Mb inversion (INV894, chr10: 52.7–59.1 Mb) on chromosome 10 but PG6244 H2 shows synteny with the DMv6.1 genome. b, The Hi-C map for the assembly of the two PG6244 haplotypes for chromosome 10. An inversion signal between the two haplotypes is marked by the black rectangles.
Extended Data Fig. 6 Pairwise Jaccard similarity analysis of 60 potato haplotypes.
The heatmap indicates the correlation among 60 potato haplotypes. Paired haplotypes of each accession are indicated at the top and left with dashed lines. The rectangle colours represent different potato groups (see legend in upper-right corner).
Extended Data Fig. 7 The characteristics of SVs, dSNPs and dSVs in the potato genome.
a, The number of homozygous SVs, heterozygous SVs and the sum of all SVs (twice the number of homozygous SVs plus the number of heterozygous SVs) across our panel of potato genomes. b, The number of homozygous dSNPs, heterozygous dSNPs and the sum of all dSNPs across our panel of potato genomes. For a and b, the boxes represent 75% and 25% quartiles, the central line indicates the median and the whiskers extend to 1.5 times the interquartile range. P values are provided in each panel representing comparisons between wild (n = 10) and cultivated potato samples (n = 19), using two-tailed Student’s t-tests. c, Genome architecture of dSVs across the potato genome. d, The distribution of dSVs and dSNPs across the potato genome.
Extended Data Fig. 8 The recombination map of two inbred lines.
a, Recombination map of the parent E86-69 and its inbred offspring E4-63. The x-axis represents the chromosomes. The y-axis represents the number of heterozygous markers (SNPs and indels). Top: heterozygosity was identified by comparing E86-69 H1 and E86-69 H2; middle: heterozygosity was identified by comparing E86-69 H2 and E4-63; bottom: heterozygosity was identified by comparing E86-69 H1 and E4-63. The recombination map and events (solid triangles) were inferred based on the inheritance pattern of heterozygous variants between the parental (E86-69 H1 and H2) and their inbred offspring haplotypes (E4-63). b, Analogous recombination map of the parental line PG6359 and its inbred offspring A6-26.
Extended Data Fig. 9 Statistical significance of dSNP enrichment in the dSV coupling phase.
a, Diagram depicting our dSNP accumulation assessment. For calculation purposes, chromosomes were segmented into fragments based on the midpoint between adjacent dSVs (irrespective whether in coupling or repulsion phase). Statistical tests were performed on each 10-kb non-overlapping window within each fragment. b, Statistical significance for the number of dSNPs flanking dSVs using the 10-kb non-overlapping windows between dSV coupling and repulsion phases. The significance threshold of P = 0.05 (two-sided Student’s t-test) is indicated. c, Diagram illustrating the calculation of the distance of a focal dSV to the nearest dSV in coupling phase (Dis-coupling) and to the nearest dSV in repulsion phase (Dis-repulsion). The focal dSV is highlighted in red. d, The frequency distribution of Dis-coupling and Dis-repulsion in potato genomes.
Extended Data Fig. 10 The schematic diagram of IPHs-v1.0 for inbred lines.
a, The number of dSVs in the GON/STN group for inferring the ideal A inbred line. b, Similarly, the number of dSVs in the PHU group for designing the ideal E inbred line. c, Validating the power of recombination (simulated single recombination event) and haplotype selection in eliminating dSVs. We simulated selfing experiments and the distribution of dSVs in offspring. Two accessions (the optimal and worst-case scenarios) from heterotic group A, PG6359 and PG6029, are displayed. d, Similarly, two accessions (the optimal and worst-case scenarios) from heterotic group E, PG6245 and RH89-039-16. The simulated progenies show that recombination could dramatically reduce the number of dSVs at the haplotype level. e, Schematic diagram indicating the generation of ideal potato haplotypes (IPHs-v1.0) based on two heterotic groups including inbred lines E4-63 and A6-26. This approach prioritizes the minimization of the dSV count as its central objective. Recombination and haplotype selection could reduce the number of dSVs during potato recurrent selection.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cheng, L., Wang, N., Bao, Z. et al. Leveraging a phased pangenome for haplotype design of hybrid potato. Nature 640, 408–417 (2025). https://doi.org/10.1038/s41586-024-08476-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-024-08476-9
This article is cited by
-
Mumemto: efficient maximal matching across pangenomes
Genome Biology (2025)
-
Genus-wide plant pangenome could inform next-generation crop design
Nature (2025)
-
Impacts of reproductive systems on grapevine genome and breeding
Nature Communications (2025)