Abstract
Aneuploidy is a hallmark of genomic instability that leads to tumor initiation, progression, and metastasis. CDC20, Bub1, and Bub3 form the mitosis checkpoint complex (MCC) that binds the anaphase-promoting complex or cyclosome (APC/C), a crucial factor of the spindle assembly checkpoint (SAC), to ensure the bi-directional attachment and proper segregation of all sister chromosomes. However, just how MCC is regulated to ensure normal mitosis during cellular division remains unclear. In the present study, we demonstrated that LNC CRYBG3, an ionizing radiation-inducible long noncoding RNA, directly binds with Bub3 and interrupts its interaction with CDC20 to result in aneuploidy. The 261–317 (S3) residual of the LNC CRYBG3 sequence is critical for its interaction with Bub3 protein. Overexpression of LNC CRYBG3 leads to aneuploidy and promotes tumorigenesis and metastasis of lung cancer cells, implying that LNC CRYBG3 is a novel oncogene. These findings provide a novel mechanistic basis for the pathogenesis of NSCLC after exposure to ionizing radiation as well as a potential target for the diagnosis, treatment, and prognosis of an often fatal disease.
Similar content being viewed by others
Introduction
Non-small cell lung cancer (NSCLC) is the most common type of human lung cancers as well as one that has the highest mortality and morbidity rate in both China [1] and the rest of the world [2]. NSCLC is usually diagnosed at an advanced stage accompanied by malignant proliferation and poor prognosis in most patients [3]. Several protein-coding genes, such as p53 and PRDM5 have been identified either as oncogenes [4] or tumor suppressors [5] that contribute to the stepwise development of NSCLC. However, protein-coding sequences occupy less than 2% of the human genome [6]. Long noncoding RNAs (lncRNAs) are operationally defined as RNA transcripts that are >200 nt in length with limited protein-coding potential [7], which have been shown to play an important role in tumorigenesis, including NSCLC [8]. There is evidence that many lncRNAs are involved in cancer development by modulating protein translocation, chromatin structures as well as transcriptional and posttranscriptional processing [9]. For example, a gain in H3K4me3 and H3K27 acetylation has been shown to activate the expression of HOXC-AS3, which regulates cell proliferation and migration both in vitro and in vivo by interacting with YBX1 and eventually leads to tumorigenesis [10]. However, the biological functions of lncRNAs in the initiation and progression of NSCLC are not well characterized.
Aneuploidy, a common form of genetic instability, predisposes cells to undergo malignant transformation and tumor development [11]. Even though it is generally believed that aneuploidy frequently results from errors of the mitotic checkpoint [12], the molecular mechanisms underlying the induction of aneuploidy by carcinogens, such as ionizing radiation are not yet fully understood. The anaphase-promoting complex or cyclosome (APC/C) is a crucial factor for the spindle assembly checkpoint (SAC) that ensures genomic stability at the end of mitosis. This is accomplished by making sure that all sister chromatids are bi-directionally attached to the mitotic spindle before activating the E3 ubiquitin ligase to initiate chromosome separation [13, 14]. While components of the SAC are responsible for detecting unattached kinetochores and for sending regulatory signals to the APC/C [15, 16], the molecular basis of these processes is not fully understood [17]. CDC20, Mad2, Mad3, Bub3, and BubR1 form the mitosis checkpoint complex (MCC) that directly binds the APC/C to inhibit substrate recognition [13, 18]. Mad2 and Mad3 cooperate to inhabit CDC20-dependent activation of the APC/C [19]. The BUB gene family encodes proteins that are part of a large multiprotein kinetochore complex and is considered to be a key component of the checkpoint regulation pathway. Failure of the monitoring system leads to genomic instability and increases the incidence of aneuploidy in gastric cancer [20]. Overexpression of CDC20 protein predicts poor prognosis in primary NSCLC cancer patients [21]. Furthermore, CDC20 may function as an oncoprotein to promote the development and progression of human cancers [22]. Therefore, all the components of SAC are of great significance for the correct chromosome segregation and genomic maintenance. The deletion and dysfunction of any part of MCC may cause aneuploidy and genetic instability.
In our previous work, we have shown that a radiation-inducible lncRNA, LNC CRYBG3, inhibits the formation of constriction rings and completion of cell division by directly binding to the monomeric G-actin and blocks the assembly of the polymeric F-actin [23]. We have further demonstrated that LNC CRYBG3 is significantly upregulated in NSCLC tissues and enhances glycolytic pathway by interacting with lactate dehydrogenase A (LDHA) [24]. Based on these findings, we propose that LNC CRYBG3 is relevant to the neoplastic development of NSCLC. In this paper, we found that LNC CRYBG3 directly interacted with Bub3 and overexpression of LNC CRYBG3 inhibited the interaction between Bub3 and CDC20 to result in activation of the APC/C complex that promoted mitosis. This aberrant mitosis led to aneuploidy and promoted the development of NSCLC. A better understanding of the role of lncRNAs functioning in NSCLC initiation and progression will improve the diagnosis, treatment, and prognosis of NSCLC, and ultimately provides novel targets for the treatment of this fatal disease.
Results
LNC CRYBG3 promotes in situ lung adenocarcinoma development in mice
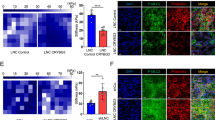
We have reported previously that LNC CRYBG3 induces genomic instability by inhibiting the formation of constriction rings [23] and enhances glycolytic pathway by interacting with LDHA [24], implying its potential in tumor initiation and promotion. The current evidence suggests that aneuploidy promotes tumorigenesis, at least at low frequency [12, 25]. As a first step, we tested the effects of LNC CRYBG3 on the incidence of NSCLC using the urethane-NSCLC mouse model. As shown in Fig. 1A, all nine mice treated with both urethane and LNC CRYBG3 died within 76 days after treatment, whereas all of the mice in the other three groups (control, urethane, and urethane + control RNA) survived at 100 days post treatment. On day 70, we conducted 18F-labeled glucose administration and PET-CT examination and showed that the absorption of 18F isotope in lung tissues was significantly higher in the urethane and LNC CRYBG3 treatment group than all the other three groups. The results implied either there were more tumor nodules or larger tumor volumes in this group of animals (Fig. 1B, C; Table S1). Next, we dissected the mice and found tumor nodules only in the lungs of the combined treatment group of urethane and LNC CRYBG3 (Table S2), which were confirmed pathologically as lung adenocarcinoma and carried the biomarkers CK7/ TTF-1, which is highly specific for lung adenocarcinoma (Fig. 1D). In addition, these tumors exhibited increased staining for the proliferation marker Ki-67 than any other groups of animals (Fig. 1D). In the group that overexpressed LNC CRYBG3 alone, there were high levels of 18F uptake in areas of the lung that corresponded to tumor-like nodules. However, pathological examination confirmed that these nodules were inflammatory nodules (Supplementary Fig. S1). These findings indicate that LNC CRYBG3 promotes the formation of primary lung tumors in mice induced by urethane. In addition, these data suggest that LNC CRYBG3 does not initiate but can promote lung adenocarcinoma formation.
A Mouse survival curves. The mice were divided into four groups: a group of control (Ctrl), a group with urethane injection (Urethane), a group injected with urethane and negative control RNA (Urethane + NC), and a group injected with urethane and LNC CRYBG3 (Urethane + LNC CRYBG3). 10 mice were observed in each group. B Quantified absorption of 18F-FDG in lung tissue. *p < 0.05; **p < 0.01. C Micro-PET/CT imagines. Mice were injected with 18F-FDG (100–150 μCi) via tail vein. One hour later, micro-PET/CT static scanning was performed for 10 min. Four mice for each group were examined. D Immunohistochemistry of lung tissues. HE hematoxylin and eosin staining. CK7 and TTF-1 were the biomarkers of lung adenocarcinoma. Ki67 was used as a biomarker of proliferation. 10 mice were observed in this group.
LNC CRYBG3 promotes NSCLC cells metastasis
Another consequence of increased genomic instability is the promotion of malignant characteristics in tumor cells. We next examined the metastasis of NSCLC both in vitro and in vivo after artificially modifying the expression level of LNC CRYBG3. We first established LNC CRYBG3 overexpression and knockdown lung adenocarcinoma A549 cell lines (sh1, sh2, sh3, and sh4). As shown in Fig. 2A, B, the expression level of LNC CRYBG3 in the overexpressed cells was significantly higher than in A549 cells that stably expressed control RNAs. Just as expected, its expression level was lower in A549 cells stably transfected with LNC CRYBG3 shRNAs compared with controls. A549 cells expressing hairpin sh1 and sh2 were selected for further studies. A549 cells with the knockdown of LNC CRYBG3 expression showed significantly reduced cell proliferation compared with control group and, by contrast, overexpression of LNC CRYBG3 promoted cell proliferation (Fig. 2E, F). Similarly, results from the colony formation assay revealed that LNC CRYBG3 had a significant impact on NSCLC cell proliferation (Fig. 2C, D). The ability of LNC CRYBG3 to modulate tumor invasiveness in A549 cells was subsequently evaluated using the transwell assay (Fig. 2G). The three images in the top panel show invasion of A549 cells in control or in cells overexpressed with negative vector control or LNC CRYBG3. The images in the middle (sh1) and bottom panel (sh2) show A549 cells with silenced LNC CRYBG3 using short hairpin RNA technique followed by re-expression using either LNC NC or LNC CRYBG3. (Fig. 2G, left). Quantification of the transwell images showed that the overexpression of LNC CRYBG3 significantly promoted cell migration in vitro and shRNAs partially abrogated the effect (Fig. 2G). To validate the effects of LNC CRYBG3 on cell metastasis in vivo, A549 cells transfected with LNC CRYBG3 or control vector were injected into the tail veins of 10 nude mice. Four weeks post injection, metastatic nodules on the surface of the lungs were detected using IVIS Spectrum CT. As shown in Fig. 2H, after the overexpression of LNC CRYBG3, the metastatic incidence of A549 cells in the lungs was significantly increased. However, shRNA-LNC CRYBG3 did not decrease the metastatic incidence of A549 cells in the lung. These findings indicate that LNC CRYBG3 promotes tumor metastasis.
A, B Establishment of A549 cell lines stably expressing LNC CRYBG3 or shRNA against LNC CRYBG3. sh1, sh2, sh3, and sh4 were four small hairpin RNAs against LNC CRYBG3. C, D The proliferation of A549 overexpressing LNC CRYBG3. E, F Cell migration quantified by a cell scratch test. G Metastatic potential of A549 cells revealed by a transwell migration assay. Image J software was used to analyze the images. All experiments were conducted at least three times independently. *p < 0.05. H In vivo metastasis test. A549 cells overexpressing negative control RNA (NC) or LNC CRYBG3 were injected into the caudal vein of each mice and 30 days later, imagines were taken and representative images were shown. n = 5 for each group.
Overexpression of LNC CRYBG3 promotes tumorigenesis and metastasis by targeting BUB3
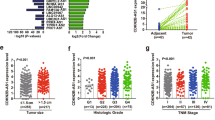
In order to reveal the mechanisms underlying the promotion of tumorigenesis and metastasis of LNC CRYBG3, we identified 1501 differentially expressed proteins in A549 cells that overexpressed LNC CRYBG3 compared with control cells using mass spectroscopy [fold change > 2; p < 0.05]. Among these 1501 proteins, 1075 were found to be significantly upregulated whereas 426 were downregulated. As shown in Fig. 3A, Kyoto Encyclopedia of Genes and Genomes analysis showed that several functional pathways were enriched and, among them, metabolic pathways were the most altered ones in A549 tumor cells that overexpressed LNC CRYBG3, which is consistent with what we observed previously [24]. The top 20 altered pathways were all highly correlated with cancer and cell cycle. The heat map showed that 14 proteins were clustered into negative regulation whereas 23 proteins were associated with positive regulation of the cell cycle (Fig. 3B). Bub3, CDC20, and cyclin B1 were significantly upregulated in LNC CRYBG3 overexpressed groups. Meanwhile, cyclin D1 showed downregulation in LNC CRYBG3 overexpressed cells, the results were further validated and confirmed using western blot (Fig. 3C). CDC20, BubR1, and Bub3 form the MCC that directly binds the APC/C to inhibit substrate recognition [13, 18]. Therefore, we investigated the correlation of gene expression and patient survival data using the Cancer Genome Atlas database and found that Bub3 expression level was highly correlated to NSCLC incidence (Fig. 3D). High expression level of Bub3 predicted a poor overall survival in 105 patients with NSCLC (p = 0.0014, Fig. 3E). We also found a positive correlation between Bub3 expression level with the Tumor, Node, Metastases (TNM) staging. The higher the Bub3 expression, the higher the TNM staging and vice versa (Fig. 3F). In addition, the expression of Bub3 was found to correlate with metastatic potential of NSCLC cells (Fig. 3G). The higher the expression of Bub3, the lower the survival and the higher the metastatic incidence. In contrast, the MCC protein, CDC20, BubR1, and Bub3 expression had no significant correlation with tumor prognosis (Supplementary Fig. S2A–D).
A Top 20 enrichment KEGG pathways of the protein expression profile obtained with global protein mass spectrum assay in LNC CRYBG3 overexpressing A549 cells lines. B The heat map of total differentially expressed cell cycle-related proteins in A549 cells overexpressing LNC CRYBG3. C Representative protein expressions that were upregulated in B were validated using western blot including Cyclin B1 and Cyclin D1. D Expression of Bub3 in primary tumors (n = 515) compared to normal tissues (n = 59) in TCGA database. E Overall survival curves of patients with NSCLC tumors of different Bub3 expression levels (n = 215). The expressions of Bub3 in NSCLC tissues were analyzed using the Cancer Genome Atlas data set. F Correlation between Bub3 expression level and individual NSCLC stages (n = 574). G Bub3 expression level visualized by immunohistology. **p < 0.01.
Overexpression of LNC CRYBG3 induces aneuploidy in immortalized lung epithelial cells
There is evidence that cellular immortalization by SV40 is not sufficient for the neoplastic transformation of BEAS-2B cells since non-carcinogen-treated BEAS-2B cells are non-malignant and do not form tumors when injected into athymic nude mice [26]. However, it is likely that p53 deficiency due to the presence of the large T-antigen may predispose damaged cells to aneuploidy [27, 28]. We synchronized BEAS-2B cells at M phase with colchicine followed by overexpression of LNC CRYBG3 and monitored the cell cycle distribution. As shown in Fig. 4B, the Cyclin B1 and phosphorylated histone H3 maintained at a high expression level in the negative RNA control (NC) group whereas Cyclin D1’s level remained low, indicating that cells in the NC group were blocked in M phase after colchicine treatment. In contrast, in the LNC CRYBG3 overexpression group, cells were able to cross the M phase and entered G0/G1 phase. LNC CRYBG3 promoted cell transition from colchicine-blocked M phase cells to G0/G1 phase. Securin is a protein which is involved in control of the metaphase–anaphase transition and anaphase onset. We found that when LNC CRYBG3 is overexpressed, Securin is then ubiquitinated by the APC to result in separase activation, loss of chromosome cohesion, and anaphase onset in spite of mitosis errors. It may can result in aneuploidy (Fig. 4C). Cell cycle assay showed, in LNC CRYBG3 group, a sub-G2/M phase peak or aneuploidy peak appeared behind the main G2/M phase peak (Fig. 4D). Using a live cell imaging system, we further confirmed the incomplete cytoplasmic division induced by LNC CRYBG3. As shown in Fig. 4E, cells transfected with LNC CRYBG3 completed karyomitosis step and then fused, started a new cell cycle with wrong number of chromosomes. This may be the main reason of how LNC CRYBG3 induced aneuploidy. We also found that overexpression of LNC CRYBG3 induced chromosomal aberration in BEAS-2B cells (Fig. 4F, G). Furthermore, overexpression of LNC CRYBG3 and knockdown of Bub3 using shRNA resulted in aneuploidy compared with the control group [both of which have a synergistic effect]. In order to confirm these findings, we examined the genomic instability of peripheral venous blood from 12 volunteers (22–24 years old, males) and exposed the whole blood samples ex vivo to 2.5 Gy of X-rays. Conventional chromosome assay showed that compared with the control group, X-ray induced chromosomal changes in peripheral blood lymphocytes and the proportion of aneuploidy significantly increased above controls (Fig. 4H–J). The qRT-PCR assay showed that the expression of LNC CRYBG3 and Bub3 in peripheral blood lymphocyte increased significantly after X-ray irradiation (p ≤ 0.05, Fig. 4K, L). Therefore, we hypothesize that LNC CRYBG3 might act as an oncogene and its overexpression promotes tumorigenesis by inducing aneuploidy.
A Demonstration of the role of BUB3 and other factors in spindle assembly checkpoint. B Effect of LNC CRYBG3 on cell cycle. Colcemid was used to synchronize BEAS-2B cells at M phase and then release the cells to various cell cycle phases. The protein makers of different phase of cell cycle, Cyclin D1 for G0/G1 phase, phosphorylated H3 (p-H3) for M phase, and Cyclin B1 G2/M phase, was detected by western blot analysis. C The relative protein level of Securin which was detected by western blot analysis. Colcemid were used to synchronize cell cycle of BEAS-2B cells at M phase in normal and LNC CRYBG3 knockdown cell lines. D The cell cycle of A549 cells transfected with negative control RNAs (NC) or LNC CRYBG3 was monitored after cells were released to various phases from synchronized M phase. The proportion of aneuploid cells was assayed by flow cytometry. All experiments were repeated at least three times independently. The quantitative data were presented as mean ± SEM. E A confocal time-lapse was used to monitor the cell mitosis of BEAS-2B cells overexpressing control RNA (NC) or LNC CRYBG3. F, G Chromosome stability were visualized and quantified by karyotype analysis after BEAS-2B cells were transfected with LNC CRYBG3 and/or Bub3 shRNA. H, I, J The proportion of aneuploidy in human peripheral blood lymphocyte after irradiation (2.5 Gy X-rays). K, L The expression of LNC CRYBG3 and Bub3 in human peripheral blood lymphocyte after irradiation (2.5 Gy X-rays), measured by qRT-PCR The animal protocol was approved by the Research Ethics Committee of Soochow Medical University, China and the written informed consent was obtained from all subjects. *p < 0.05; **p < 0.01; **p < 0.001.
Knocking down Bub3 together with the overexpression of LNC CRYBG3 significantly promotes tumor formation and metastasis of immortalized lung epithelial cells xenografted into SCID mice
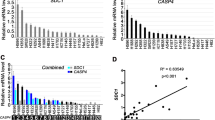
We examined the expression level of LNC CRYBG3 and Bub3 mRNA in the lung tumor tissue of 23 patients and four normal human lung tissues. As shown in Fig. 5A, B, the expression levels of both LNC CRYBG3 and Bub3 mRNA in tumor tissues were significantly higher compared with normal tissues (P < 0.01). The expression level of LNC CRYBG3 and Bub3 mRNA was positively correlated (Fig. 5C). To further confirm the involvement of Bub3 in tumor development mediated by LNC CRYBG3, we established an immortalized lung epithelial cell line HSAEC1-KT that stably expressed Bub3 shRNA (Fig. 5D) and then transfected with LNC CRYBG3 overexpressing adenovirus. These cells were then inoculated subcutaneously into NOD/SCID mice. Thirty days after injection, tumors measuring ~1 cm in diameter in size were detected and excised in animals carrying the shBub3 plus LNC CRYBG3 transcripts. Although tumor nodules were also found in animals injected with HSAEC1-KT cells that stably expressed either Bub3 shRNA or LNC CRYBG3 transcripts, their sizes were smaller (Fig. 5E, F). The established cell lines were injected into the tail veins of NOD/SCID mice. Four weeks post injection, metastatic nodules on the surface of the lungs and other places were detected using IVIS Spectrum CT. As is shown in Fig. 5H, all tumors from the LNC CRYBG3 overexpression group were highly metastatic and no significant metastasis was observed in the group with knockdown of Bub3 alone. At the same time, the LNC CRYBG3 overexpression group and knockdown of Bub3 were also highly metastatic. All these results indicated that regardless of the status of Bub3, all tumors from the LNC CRYBG3 overexpression group were highly metastatic. In addition, we also found that overexpression of LNC CRYBG3 increased the expression of Bub3 protein (Fig. 5G). These findings indicate that the overexpression of LNC CRYBG3 combined with Bub3 knockdown promotes the progression of NSCLC.
The absolute expression levels of LNC CRYBG3 (A) and Bub3 (B) were determined in 23 clinical lung cancer tissues and four normal lung tissues using quantitative RT-PCR. The data are expressed as the mean ± SEM. C The relationship between the expression of LNC CRYBG3 and Bub3. D Bub3 expression in an established A549 cell line expressing Bub3 shRNA (sh1 and sh2) was detected by western blotting. The experiment was independently repeated three times. E HSAEC1-KT cells stably expressing Bub3 shRNA with or without transfection of LNC CRYBG3 overexpressing lentivirus were inoculated into NOD/SCID mice (n = 5 for each group). Thirty days after the injection, the xenograft tumors were harvested in each group for hematoxylin and eosin staining and other analyses. F The weight of xenograft tumors from different groups. G The expression of Bub3 in xenograft tumors. h In vivo cell metastasis experiment (n = 5 for each group, but four mice of the shBub3 group died). *p < 0.05; **p < 0.01; ***p < 0.001.
LNC CRYBG3 interacts with Bub3 and inhibits the mitotic checkpoint complex
We performed RNA pulldown and RNA immunoprecipitation (RIP) assay and found that there was a direct interaction between Bub3 and LNC CRYBG3 (Fig. 6A, B). Immunoprecipitation (IP) with the antibody to Bub3 protein showed that the interaction between APC and CDC20 to Bub3 was inhibited by LNC CRYBG3 overexpression but not the interaction between APC and CDC20 (Fig. 6C). Based on the predicted secondary structure of LNC CRYBG3, we synthesized three truncated fragments of LNC CRYBG3 (Fig. 6D). These fragments were used to repeat the experiments and the nucleotide sequence encompassing 261–317 (S3) showed similar inhibitory effects on the interaction between Bub3 and CDC20 (Fig. 6D), implying that this segment was essential for the binding of LNC CRYBG3 with Bub3. We further demonstrated the suppressive effects of LNC CRYBG3 on the interaction of Bub3 with other MCC components. As shown in Fig. 6C, CDC20 interacted with APC and Bub3, but LNC CRYBG3 had little effect on the binding of CDC20 with APC. Furthermore, APC interacted with CDC20 and Bub3, however, LNC CRYBG3 showed no effect on the interaction of APC with either CDC20 or Bub3. Similarly, BubR1 interacted with APC, CDC20, and Bub2 but LNC CYBG3 had little effect on their interactions. Finally, MAD1L2 interacted with APC or Bub3 but CDC20, which was inhibited by LNC CRYBG3. Using immunofluorescent assay, we confirmed that Bub3 co-localized with CDC20 and this interaction decreased after LNC CRYBG3 overexpression (Fig. 6E, G). In contrast, the colocalization of Bub3 and CENP-A increased after LNC CRYBG3 overexpression (Fig. 6F, H). We also used the three truncated fragments of LNC CRYBG3 described above to further confirm the critical segment essential for the binding activities. Consistent with the data shown in Fig. 6D, we found that only the S3 segment (261–371 nucleotides) was inhibitory in the colocalization of Bub3 and CDC20 (Fig. 6J, K) as well as to increase the colocalization of Bub3 and CENP-A (Fig. 6I, L). These results imply that LNC CRYBG3 disturbs the SAC through interfering the interaction between Bub3 and CDC20. Therefore, once LNC CRYBG3 is overexpressed, Bub3 will be released from the mitotic checkpoint complex regardless of possible mitotic errors and consequently leads to aneuploidy.
A RNA pulldown analysis was used to determine the Bub3-LNC CRYBG3 interaction in A549 cells. B Histogram of LNC CRYBG3 enrichment after RNA immunoprecipitation (IP) assays. Bub3 antibodies were used. C Immunoprecipitation followed by western blotting was used to detect the interaction of LNC CRYBG3 with MCC components. All experiments were independently repeated at least three times. D Three truncated fragments of LNC CRYBG3 (S1, S2, and S3) were synthesized based on the predicted secondary structure of LNC CRYBG3 and then used for immunoprecipitation to detect the interaction of Bub3 and CDC20. The quantitative data were presented as mean ± SEM. *p < 0.05. E, F The colocalization of Bub3 and CDC20 after overexpression of LNC CRYBG3. G, H The colocalization of Bub3 and CENP-A after the overexpression of LNC CRYBG3. J, K The colocalization of Bub3 and CDC20 after overexpression of truncated body of LNC CRYBG3. I, L The colocalization of Bub3 and CENP-A after the overexpression of truncated body of LNC CRYBG3. Ctrl, A549 cells without transfection; NC, A549 cells transfected with negative control RNA; LNC CRYBG3, A549 cells transfected with LNC CRYBG3 overexpressing adenovirus. All experiments were repeated at least three times independently. The quantitative data were presented as mean ± SEM. *p < 0.05; **p < 0.01.
Discussion
In the present study, we examined the function of the lncRNA LNC CRYBG3 in promoting aneuploidy and tumorigenic phenotypes in NSCLC. We also elucidated the underlying mechanisms by showing that LNC CRYBG3 directly binds with Bub3 to negatively regulated the function of MCC in SAC. These findings demonstrate that LNC CRYBG3 functions as a novel oncogene and could be a potential target for the diagnosis, treatment, and prognosis of NSCLC.
There is a positive correlation between the expression of LNC CRYBG3 and Bub3. Both overexpress in clinical lung cancer tissues compared to normal tissues and both are induced by ionizing radiation, implying that both of them are key players in cellular response to ionizing radiation and tumorigenesis. However, how these two factors regulate each other’s expression remains unknown. The present study did not focus on their expression but on their interaction. RNA pulldown and the RIP assay revealed that LNC CRYBG3 interacted with the MCC components, especially Bub3. LNC CRYBG3 binds to Bub3 to interrupt its interaction with CDC20. We demonstrated the 261–317 residual of LNC CRYBG3 is critical for its interaction with Bub3. Even though there are few published studies on Bub3, it plays an essential role in mitosis as an important component of the SAC to promote the establishment of correct kinetochore-microtubule attachments during oocyte meiosis [29, 30]. At metaphase, when all kinetochores have attached and are under tension, the checkpoint is then turned off to release Bub3 and BubR1 from the complex; thus freeing CDC20 for APC/C activation. The activated APC/CCDC20 complex then targets and leads to the degradation of securin and cyclin B1, resulting in sister chromatid separation and mitotic exit, respectively [14].
As generally known that prior to anaphase, sister chromatids are kept together by a protein bridge formed by a multiprotein complex called cohesin [31]. Anaphase is triggered by the proteolysis of one of the cohesin subunits, Scc1, by a protease called separase [32]. Separase is normally kept inactive by an associated inhibitor termed securin [33]. After the proper attachment of all sister chromatids to the mitotic spindle, the spindle checkpoint is satisfied. Securin is then ubiquitinated by the APC [25], a multi-subunit E3 ubiquitin ligase [34]. The destruction of ubiquitinated securin by the proteasome then leads to separase activation, Scc1 cleavage, loss of chromosome cohesion, and anaphase onset [31, 32] (Fig. 7). Nevertheless, if the spindle checkpoint is abnormal, mitotic mistakes may not be effectively repaired resulting in segregation errors of sister chromatids, which eventually lead to aneuploidy [35]. In this paper, we found that LNC CRYBG3 interferes with SAC activities through inhibiting the interaction between Bub3 and CDC20. Once LNC CRYBG3 is overexpressed, Bub3 is released from MCC in spite of mitosis errors to result in aneuploidy.
Molecular mechanism of chromosome segregation. At the metaphase–anaphase transition, APC/CCDC20 ubiquitinates securin. Degradation of securin activates separase. Separase then cleaves the Scc1 subunit of cohesion, allowing chromosome segregation. When sister chromatids are not properly attached to the mitotic spindle, the spindle checkpoint promotes the assembly of checkpoint protein complexes that inhibit the activity of APC/C, leading to the stabilization of securin, preservation of sister chromatid cohesion, and a delay in the onset of anaphase. After overexpression of LNC CRYBG3, Bub3 will be released from the mitotic checkpoint complex (MCC) even in the presence of mitotic errors. Although cells can escape from the SAC, mitotic errors still exist leading to aneuploidy.
In 1902 and 1903, the German zoologist and cytologist Boveri proposed aneuploidy as a cause of tumorigenesis because of the prevalence of aneuploidy in cancer cells and its relatively low incidence in normal cells [36,37,38]. Boveri found that when sea urchin embryos were manipulated to undergo mitosis in the presence of multipolar spindles, they produced aneuploid progeny suggesting that tumors derived from normal cells through aberrant mitosis. In 1999, David Rasnick proposed that cancer is the phenotype of cells above a certain threshold of aneuploidy [39] by autocatalytic genetic instability of aneuploid cells, or by tetraploidization followed by gradual loss of chromosomes. Thus, the initiation step of carcinogenesis produces aneuploidy below the threshold for cancer, and the promotion step increases the level of aneuploidy above this threshold. Once reaching the threshold, it will lead to tumorigenesis.
Aneuploidy, including both numerical and structural chromosome abnormalities, is a common form of genome instability, which is a hallmark of cancer [38]. Our results indicate that LNC CRYBG3 can shorten the neoplastic transformation process in urethane-induced primary lung cancer in mice and to promote the metastasis of A549 cells due to its induction of aneuploidy. Taken together, our findings demonstrate that LNC CRYBG3 is an oncogene and might be used as a potential therapeutic target for the diagnosis, treatment, and prognosis of NSCLC. [40,41,42]
Materials and methods
Cell culture
All cell lines used in the current study were purchased from the ATCC (Washington, USA). Cell lines were routinely tested by using Mycoplasma-free Mycoplasma Detection Kit (Beyotime, China). All the cell lines were authenticated using short tandem repeat profiling and LDH, G6PD, MDH, PEP-B isozyme detection by the Cell Bank of Typical Culture Preservation Committee of Chinese Academy of Sciences. In this study, a panel of lung cancer cell line A549, immortalized lung epithelial cell line HSAEC1-KT, and immortalized human bronchial epithelial cell line BEAS-2B were used. A549 cells were maintained in RPMI-1640 medium (Sigma, St. Louis, MO, USA) HSAEC1-KT and BEAS-2B cells were grown in DMEM medium (Sigma, St. Louis, MO, USA). All the media were supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), 1% penicillin sodium, and 100 μg/mL streptomycin and incubated at 37 °C in 5% CO2 in a humidified incubator (Thermo Scientific, Asheville, NC, USA). Cells were cultured to 80% confluence in 100 mm diameter tissue culture dishes (Corning, NY, USA) before being subjected to other treatments. All cell lines were used within 10–20 passages of their original stock.
Tissue collection and ethics statement
A total of 23 paired human non-small lung cancer tissues were used in this paper and they were collected from the Department of Lung Cancer, Shanghai Jiao-tong University. Normal lung tissues were collected at the Judicial Expertise Center of Soochow University [43]. All human specimens were approved for research use by the Collaborative Center for Research of the Ethical Review Committee of the World Health Organization, which is authorized by the Shanghai Municipal Government. In addition, the current study was also approved by the Ethics Committee of Soochow University (License no. ECSU-2019000108).
Animal experiment
All the animal experiments were performed as described in “Results” sections. The method of urethane-induced lung tumors in mice refers to Gurley et al. article [44]. All mice were maintained in the SPF Animal Laboratory, Soochow University. And all animal experiments were carried out with the approval of the Soochow University Experimental Animal Care and Use Committee. (Approval no. ECSU-2019000107) Animals will be housed in solid bottom polycarbonate cages on ventilated animal racks (~4–5 mice/cage) under temperature, humidity, and light-controlled conditions. Food and water will be available ad libitum.
Immunoblotting and co-immunoprecipitation assays
The cell lysates were extracted from cultured cells with RIPA lysis buffer (or co-IP lysis buffer) containing protease inhibitors (Byotime, China), and cleared by centrifugation at 4 °C for 15 min at 12,000 × g. The protein samples were separated by 10 and 12% SDS-PAGE and transferred to nitrocellulose filter membranes (Millipore, USA). After blocking in phosphate-buffered saline (PBS)/Tween-20 containing 5% nonfat milk, the membranes were incubated with the primary antibodies (Antibodies used in this study included anti-actin (mouse monoclonal antibody, 1:1000; Sigma-Aldrich); and anti-Bub3 (rabbit monoclonal antibody, 1:1,000; Abcam, Cat. ab133699, USA); and anti-BUB1 (rabbit polyclonal antibody, 1:5,000; Abcam, Cat. Ab70372, USA); and anti-MAD2L1 (rabbit monoclonal antibody, 1:1,000; CST, Cat. #4636, USA); and anti-Cyclin D1(rabbit monoclonal antibody, 1:1,000; CST, Cat. #2978, USA); and anti-CDC20 (rabbit monoclonal antibody, 1:1,000; CST, Cat. #14866, USA)). Subsequent visualization was conducted using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo). In co-IP assay, 2,500 μg protein lysates were diluted in IP lysis buffer and incubated with 2 μg normal rabbit or mouse IgG for 2 h, followed by 2 h of incubation with 50 μl Protein A/G magnetic beads (Bio-tool, China) to precipitate proteins that interacted non-specifically with IgG and/or Protein A/G magnetic beads. This pre-cleared lysate was then incubated with Bub3, CDC20, MAD2L1, BUB1, APC antibody (1 μg) at room temperature overnight. Protein A/G magnetic beads were added and incubated at room temperature for 6 h, which were then washed with IP lysis buffer, and proteins in the immunocomplexes were then extracted in SDS sample buffer and used for immunoblotting to identify Bub3, CDC20, MAD2L1, BUB1, and APC proteins.
Immunofluorescence
All the immunofluorescence assays in this study were performed as previously described [45]. Cells cultured on slides were treated with the compounds and fixed with 4% paraformaldehyde for 10 min at room temperature, permeabilized for 20 min in ethanol at −20 °C, washed with PBS for 30 min, and treated with 0.5% Triton for 10 min. The samples were then blocked for 1 h with 5% skim milk, and stained with anti-Bub3 (rabbit monoclonal antibody, 1:1,000; Abcam, USA), anti-CDC20 (rabbit monoclonal antibody, 1:1,000; CST, USA), CENP-A (rabbit monoclonal antibody, 1:1,000; Abcam, USA) for 2 h. The bound antibody was visualized using Alexa Fluor® 488 anti-mouse antibody (Abcam, NY, USA) and cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Invitrogen, CA, USA). Slides were observed using a confocal laser scanning microscope (FV1200; Olympus, Tokyo, Japan) [46]. Data were analyzed using the Image-j software. Measurements were exported in Excel (Microsoft) and graphed with GraphPad Prism 6.0 (GraphPad Software).
Cell cycle assay
After treatment, cells were harvested and fixed with 70% pre-chilled ethanol for >24 h at −20 °C. Prior to analysis, the fixed samples were washed with PBS twice and treated with 100 μg/mL RNase A and 50 μg/mL propidium iodide (BD Biosciences, San Jose, CA, USA) for 30 min at room temperature. Cell cycle distribution was measured using the FACS Verse flow cytometry (Becton Dickinson) and analyzed using CellQuest (Becton Dickinson) [47].
Gene silencing, overexpression
LNC CRYBG3 shRNA lentivirus particles (Sangon Shanghai, China) were transfected into cells and subsequently screened with medium contains 2 mg/mL puromycin (Invitrogen) and double-checked with RT-PCR. Bub3 shRNA lentivirus particles (Sangon) were also transfected into cells and subsequently screened with medium contains 2 mg/mL puromycin (Invitrogen) and double-checked with western blot. We used LNC CRYBG3 adenovirus particles (Sangon) to overexpress LNC CRYBG3 in cells. Luciferase assay was performed as described previously [48]. A549 cells on a 12-well plate were cotransfected with 300 ng DNA followed by the Luciferase Reporter Assay System (Promega) with Multiscan Spectrum (BioTek Synergy 2) to measure the Luciferase activity after 48 h.
RNA pulldown analysis
Cells were harvested and resuspended in 1 mL ice-cold RIP buffer containing RNase and protease inhibitors. The cell pellets were sheared and centrifuged at 15,000 × g for 15 min at 4 °C to clear the cell lysate. Folded RNA was mixed with cell lysate and incubated at room temperature for 2 h. A total of 60 μl washed streptavidin agarose beads were added to each binding reaction and further incubated at RT for 1 h. Beads were boiled in Laemmli loading buffer and samples were run on SDS-PAGE and analyzed by western blot assay [49].
RNA immunoprecipitation (RIP) assays
RIP experiments were performed using a Magna RIP™ RNA-Binding Protein IP Kit (Cat. 17-701, Millipore, USA) according to the manufacturer’s instructions. The antibodies for RIP assays of Bub3 (Cat. ab133699) were from Abcam [50].
Colony formation assay
Cells were plated into six-well plates (5 × 102 cells/well) and cultured as described above. On designated time after plating (for day 1, 2, 3, and 4), cells were fixed with methanol for 10 min and stained with 0.5% crystal violet dye (in methanol: de-ionized water, 1:5) for 10 min. After three washes with de-ionized water to remove excess crystal violet dye, the crystal violet dye was released from the cells by incubation with 1% SDS for 2 h before optical density (OD) 570 nm measurement.
Hematoxylin and eosin staining
After the mouse tumor tissues were dissected out, they were rinsed in PBS and fixed with 4% formalin (V/V) and embedded in paraffin. Tissues were sectioned into six micron thin sections, deparaffinized with xylene and submerged into EDTA antigenic retrieval buffer for antigenic retrieval. After stainng with hematoxylin–eosin, the tissue sections were dehydrated through increasing concentrations of ethanol and xylene. Digital images of tumor tissues were acquired by Nanozoomer (Hamamatsu Photonics).
Mass spectrometry analysis
All LC-MS/MS analyses were performed on a Q-Exactive mass spectrometer (Thermo, USA) equipped with a Nanospray Flex source (Thermo, USA). The peptides mixtures were loaded by a capillary C18 trap column (3 cm × 100 µm, C18, 3 µm, 150 Å) and separated by a C18 column (15 cm × 75 µm, C18, 3 µm, 120 Å) on an ChromXP Eksigent system (AB Sciex). The flow rate was 300 nL/min and linear gradient was 70 min (0–0.5 min, 95–92% A; 0.5–48 min, 92–74% A; 48–61 min, 74–62% A; 61–61.1 min, 62–15% A; 61.1–67 min, 15% A; 67–67.1, 15–95% A; 67.1–70 min, 95% A. Mobile phase A = 2% ACN/0.1% FA and B = 95% ACN/0.1% FA). Full MS scans were acquired in the mass range of 300–1600 m/z with a mass resolution of 70000 and the AGC target value was set at 1e6. The ten most intense peaks in MS were fragmented with higher-energy collisional dissociation with collision energy of 30. MS/MS spectra were obtained with a resolution of 17500 with an AGC target of 200000 and a max injection time of 50 ms. The Q-E dynamic exclusion was set for 15.0 s and run under positive mode. The MS data were performed and analyzed with the help of the commercial biotechnology company Oebiotech (Shanghai, China).
Wound healing assay and transwell assays
The migration of A549 cells was tested by wound healing assay. Cells were plated into a six-well plate with FBS-free media for 12 h. Then, using a pipette tip to create a wound area in the bottom of the well. After 24 h, wounds were imaged under microscope (×40, CKX41F, Olympus, Tokyo, Japan). Data were analyzed using the Image-j software. Measurements were exported in Excel (Microsoft) and graphed with GraphPad Prism 6.0 (GraphPad Software). For the transwell assay, refer to Senger D’s article [51].
Statistical analysis
All experiments were independently repeated at least three times and all data were presented as mean ± SE. Student’s t tests were used for statistical analysis. Probability (p) values less than 0.05 were considered to be statistically significant.
Conclusion
In the present study, we identified the first lncRNA, LNC CRYBG3, that binds with Bub3 to interrupt its interaction with CDC20, which leads to the malfunction of MCC and, consequently, the occurrence of aneuploidy. Therefore, LNC CRYBG3 functions as a novel oncogene and its overexpression promotes tumorigenesis and metastasis of NSCLC. These findings provide a roadmap for the initiation, progression, and malignancy of NSCLC as well as a potential target for the diagnosis, treatment, and prognosis of the disease.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Change history
05 March 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1038/s41388-024-02997-3
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Siegel R, Naishadham D, Jemal A. Cancer statistics. 2013 CA. 2013;63:11–30.
Wang Y, Gu J, Roth JA, Hildebrandt MA, Lippman SM, Ye Y, et al. Pathway-based serum microRNA profiling and survival in patients with advanced stage non-small cell lung cancer. Cancer Res. 2013;73:4801–9.
Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7.
Duan Z, Person RE, Lee HH, Huang S, Donadieu J, Badolato R, et al. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol Cell Biol. 2007;27:6889–902.
Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7.
Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–81.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–504.
Zhang E, He X, Zhang C, Su J, Lu X, Si X, et al. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018;19:154.
Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203.
Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin cell Biol. 2006;18:658–67.
Herzog F, Primorac I, Dube P, Lenart P, Sander B, Mechtler K, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–81.
Lischetti T, Nilsson J. Regulation of mitotic progression by the spindle assembly checkpoint. Mol Cell Oncol. 2015;2:e970484.
Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–17.
Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–31.
Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol cell Biol. 2007;8:379–93.
Sudakin V, Chan G, Yen T. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36.
Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol cell. 2002;13:755–66.
Heike G, Shinsuke T, Parsons WJ, Natalja P, Alfred B, Helmut Erich G, et al. Overexpression of the mitotic checkpoint genes BUB1, BUBR1, and BUB3 in gastric cancer-association with tumour cell proliferation. J Pathol. 2003;24:16–22.
Kato T, Daigo Y, Aragaki M, Ishikawa K, Sato M, Kaji M. Overexpression of CDC20 predicts poor prognosis in primary non-small cell lung cancer patients. J Surgical Oncol. 2012;106:423–30.
Zhiwei W, Lixin W, Jiateng Z, Hiroyuki I, Pengda L, Sarkar FH, et al. Cdc20: a potential novel therapeutic target for cancer treatment. Curr Pharm Des. 2013;19.
Pei H, Hu W, Guo Z, Chen H, Ma J, Mao W, et al. Long noncoding RNA CRYBG3 blocks cytokinesis by directly binding G-actin. Cancer Res. 2018;78:4563–72.
Chen H, Pei H, Hu W, Ma J, Zhang J, Mao W, et al. Long non-coding RNA CRYBG3 regulates glycolysis of lung cancer cells by interacting with lactate dehydrogenase A. J Cancer. 2018;9:2580–8.
Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–27.
Park YH, Kim D, Dai J, Zhang Z. Human bronchial epithelial BEAS-2B cells, an appropriate in vitro model to study heavy metals induced carcinogenesis. Toxicol Appl Pharmacol. 2015;287:240–5.
Holliday R. Chromosome error propagation and cancer. Trends Genet. 1989;5:42–45.
Rodrigues CF, Urbano AM, Matoso E, Carreira I, Almeida A, Santos P, et al. Human bronchial epithelial cells malignantly transformed by hexavalent chromium exhibit an aneuploid phenotype but no microsatellite instability. Mutat Res. 2009;670:42–52.
Logarinho E, Resende T, Torres C, Bousbaa H. The human spindle assembly checkpoint protein Bub3 is required for the establishment of efficient kinetochore-microtubule attachments. Mol Biol Cell. 2008;19:1798–813.
Overlack K, Bange T, Weissmann F, Faesen AC, Maffini S, Primorac I, et al. BubR1 promotes Bub3-dependent APC/C inhibition during spindle assembly checkpoint signaling. Curr Biol. 2017;27:2915–27 e2917.
Nasmyth K. Segregating sister genomes: the molecular biology of chromosome separation. Science. 2002;297:559–65.
Nasmyth K, Peters JM, Uhlmann F. Splitting the chromosome: cutting the ties that bind sister chromatids. Science. 2000;288:1379–85.
Zou H, McGarry TJ, Bernal T, Kirschner MW. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science. 1999;285:418–22.
Yu H, Peters JM, King RW, Page AM, Hieter P, Kirschner MW. Identification of a cullin homology region in a subunit of the anaphase-promoting complex. Science. 1998;279:1219–22.
Collin P, Nashchekina O, Walker R, Pines J. The spindle assembly checkpoint works like a rheostat rather than a toggle switch. Nat Cell Biol. 2013;15:1378.
McWhorter JE. Malignant epithelial tumors of the neck of unknown origin. Ann Surg. 1929;90:1–14.
Manchester KL. Theodor Boveri and the origin of malignant tumours. Trends cell Biol. 1995;5:384–7.
Ricke RM, van Ree JH, van Deursen JM. Whole chromosome instability and cancer: a complex relationship. Trends Genet. 2008;24:457–66.
Rasnick D, Duesberg PH. How aneuploidy affects metabolic control and causes cancer. Biochem J. 1999;340:621–30.
White NM, Rozycki EB, Dang HX, Maher CA. Abstract 3498: characterization of a novel metastatic lung cancer associated lncRNA. Cancer Res. 2017;77:3498–3498.
Sun M, Liu X-H, Lu K-H, Nie F-Q, Xia R, Kong R, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298.
Sun M, Liu XH, Wang KM, Nie FQ, Kong R, Yang JS, et al. Downregulation of BRAF activated non-coding RNA is associated with poor prognosis for non-small cell lung cancer and promotes metastasis by affecting epithelial-mesenchymal transition. Mol Cancer. 2014;13:68–68.
Pei H, Guo Z, Wang Z, Dai Y, Zheng L, Zhu L, et al. RAC2 promotes abnormal proliferation of quiescent cells by enhanced JUNB expression via the MAL-SRF pathway. Cell Cycle. 2018;17:1115–23.
Gurley KE, Moser RD, Kemp CJ (2015). Induction of lung tumors in mice with urethane. Cold Spring Harb Protoc. 2015. https://doi.org/10.1101/pdb.prot077446.
Guo Z, Pei H, Nie J, Hu W, Zhang J, Ding J, et al. Anti-cancer effects of CQBTO, a chloroquine, and benzo(e)triazine oxide conjugate. Chem Biol Drug Des. 2019;93:874–82.
Hu W, Pei H, Li H, Ding N, He J, Wang J, et al. Effects of shielding on the induction of 53BP1 foci and micronuclei after Fe ion exposures. J Radiat Res. 2014;55:10–16.
Pei H, Zhang J, Nie J, Ding N, Hu W, Hua J, et al. RAC2-P38 MAPK-dependent NADPH oxidase activity is associated with the resistance of quiescent cells to ionizing radiation. Cell Cycle. 2017;16:113–22.
Hu W, Xu S, Yao B, Hong M, Wu X, Pei H, et al. MiR-663 inhibits radiation-induced bystander effects by targeting TGFB1 in a feedback mode. RNA Biol. 2014;11:1189–98.
Ma MZ, Zhang Y, Weng MZ, Wang SH, Hu Y, Hou ZY, et al. Long noncoding RNA GCASPC, a target of miR-17-3p, negatively regulates pyruvate carboxylase-dependent cell proliferation in gallbladder cancer. Cancer Res. 2016;76:5361–71.
Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–65.
Senger DR, Perruzzi CA, Streit M, Koteliansky VE, de Fougerolles AR, Detmar M. The alpha(1)beta(1) and alpha(2)beta(1) integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am J Pathol. 2002;160:195–204.
Acknowledgements
We are grateful to Dr. Shengfang Ge of the Shanghai Jiao Tong University for his valuable suggestion.
Funding
This work was supported by the China National Natural Science Foundation awards (Nos. 82073480, 81773463, 81872622), National Key R&D Program of China (No. 2018YFC0115704), and the US National Institutes of Health grant R01-ES 011804-10.
Author information
Authors and Affiliations
Contributions
Conception and design: GZ, HP, TKH, and ZG. Development of methodology: HP, WM, AW, and WH. Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): HP, ZG, BL, JN, and NL. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): ZG, HP, GZ, WH, BL, TKH, and LC. Writing, review, and/or revision of the manuscript: ZG, GZ, HP, and TKH. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): HP, ZG, WM, BL, AW, JN, and GZ. Study supervision: GZ and TKH.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Consent for publication
We have obtained consents to publish this paper from all the participants.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Soochow University and written informed consent was obtained from all patients.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1038/s41388-024-02997-3"
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Guo, Z., Dai, Y., Hu, W. et al. RETRACTED ARTICLE: The long noncoding RNA CRYBG3 induces aneuploidy by interfering with spindle assembly checkpoint via direct binding with Bub3. Oncogene 40, 1821–1835 (2021). https://doi.org/10.1038/s41388-020-01601-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-020-01601-8
This article is cited by
-
Mitotic phosphorylation of tumor suppressor DAB2IP maintains spindle assembly checkpoint and chromosomal stability through activating PLK1-Mps1 signal pathway and stabilizing mitotic checkpoint complex
Oncogene (2022)
-
Regulatory mechanisms of mechanotransduction in genome instability
Genome Instability & Disease (2022)