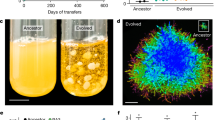
Abstract
Whole-genome duplication (WGD) is widespread across eukaryotes and can promote adaptive evolution1,2,3,4. However, given the instability of newly formed polyploid genomes5,6,7, understanding how WGDs arise in a population, persist, and underpin adaptations remains a challenge. Here, using our ongoing Multicellularity Long Term Evolution Experiment (MuLTEE)8, we show that diploid snowflake yeast (Saccharomyces cerevisiae) under selection for larger multicellular size rapidly evolve to be tetraploid. From their origin within the first 50 days of the experiment, tetraploids persisted for the next 950 days (nearly 5,000 generations, the current leading edge of our experiment) in 10 replicate populations, despite being genomically unstable. Using synthetic reconstruction, biophysical modelling and counter-selection, we found that tetraploidy evolved because it confers immediate fitness benefits under this selection, by producing larger, longer cells that yield larger clusters. The same selective benefit also maintained tetraploidy over long evolutionary timescales, inhibiting the reversion to diploidy that is typically seen in laboratory evolution experiments. Once established, tetraploidy facilitated novel genetic routes for adaptation, having a key role in the evolution of macroscopic multicellular size via the origin of evolutionarily conserved aneuploidy. These results provide unique empirical insights into the evolutionary dynamics and impacts of WGD, showing how it can initially arise due to its immediate adaptive benefits, be maintained by selection and fuel long-term innovations by creating additional dimensions of heritable genetic variation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
199,00 € per year
only 3,90 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Underlying data used to make the figures as well as raw experimental data are available on GitHub (https://github.com/ktong25/WGD_in_MuLTEE). Raw Illumina sequencing reads are available at the NIH Sequence Read Archive under the accession numbers PRJNA943273 (for MuLTEE evolved isolates) and PRJNA1093477 (for donut and spread strains). All raw microscopy images are publicly available at the BioImage Archive under the accession number S-BIAD1559. The yeast genome (S288C reference genome, R64-3-1 build) used for genome sequence analysis was downloaded from the Saccharomyces Genome Database.
Code availability
All code used for image analysis, genome sequence analysis, biophysical modelling, data analysis and figure generation are available on GitHub (https://github.com/ktong25/WGD_in_MuLTEE).
References
Otto, S. P. The evolutionary consequences of polyploidy. Cell 131, 452–462 (2007).
Selmecki, A. M. et al. Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352 (2015).
Van de Peer, Y., Mizrachi, E. & Marchal, K. The evolutionary significance of polyploidy. Nat. Rev. Genet. 18, 411–424 (2017).
Vittoria, M. A., Quinton, R. J. & Ganem, N. J. Whole-genome doubling in tissues and tumors. Trends Genet. 39, 954–967 (2023).
Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846 (2005).
Storchova, Z. et al. Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541–547 (2006).
Buggs, R. J. et al. Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 22, 248–252 (2012).
Bozdag, G. O. et al. De novo evolution of macroscopic multicellularity. Nature 617, 747–754 (2023).
Fox, D. T., Soltis, D. E., Soltis, P. S., Ashman, T. L. & Van de Peer, Y. Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol. 30, 688–694 (2020).
Mayrose, I. et al. Recently formed polyploid plants diversify at lower rates. Science 333, 1257 (2011).
Levin, D. A. Why polyploid exceptionalism is not accompanied by reduced extinction rates. Plant Syst. Evol. 305, 1–11 (2018).
Clo, J. & Kolar, F. Short- and long-term consequences of genome doubling: a meta-analysis. Am. J. Bot. 108, 2315–2322 (2021).
Gerstein, A. C., Chun, H. J., Grant, A. & Otto, S. P. Genomic convergence toward diploidy in Saccharomyces cerevisiae. PLoS Genet. 2, e145 (2006).
Gerstein, A. C. & Sharp, N. P. The population genetics of ploidy change in unicellular fungi. FEMS Microbiol. Rev. 45, fuab006 (2021).
Todd, R. T., Forche, A. & Selmecki, A. Ploidy variation in fungi: polyploidy, aneuploidy, and genome evolution. Microbiol. Spectr. 5, 5–4 (2017).
Lu, Y. J., Swamy, K. B. & Leu, J. Y. Experimental evolution reveals interplay between Sch9 and polyploid stability in yeast. PLoS Genet. 12, e1006409 (2016).
Bomblies, K. When everything changes at once: finding a new normal after genome duplication. Proc. Biol. Sci. 287, 20202154 (2020).
Doyle, J. J. & Coate, J. E. Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 180, 1–52 (2019).
Levin, D. A. Polyploidy and novelty in flowering plants. Am. Nat. 122, 1–25 (1983).
Mortier, F. et al. Understanding polyploid establishment: temporary persistence or stable coexistence? Oikos https://doi.org/10.1111/oik.09929 (2024).
Van de Peer, Y., Ashman, T. L., Soltis, P. S. & Soltis, D. E. Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell 33, 11–26 (2021).
Storchova, Z. Ploidy changes and genome stability in yeast. Yeast 31, 421–430 (2014).
Gerstein, A. C. et al. Polyploid titan cells produce haploid and aneuploid progeny to promote stress adaptation. mBio 6, e01340-15 (2015).
Hirakawa, M. P., Chyou, D. E., Huang, D., Slan, A. R. & Bennett, R. J. Parasex generates phenotypic diversity de novo and impacts drug resistance and virulence in Candida albicans. Genetics 207, 1195–1211 (2017).
Scott, A. L., Richmond, P. A., Dowell, R. D. & Selmecki, A. M. The influence of polyploidy on the evolution of yeast grown in a sub-optimal carbon source. Mol. Biol. Evol. 34, 2690–2703 (2017).
Bielski, C. M. et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195 (2018).
Fujiwara, T. et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437, 1043–1047 (2005).
Ratcliff, W. C., Fankhauser, J. D., Rogers, D. W., Greig, D. & Travisano, M. Origins of multicellular evolvability in snowflake yeast. Nat. Commun. 6, 6102 (2015).
Jacobeen, S. et al. Cellular packing, mechanical stress and the evolution of multicellularity. Nat. Phys. 14, 286–290 (2018).
Bozdag, G. O., Libby, E., Pineau, R., Reinhard, C. T. & Ratcliff, W. C. Oxygen suppression of macroscopic multicellularity. Nat. Commun. 12, 2838 (2021).
Day, T. C. et al. Morphological entanglement in living systems. Phys. Rev. 14, 011008 (2024).
Day, T. C. et al. Cellular organization in lab-evolved and extant multicellular species obeys a maximum entropy law. eLife 11, e72707 (2022).
Harrison, B. D. et al. A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol. 12, e1001815 (2014).
Ramsey, J. S. & Schemske, D. W. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Ann. Rev. Ecol. Syst. 29, 467–501 (1998).
Gerstein, A. C., McBride, R. M. & Otto, S. P. Ploidy reduction in Saccharomyces cerevisiae. Biol. Lett. 4, 91–94 (2008).
Voordeckers, K. et al. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 11, e1005635 (2015).
Galitski, T., Saldanha, A. J., Styles, C. A., Lander, E. S. & Fink, G. R. Ploidy regulation of gene expression. Science 285, 251–254 (1999).
Rebolleda-Gomez, M. & Travisano, M. The cost of being big: local competition, importance of dispersal, and experimental evolution of reversal to unicellularity. Am. Nat. 192, 731–744 (2018).
Treinin, M. S. G. Mitochondrial activity is required for the expression of IME1, a regulator of meiosis in yeast. Curr. Genet. 23, 223–227 (1993).
Gilchrist, C. & Stelkens, R. Aneuploidy in yeast: segregation error or adaptation mechanism? Yeast 36, 525–539 (2019).
Vande Zande, P., Zhou, X. & Selmecki, A. The dynamic fungal genome: polyploidy, aneuploidy and copy number variation in response to stress. Annu. Rev. Microbiol. 77, 341–361 (2023).
O’Donnell, S. et al. Telomere-to-telomere assemblies of 142 strains characterize the genome structural landscape in Saccharomyces cerevisiae. Nat. Genet. 55, 1390–1399 (2023).
De Chiara, M. et al. Domestication reprogrammed the budding yeast life cycle. Nat. Ecol. Evol. 6, 448–460 (2022).
Albertin, W. et al. Evidence for autotetraploidy associated with reproductive isolation in Saccharomyces cerevisiae: towards a new domesticated species. J. Evol. Biol. 22, 2157–2170 (2009).
Yona, A. H. et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc. Natl Acad. Sci. USA 109, 21010–21015 (2012).
Li, R. & Zhu, J. Effects of aneuploidy on cell behaviour and function. Nat. Rev. Mol. Cell Biol. 23, 250–265 (2022).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Cherry, J. M. et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res. 40, D700–D705 (2012).
Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. Preprint at https://arxiv.org/abs/1303.3997 (2013).
Van der Auwera, G. A. & O’Connor, B. D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra (O’Reilly Media, 2020).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Todd, R. T., Braverman, A. L. & Selmecki, A. Flow cytometry analysis of fungal ploidy. Curr. Protoc. Microbiol. 50, e58 (2018).
Acknowledgements
We thank the Microbial Sequencing Center at University of Pittsburgh for sequencing the genomes; the Partnership for an Advanced Computing Environment (PACE) at Georgia Institute of Technology for providing the research cyberinfrastructure resources and services; and all members of the Ratcliff laboratory, and H. Chen and F. Mortier of Yves Van de Peer’s laboratory for feedback on the study. This work was supported by US National Institutes of Health (NIH) grant R35-GM138030 to W.C.R., Human Frontiers Science Program grant RGY0080/2020 to W.C.R., Packard Fellowship for Science and Engineering to W.C.R., NIH grant R35-GM138354 to P. J. Yunker, NIH grant R01AI171100 to A.S.K., and Schmidt Sciences Polymath award G-22-63292 to A.S.K.
Author information
Authors and Affiliations
Contributions
K.T. and W.C.R. conceived the project. K.T., W.C.R., P.L.C., S.D. and A.S.K. designed the experiments. G.O.B. performed the MuLTEE, analysed the point mutations in the evolved isolates and identified the genomic signature of tetraploidy. V.C., D.J.H. and K.T. genetically engineered the tetraploid snowflake yeast. S.G., H.L.Y. and P.L.C. performed the evolution experiments with selection against larger size and measured the evolved populations. S.D. and K.T. isolated donut and spread strains, characterized their phenotypes and analysed their point mutations. T.C.D. performed the biophysical simulations. D.T.L. performed the genomic DNA extraction and helped with genome analysis. S.D. and K.T. performed the mutation accumulation experiment. K.T. performed the rest of the experiments, analysed data and made figures. K.T. prepared the first draft of the manuscript. All authors contributed to the revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Omaya Dudin, Gianni Liti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Images of the ancestors and evolved isolates in the MuLTEE.
a,b, Representative cluster-level (a) and cell-level (b) images of PM/PA t0 and PM/PA1-5 t200, t400, t600, and t1000 isolates. The images of PM/PA t0 are reused for five replicate populations. Scale bars, 200 μm (a) and 10 μm (b).
Extended Data Fig. 2 Phenotypic characterization of the ancestors and evolved isolates in the MuLTEE.
a-d, Violin plots showing the distributions of cluster radius (a,b, where b is weighted by cluster volume), cell volume (c), and cell aspect ratio (d) in PM/PA t0 and PM/PA1-5 t200, t400, t600, and t1000 isolates (on average, n = 789 clusters (a,b) and 1,288 cells (c,d) measured per sample). The distributions of PM/PA t0 are reused for five replicate populations. For a,b, we measured cluster radius at 4 h (exponential phase) and 24 h (stationary phase) after transferring the culture to fresh media, and the 24-hour measurements (corresponding to the states of the cultures right before settling selection) are used throughout the paper unless otherwise noted. For b, filled circles show biomass-weighted mean cluster radius (the 24-hour values are the same as the values in Fig. 1c). For c,d, boxes, IQR; center lines, median; whiskers, values within 1.5 × IQR of the first and third quartiles.
Extended Data Fig. 3 Imaging-based method for measuring ploidy level of snowflake yeast.
a, Overview of imaging and image analysis workflow. Snowflake yeast clusters are crushed into a single layer of cells and imaged at the brightfield channel and fluorescent channel, with the latter showing the nuclear DNA stained by propidium iodide (PI). The nuclei in the fluorescent image are segmented and filtered to get single round nuclei, outlined in cyan. The fluorescent image is also nuclei-cleared and brightness/contrast-enhanced to show the background fluorescence in the cytoplasm, and the cytoplasm is segmented, shown in white, for background subtraction. The total fluorescence intensity of PI in each nucleus is quantified and background-subtracted. Scale bar, 20 µm. b, Distribution of the nuclear PI intensity (arbitrary unit) of the engineered diploid and tetraploid mixotrophic clusters (n = 14,276 and 10,031 nuclei, respectively), as a validation for this ploidy measurement method. Since asynchronous, exponential-phase cultures are used for ploidy measurements, each strain shows two peaks that correspond to G1- and G2-phase nuclei of the actively-dividing cells, and the G2 peak has double the fluorescent intensity of the G1 peak. Also, the G2 peak of diploid clusters aligns nicely with the G1 peak of tetraploid clusters.
Extended Data Fig. 4 Copy number variation of the ancestors and evolved isolates in the MuLTEE.
a,b, Estimated copy number of each 1-kb non-overlapping bin in each chromosome in PM/PA t0 and PM/PA1-5 t200, t400, t600, and t1000 isolates (a, PM; b, PA). Estimated bin copy numbers above 12 are shown as 12, indicated by little triangles. Red horizontal line, baseline ploidy of each strain (i.e., 2 for PM/PA t0 and 4 for all evolved isolates). Red arrowhead, incidence of segmental aneuploidy.
Extended Data Fig. 5 Genetic construction and phenotypic characterization of diploid and tetraploid clusters.
a, Procedure for engineering isogenic grande diploid and tetraploid clusters, from each of which four independent petite mutants were isolated. Isolating multiple petite mutants is important because petite mutations are not isogenic and may confound ploidy-phenotype map. Grande and petite clusters correspond to mixotrophic and anaerobic conditions, respectively. b-e, Violin plots showing the distributions of cluster radius (b,c, where c is weighted by cluster volume), cell volume (d), and cell aspect ratio (e) in engineered diploid and tetraploid clusters under mixotrophic and anaerobic conditions (on average, n = 922 clusters (b,c) and 2,458 cells (d,e) measured per sample). Four biological replicates were measured for the mixotrophic condition, and the four independent petite mutants (each with one biological replicate) were measured for the anaerobic condition. For b,c, we measured cluster radius at 4 h (exponential phase) and 24 h (stationary phase) after transferring the culture to fresh media, and the 24-hour measurements are used throughout the paper unless otherwise noted. For c, filled circles show biomass-weighted mean cluster radius (the 24-hour values are the same as the values in Fig. 3e). For d,e, boxes, IQR; center lines, median; whiskers, values within 1.5 × IQR of the first and third quartiles. f,g, Comparison of the biomass-weighted mean cluster radius (f) and mean cell aspect ratio (g) of the engineered petite tetraploid clusters (mean of the four independent petite mutants, the same values as those in Fig. 1c,e) to the PA t0 and PA1-5 t50, t100, t150, and t200 populations (data from Bozdag et al.8).
Extended Data Fig. 6 Label-free method for distinguishing engineered diploid and tetraploid clusters in competition assays.
a, Brightfield image of a snowflake yeast cluster (an engineered tetraploid mixotrophic cluster is shown) (top), whose bright cells in the cluster edges are detected (bottom). Scale bar, 30 µm. On average, 381 clusters were segmented per sample using this approach. b, Mean area of the five largest cells detected in a cluster can be used to distinguish between the engineered tetraploid and diploid clusters under both mixotrophic and anaerobic conditions, with the dashed line indicating the manually-chosen decision boundary.
Extended Data Fig. 7 Experimental evolution of the MuLTEE ancestors and t1000 isolates with selection against larger size.
a, Experimental setup. We evolved PM/PA t0 and PM/PA1-5 t1000 isolates, each with four replicate populations (A, B, C, D), under selection against larger size for 70 days (~500 generations) by growing them on agar in 24-well plates with daily dilution. b, Distributions of cluster radius (weighted by cluster volume) in the ancestral (“anc”) and evolved populations (on average, n = 406 clusters measured per population). Vertical thick solid line, biomass-weighted mean cluster radius of each population. Vertical dashed line, biomass-weighted mean cluster radius of PM/PA t0. Color codes for the evolved ploidy in each t0 population and in each t1000 population are the same as those in Fig. 3h,i, respectively, and the ancestral populations are colored in gray.
Extended Data Fig. 8 Experimental evolution of the MuLTEE ancestors, t1000 isolates, and engineered tetraploids with minimal selection.
We evolved PM/PA t0, PM/PA1-5 t1000 isolates, and engineered mixotrophic and anaerobic tetraploids, each with three replicate populations (A, B, C), with minimal selection for 56 days or 28 bottlenecks, by growing them on agar with picking and streaking single colonies every two days. a, Distributions of cellular DNA contents in the ancestral (“anc”) and evolved populations (on average, n = 15,848 cells measured per population). Color code for ploidy change categories is the same as that in Fig. 3j, and the ancestral populations are colored in light gray.
Extended Data Fig. 9 Point mutation changes in donut-to-spread transitions.
Two donut-to-spread transitions with near-triploidization were excluded, and mutation allele frequency refers to the corrected allele frequency, calculated by dividing estimated allele copy number with copy number of the chromosome that carries the mutation. LOH, loss of heterozygosity. a,b, Number of mutations in each category of change in allele frequency in all donut-to-spread transitions combined, colored by change in allele copy number (a), and how change in allele frequency is associated with change in chromosome copy number (b). a,b share the color code. c, Number of mutations in each category of change in allele frequency in each donut-to-spread transition, colored by mutation impact. d, Percentage of mutations in each mutation impact category in each donut background, whose total number of mutations is indicated in the brackets. c,d share the color code. e, Comparison of the distribution of mutation impacts, between the mutations that were gained in all donut-to-spread transitions combined and the mutations randomly introduced into yeast genome, as well as between the mutations that underwent loss, LOH, increase, decrease, or maintenance in terms of allele frequency in all donut-to-spread transitions combined and the mutations in all donut backgrounds combined. Number of mutations is indicated in the brackets. P values were calculated by two-sided chi-squared test. f, For each donut-to-spread transition, the percentage of high/moderate-impact mutations in the mutations that increased or decreased in allele frequency is on average not significantly larger than the percentage of high/moderate-impact mutations in the donut background (for increase and decrease, respectively, P = 0.891 and 0.442, t22 = −1.27 and 0.147, one-tailed one-sample Student’s t-test), and is largely explained by random sampling of mutations in the donut background (simulation with random seed = 1). Values are mean ± s.d. (n = 23 donut-to-spread transitions).
Supplementary information
Supplementary Information
Supplementary Table 1 and Supplementary Note 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tong, K., Datta, S., Cheng, V. et al. Genome duplication in a long-term multicellularity evolution experiment. Nature 639, 691–699 (2025). https://doi.org/10.1038/s41586-025-08689-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-025-08689-6