Abstract
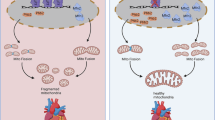
Mitochondrial dynamics and mitophagy are intimately linked physiological processes that are essential for cardiac homeostasis. Here we show that cardiac Krüppel-like factor 9 (Klf9) is dysregulated in human and rodent cardiomyopathy. Both global and cardiac-specific Klf9-deficient mice displayed hypertrophic cardiomyopathy. Klf9 knockout led to mitochondrial disarray and fragmentation, impairing mitochondrial respiratory function in cardiomyocytes. Furthermore, cardiac Klf9 deficiency inhibited mitophagy, thereby causing accumulation of dysfunctional mitochondria and acceleration of heart failure in response to angiotensin II treatment. In contrast, cardiac-specific Klf9 transgene improved cardiac systolic function. Mechanistically, Klf9 knockout decreased the expression of PGC-1α and its target genes involved in mitochondrial energy metabolism. Moreover, Klf9 controlled the expression of Mfn2, thereby regulating mitochondrial dynamics and mitophagy. Finally, adeno-associated virus–mediated Mfn2 rescue in Klf9-CKO hearts improved cardiac mitochondrial and systolic function. Thus, Klf9 integrates cardiac energy metabolism, mitochondrial dynamics and mitophagy. Modulating Klf9 activity may have therapeutic potential in the treatment of heart failure.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 digital issues and online access to articles
118,99 € per year
only 9,92 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
All data are available in the main paper, extended data and supplementary data. RNA-seq data are deposited in the Gene Expression Omnibus under accession number GSE278326.
References
Huss, J. M. & Kelly, D. P. Nuclear receptor signaling and cardiac energetics. Circ. Res. 95, 568–578 (2004).
Ni, H. M., Williams, J. A. & Ding, W. X. Mitochondrial dynamics and mitochondrial quality control. Redox Biol. 4, 6–13 (2015).
Youle, R. J. & van der Bliek, A. M. Mitochondrial fission, fusion, and stress. Science 337, 1062–1065 (2012).
Song, M., Mihara, K., Chen, Y., Scorrano, L. & Dorn, G. W. II. Mitochondrial fission and fusion factors reciprocally orchestrate mitophagic culling in mouse hearts and cultured fibroblasts. Cell Metab. 21, 273–286 (2015).
Chen, M. H., Colan, S. D. & Diller, L. Cardiovascular disease: cause of morbidity and mortality in adult survivors of childhood cancers. Circ. Res. 108, 619–628 (2011).
Papanicolaou, K. N. et al. Mitofusins 1 and 2 are essential for postnatal metabolic remodeling in heart. Circ. Res. 111, 1012–1026 (2012).
Song, M. & Dorn, G. W. II. Mitoconfusion: noncanonical functioning of dynamism factors in static mitochondria of the heart. Cell Metab. 21, 195–205 (2015).
Billia, F. et al. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc. Natl Acad. Sci. USA 108, 9572–9577 (2011).
Finck, B. N. & Kelly, D. P. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 116, 615–622 (2006).
Arany, Z. et al. Transcriptional coactivator PGC-1α controls the energy state and contractile function of cardiac muscle. Cell Metab. 1, 259–271 (2005).
Leone, T. C. et al. PGC-1α deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 3, e101 (2005).
Avci, H. X. et al. Thyroid hormone triggers the developmental loss of axonal regenerative capacity via thyroid hormone receptor α1 and Krüppel-like factor 9 in Purkinje cells. Proc. Natl Acad. Sci. USA 109, 14206–14211 (2012).
Morita, M. et al. Functional analysis of basic transcription element binding protein by gene targeting technology. Mol. Cell. Biol. 23, 2489–2500 (2003).
Simmen, R. C. et al. Subfertility, uterine hypoplasia, and partial progesterone resistance in mice lacking the Krüppel-like factor 9/basic transcription element-binding protein-1 (Bteb1) gene. J. Biol. Chem. 279, 29286–29294 (2004).
Cayrou, C., Denver, R. J. & Puymirat, J. Suppression of the basic transcription element-binding protein in brain neuronal cultures inhibits thyroid hormone-induced neurite branching. Endocrinology 143, 2242–2249 (2002).
Denver, R. J. et al. Basic transcription element-binding protein (BTEB) is a thyroid hormone-regulated gene in the developing central nervous system. Evidence for a role in neurite outgrowth. J. Biol. Chem. 274, 23128–23134 (1999).
Cui, A. et al. Dexamethasone-induced Krüppel-like factor 9 expression promotes hepatic gluconeogenesis and hyperglycemia. J. Clin. Invest. 129, 2266–2278 (2019).
Fan, H. et al. Cold-inducible Klf9 regulates thermogenesis of brown and beige fat. Diabetes 69, 2603–2618 (2020).
Imataka, H. et al. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 11, 3663–3671 (1992).
Nomura, S. et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat. Commun. 9, 4435 (2018).
Rysa, J., Leskinen, H., Ilves, M. & Ruskoaho, H. Distinct upregulation of extracellular matrix genes in transition from hypertrophy to hypertensive heart failure. Hypertension 45, 927–933 (2005).
Yang, G., Chen, S., Ma, A., Lu, J. & Wang, T. Identification of the difference in the pathogenesis in heart failure arising from different etiologies using a microarray dataset. Clinics (Sao Paulo) 72, 600–608 (2017).
Lehman, J. J. et al. Peroxisome proliferator-activated receptor γ coactivator-1 promotes cardiac mitochondrial biogenesis. J. Clin. Invest. 106, 847–856 (2000).
Wu, N. N. et al. Physical exercise and selective autophagy: benefit and risk on cardiovascular health. Cells 8, 1436 (2019).
He, C. & Klionsky, D. J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93 (2009).
Fan, G., Chen, M. J. & Wei, J. Involvement of phosphatase and tensin homolog-induced putative kinase 1/Parkin-mediated autophagy in angiotensin II-induced cardiac hypertrophy in C57BL/6 mice. J. Int. Med. Res. 48, 300060519896143 (2020).
Xiong, W. et al. PTEN induced putative kinase 1 (PINK1) alleviates angiotensin II-induced cardiac injury by ameliorating mitochondrial dysfunction. Int. J. Cardiol. 266, 198–205 (2018).
Liu, S. et al. Transcription factor Klf9 controls bile acid reabsorption and enterohepatic circulation in mice via promoting intestinal Asbt expression. Acta Pharmacol. Sin. 43, 2362–2372 (2022).
Chen, Y. & Dorn, G. W. II. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340, 471–475 (2013).
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008).
Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. & Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042–1052 (2011).
Liao, X. et al. Kruppel-like factor 4 is critical for transcriptional control of cardiac mitochondrial homeostasis. J. Clin. Invest. 125, 3461–3476 (2015).
Fisch, S. et al. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proc. Natl Acad. Sci. USA 104, 7074–7079 (2007).
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFA0800601 and 2022YFA0806102); the National Natural Science Foundation of China (82230027, 81825004, 81730024 and 82200975); the Scientific and Technological Research Project of Xinjiang Production and Construction Corps (2021AB028); and the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-050).
Author information
Authors and Affiliations
Contributions
L. Zhang and M. Zhang participated in the animal studies using Klf9-KO, Klf9-CKO and Klf9-TG mice and in vitro studies using NRCMs and MEFs and contributed to data analysis. Jinlong Huang and Jincan Huang prepared adenoviruses encoding shRNA specific to Mfn1 or Mfn2. H.F., Yujie Zhang, Yinliang Zhang, C.W. and X.X. performed in vitro experiments. H.C. contributed to discussion and editing of the manuscript. D.J. provided the clinical heart samples of patients with HCM and healthy donors. Y.C., J.Z. and J.T. designed the research and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
All authors declare no competing interests. There are no conflicts of interest in the submission of this manuscript, which was approved by all authors for publication. We also confirm that this work is original. It has not been published elsewhere and is not currently under consideration for publication elsewhere.
Peer review
Peer review information
Nature Cardiovascular Research thanks Roberta Gottlieb and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Loss of Klf9 function predisposes mice to hypertrophic cardiomyopathy.
a, Left, Western blotting analysis of Klf9 in WT and Klf9 KO hearts. Right, Quantification of Klf9 protein levels (n = 3). b, Left, B- and M-mode echocardiographic analysis of WT and Klf9 KO mice. Right, Quantitative group data showing the changes of FS, EF and LV mass (n = 6). c, Ratios of hearts or lungs weight to tibia length of WT and Klf9 KO mice (n = 6). d, H&E staining of WT and Klf9 KO hearts. e, Left, WGA staining of WT and Klf9 KO hearts. Right, Quantitative data of cardiomyocyte cross-sectional area (CSA) (n = 6). f, qRT-PCR analysis of mRNA levels of ANF, BNP, and β-MHC in WT and Klf9 KO hearts (n = 6). g, Left, Masson’s trichrome staining of WT and Klf9 KO hearts. Right, Quantification of blue-staining replacement fibrosis (n = 6). h, Left, TUNEL staining of WT and Klf9 KO hearts. Right, Quantitative data for apoptosis positive cells (n = 6). i, Heart rates, systolic and diastolic blood pressures of WT and Klf9 KO the mice (n = 6). Data are shown as mean ± SEM; 2-tailed Student’s t test (a, b, c, e, f, g, h, i).
Extended Data Fig. 2 Loss of Klf9 function impairs the cardiac mitochondrial function.
a, Volcano plot indicating the genes that were differentially expressed in WT versus Klf9 KO hearts. In red are the genes significantly up-regulated and in blue are the genes significantly down-regulated. b, KEGG pathway enrichment analysis of downregulated genes in Klf9 KO hearts. Orange dots represent the number of genes of each annotated pathway and the length of each column is based on the p values. c, Volcano plots showing the GSEA of GO cellular component categories of differentially expressed genes (DEGs). Gene sets related to mitochondria are marked. Sizes of the dots represent the number of genes in the gene sets and the transparency represent the FDR of the enrichment. d, Top hits of GSEA analysis from c showing the Klf9 positive correlation with the mitochondria. NES, normalized enrichment score; FDR, false discovery rate. e, GSEA analysis of disease category of DEGs. f-i, Heat-map, qRT-PCR (n = 6) and Western blotting analysis (n = 3) of the expression of genes involved in FA oxidation, oxidative phosphorylation, mitochondrial biogenesis, mitochondrial dynamics, autophagy and mitophagy. j, ChIP assay of heart extracts showing the enrichment of Klf9 proteins at the promoter region of PGC-1α (n = 4). k, TEM analysis of hearts from the indicated mice. l, Quantitative data of mitochondrial size distribution and content based on TEM images (n = 6). m, n, Left, Flow cytometric analyses of size (forward scatter, FSC) (m) and transmembrane electrical potential (DiOC6 intensity) (n) of isolated cardiac mitochondria from the indicated hearts. Right, Quantitative data of FSC and DiOC6 intensity (n = 6). o, Left, Oxygen consumption rates (OCR) showing the respiration of isolated mitochondria from the indicated hearts. Right, Quantification of basal and FCCP induced maximal respiration (n = 9). p, Myocardial ATP levels in the indicated hearts (n = 12). q, Left, Mitochondrial ROS production of isolated mitochondria from the indicated hearts measured with MitoSOX. Right, Quantitative data of ROS intensity (n = 6). Data are shown as mean ± SEM; 2-tailed Student’s t test (g, h, i, l, m, n, p, q), 2-way ANOVA (j, o).
Extended Data Fig. 3 PGC-1α rescue increases mitochondrial energy metabolism and improves the cardiac function in Klf9-CKO mice at baseline.
a, Schematic illustration of the experimental design to study the effects of PGC-1α rescue on cardiac and mitochondrial function of Klf9-CKO hearts. b, Left, Western blotting analysis of Klf9 and PGC-1α protein levels in the hearts from Ctrl and AAV-GFP or AAV-PGC-1α injected Klf9-CKO mice. Right, Quantification of Klf9 and PGC-1α protein levels (n = 3). c, B- and M-mode echocardiographic analysis of Ctrl and AAV-GFP or AAV-PGC-1α injected Klf9-CKO mice. d, Quantitative group data showing the changes of FS, EF and LV mass (n = 6) of Ctrl and AAV-GFP or AAV-PGC-1α injected Klf9-CKO mice. e, Ratios of hearts or lungs weight to tibia length of Ctrl and AAV-GFP or AAV-PGC-1α injected Klf9-CKO mice (n = 6). f, Left, WGA staining of hearts from above mice. Right, Quantitative data for cardiomyocyte CSA (n = 6). g, Left, Masson’s trichrome staining of hearts from above mice. Right, Quantification of blue-staining replacement fibrosis (n = 6). h, H&E staining of hearts from the hearts from above mice. i, qRT-PCR analysis of genes involved in mitochondrial metabolism in hearts from above mice (n = 6). j, TEM analysis of hearts from above mice (The red arrows indicate the fragment of mitochondria). k, Quantitative data of mitochondrial size distribution (Left) and content (Right) based on TEM images (n = 6). l, Left, Flow cytometric analyses of transmembrane electrical potential (DiOC6) of isolated cardiac mitochondria from above hearts. Right, Quantitative data of DiOC6 intensity (n = 6). m, Left, OCR showing the respiration of isolated mitochondria treated with oligomycin (Oligo), FCCP and rotenone/antimycin A (Rot-AA). Right, Quantification of basal and FCCP induced maximal respiration (n = 9). n, Quantification of myocardial ATP levels of above mice hearts (n = 6). o, Left, Mitochondrial ROS production of isolated mitochondria from above hearts measured with MitoSOX. Right, Quantitative data of ROS levels (n = 6). Data are shown as mean ± SEM; 2-way ANOVA (b, d, e, f, g, i, k, l, m, n, o).
Extended Data Fig. 4 Klf9 deficiency accelerates heart failure and mitochondrial senescence with aging.
a, Representative gross morphology of 24-month-old Ctrl and Klf9-CKO mice. b, Left, B- and M-mode echocardiographic analysis of 12-month-old Ctrl and Klf9-CKO mice. Right, Quantitative data showing the changes of FS, EF and LV mass (n = 6). c, Ratios of hearts or lungs weight to tibia length of 12-month-old Ctrl and Klf9-CKO mice (n = 6). d, Left, WGA staining of 12-month-old Ctrl and Klf9-CKO hearts. Right, Quantitative data for cardiomyocyte CSA (n = 6). e, Left, Masson’s trichrome staining of 12-month-old Ctrl and Klf9-CKO hearts. Right, Quantification of blue-staining replacement fibrosis (n = 6). f, Left, TUNEL staining of 12-month-old Ctrl and Klf9-CKO hearts. Right, Quantitative data for apoptosis positive cells (n = 6). g, TEM of 12-month-old Ctrl and Klf9-CKO hearts. h, i, Quantitative data of mitochondrial size distribution (h) and content (i, n = 6) based on TEM images. j, Quantification of mitochondrial protein content of 12-month-old Ctrl and Klf9-CKO hearts (n = 20). k-m, Left, Flow cytometric analysis of size (FSC) (k), transmembrane electrical potential (DiOC6) (l) and ROS production (MitoSOX) (m) of isolated cardiac mitochondria from 12-month-old Ctrl and Klf9-CKO hearts. Right, Quantitative data of FSC, DiOC6 and MitoSOX intensity (n = 6). n, qRT-PCR analysis of expression levels of genes involved in mitochondrial biogenesis in 12-month-old Ctrl and Klf9-CKO hearts (n = 6). o, Left, Western blotting analysis of proteins involved in the mitochondrial unfolded protein response (mtUPR) in 12-month-old Ctrl and Klf9-CKO hearts. Right, Quantification of AFG3L2, LONP1 and HSP60 protein levels (n = 3). Data are shown as mean ± SEM; 2-tailed Student’s t test (b, c, d, e, f, i, j, k, l, m, n, o).
Extended Data Fig. 5 Klf9 is required for cardiac functional adaptation to hemodynamic pressure overload.
a, Ratios of hearts or lungs weight to tibia length (n = 6). b, qRT-PCR analysis of mRNA levels of ANF, BNP and β-MHC in hearts from Ctrl and Klf9-CKO mice treated with saline or ANGII (n = 6). c, Left, WGA staining of hearts from Ctrl and Klf9-CKO mice treated with saline or ANGII. Right, Quantitative data for cardiomyocyte CSA (n = 6). d, Left, Masson’s trichrome staining of hearts from Ctrl and Klf9-CKO mice treated with saline or ANGII. Right, Quantification of blue-staining replacement fibrosis (n = 6). e, Left, TUNEL staining of hearts from Ctrl and Klf9-CKO mice treated with saline or ANGII. Right, Quantitative data for apoptosis positive cells (n = 6). f, qRT-PCR analysis of mRNA levels of genes involved in mitochondrial energy metabolism in hearts from above mice (n = 6). Data are shown as mean ± SEM; 2-way ANOVA (a, b, c, d, e).
Extended Data Fig. 6 Cardiac-specific Klf9 transgene prevents cardiac hypertrophy, fibrosis and cardiomyocytes apoptosis in ANGII-treated mice.
a, Ratios of hearts or lungs weight to tibia length (n = 6). b, qRT-PCR analysis of mRNA levels of ANF, BNP and β-MHC in hearts from Ctrl and Klf9-TG mice treated with saline or ANGII (n = 6). c, Left, WGA staining of hearts from Ctrl and Klf9-TG mice treated with saline or ANGII. Right, Quantitative data for cardiomyocyte CSA (n = 6). d, Left, Masson’s trichrome staining of hearts from Ctrl and Klf9-TG mice treated with saline or ANGII. Right, Quantification of blue-staining replacement fibrosis (n = 6). e, Left, TUNEL staining of hearts from Ctrl and Klf9-TG mice treated with saline or ANGII. Right, Quantitative data for apoptosis positive cells (n = 6). Data are shown as mean ± SEM; 2-way ANOVA (a, b, c, d, e).
Extended Data Fig. 7 Klf9 regulates ANGII induced hypertrophy of NRCMs.
a, Confocal imaging of NRCMs infected with Ad-shCtrl or Ad-shKlf9 for 24 hours, followed by treatment with ANGII (1 μM) or PBS for 48 hours. α-Actinin staining was performed to identify cells. b, Quantification of cell size of 30 NRCMs in each group (n = 22). c, Quantification of myocardial ATP levels in NRCMs as described in B (n = 12, n value refers to independent experiments). d, qRT-PCR analysis of mRNA levels of hypertrophic genes in NRCMs in A (n = 6, n value refers to independent experiments). e, Confocal imaging of NRCMs infected with Ad-GFP or Ad-Klf9 for 24 hours, followed by treatment with ANGII (1 μM) or PBS for 48 hours. α-Actinin staining was performed to identify cells. f, Quantification of cell size of 30 NRCMs in each group (n = 22). g, Quantification of myocardial ATP levels of NRCMs infected with Ad-GFP or Ad-Klf9, followed by treatment with ANGII or PBS (n = 12, n value refers to independent experiments). h, qRT-PCR analysis of mRNA levels of hypertrophic genes in NRCMs in E (n = 6, n value refers to independent experiments). Data are shown as mean ± SEM; 2-way ANOVA (b, c, d, f, g, h).
Extended Data Fig. 8 Mfn2 rescue improves the cardiac and mitochondrial function in Klf9-CKO mice at baseline.
a, Schematic illustration of the experimental design to study the effects of Mfn2 rescue on cardiac and mitochondrial function of Klf9-CKO hearts. b, Left, Western blotting analysis of Klf9 and Mfn2 protein levels in hearts from Ctrl and Klf9-CKO mice infected with AAV-GFP or AAV-Mfn2. Right, Quantification of protein levels of Klf9 and Mfn2 (n = 3). c, B- and M-mode echocardiographic analysis of Ctrl and Klf9-CKO mice infected with AAV-GFP or AAV-Mfn2. d, Quantitative group data showing the changes of FS, EF and LV mass of Ctrl and Klf9-CKO mice infected with AAV-GFP or AAV-Mfn2 (n = 6). e, Ratios of hearts or lungs weight to tibia length of Ctrl and Klf9-CKO mice infected with AAV-GFP or AAV-Mfn2 (n = 6). f, H&E staining of hearts from above mice. g, Left, WGA staining of hearts from above mice. Right, Quantitative data for cardiomyocyte CSA (n = 6). h, Left, Masson’s trichrome staining of hearts from above mice. Right, Quantification of blue-staining replacement fibrosis (n = 6). i, Respective TEM of hearts above mice. j, Quantitative data for mitochondrial size distribution and content (n = 6) based on TEM images. k, Left, Flow cytometric analyses of transmembrane electrical potential (DiOC6). Right, Quantitative data of DiOC6 intensity (n = 6, n value refers to independent experiments). l, Left, OCR showing the respiration of isolated cardiac mitochondria from above mice. Right, Quantification of basal and FCCP induced maximal respiration (n = 9). m, Quantification of myocardial ATP levels of hearts from above mice (n = 6, n value refers to independent experiments). n, Left, ROS production (MitoSOX) of cardiac mitochondria isolated from above mice. Right, Quantitative data of MitoSOX intensity (n = 6, n value refers to independent experiments). Data are shown as mean ± SEM; 2-way ANOVA (b, d, e, g, h, j, k, l, m, n).
Extended Data Fig. 9 Mfn2 rescue prevents the cardiac hypertrophy, fibrosis and cardiomyocytes apoptosis in ANGII-treated Klf9-CKO mice.
a, Left, Western blotting analysis of Klf9 and Mfn2 in hearts from Ctrl and Klf9-CKO mice infected with AAV-GFP or AAV-Mfn2 and treated with ANGII. Right, Quantification of protein levels of Klf9 and Mfn2 (n = 3). b, Left, WGA staining of hearts from Ctrl and Klf9-CKO mice infected with AAV-GFP or AAV-Mfn2 and treated with ANGII. Right, Quantitative data for cardiomyocyte CSA (n = 6). c, Ratios of hearts or lungs weight to tibia length (n = 6). d, qRT-PCR analysis of mRNA levels of ANF, BNP and β-MHC in hearts from above mice (n = 6). e, Left, Masson’s trichrome staining of hearts from above mice. Right, Quantification of blue-staining replacement fibrosis (n = 6). f, Left, TUNEL staining of hearts from above mice. Right, Quantitative data for apoptosis positive cells (n = 6). Data are shown as mean ± SEM; 2-way ANOVA (a, b, c, d, e, f).
Extended Data Fig. 10 Proposed model of Klf9 regulating mitochondrial energy metabolism, mitochondrial dynamics and mitophagy in cardiomyocytes.
Cardiac Klf9 is dysregulated in hypertrophic hearts, and Klf9 serves as a critical transcriptional regulator of mitochondrial homeostasis in the heart. Klf9 controls mitochondrial energy metabolism via induction of PGC-1α, while it regulates mitochondrial dynamics and mitophagy through Mfns and Mfn2/Parkin, respectively.
Supplementary information
Supplementary information
Supplementary Tables 1–4, Supplementary Figs. 1–7 and full unedited gel of western blotting.
Supplementary Tables 1–4.
Supplementary Table 1. Clinical characteristics of patients with HCM in Excel format. Supplementary Table 2. qPCR primer sequences in Excel format. Supplementary Table 3. ChIP–qPCR primer sequences in Excel format. Supplementary Table 4. Plasmid construction primer sequences in Excel format.
Source data
Source data
Statistical Source Data for Figs. 1–8 and Extended Data Figs. 1–9.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Zhang, M., Huang, J. et al. Klf9 is essential for cardiac mitochondrial homeostasis. Nat Cardiovasc Res 3, 1318–1336 (2024). https://doi.org/10.1038/s44161-024-00561-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s44161-024-00561-6
This article is cited by
-
Bavachin ameliorates cisplatin-induced nephrotoxicity by enhancing mitochondrial β-oxidation and lipid metabolism through MFN2
Molecular Medicine (2025)
-
KLF9-Mediated Transcriptional Promotion of HMGB2 Accelerates Cardiomyocyte Apoptosis, Inflammation, and Ferroptosis in Myocardial Ischemia/Reperfusion Injury
Cardiovascular Toxicology (2025)