Abstract
Elevated postprandial glycemic responses (PPGRs) are associated with type 2 diabetes and cardiovascular disease. PPGRs to the same foods have been shown to vary between individuals, but systematic characterization of the underlying physiologic and molecular basis is lacking. We measured PPGRs using continuous glucose monitoring in 55 well-phenotyped participants challenged with seven different standard carbohydrate meals administered in replicate. We also examined whether preloading a rice meal with fiber, protein or fat (‘mitigators’) altered PPGRs. We performed gold-standard metabolic tests and multi-omics profiling to examine the physiologic and molecular basis for interindividual PPGR differences. Overall, rice was the most glucose-elevating carbohydrate meal, but there was considerable interindividual variability. Individuals with the highest PPGR to potatoes (potato-spikers) were more insulin resistant and had lower beta cell function, whereas grape-spikers were more insulin sensitive. Rice-spikers were more likely to be Asian individuals, and bread-spikers had higher blood pressure. Mitigators were less effective in reducing PPGRs in insulin-resistant as compared to insulin-sensitive participants. Multi-omics signatures of PPGR and metabolic phenotypes were discovered, including insulin-resistance-associated triglycerides, hypertension-associated metabolites and PPGR-associated microbiome pathways. These results demonstrate interindividual variability in PPGRs to carbohydrate meals and mitigators and their association with metabolic and molecular profiles.
Similar content being viewed by others
Main
One in three adults in the United States has prediabetes, and 70% of these will develop type 2 diabetes (T2D), posing a substantial public health burden1 via complications such as kidney disease, vision loss, neuropathy, cardiovascular disease (CVD) and cancer2,3,4,5,6.
High postprandial glycemic responses (PPGRs) are a hallmark of prediabetes and T2D and are risk factors for T2D, CVD and all-cause mortality independent of fasting blood glucose (FBG) and HbA1c7,8,9. However, our understanding of glucose dysregulation, especially regarding PPGRs, remains incomplete. Interindividual variability in PPGRs to the same foods has been described7,10. A variety of factors have been identified as contributors to PPGRs, including glycemic index, the total carbohydrate amount, carbohydrate characteristics (starch and simple carbohydrates), food processing, meal macronutrient composition, meal timing, ethnicity and the microbiome11,12,13,14,15,16. However, the contribution of individual physiologic and metabolic factors to PPGRs and whether they interact with food characteristics have not been well studied.
A gap in understanding how underlying metabolism and physiology affect PPGRs exists largely because quantifying metabolic functions, such as insulin resistance, insulin secretion and the incretin effect, is laborious and costly and has not been extensively employed. However, these metabolic traits differ between individuals17,18,19 and likely explain, in part, the observed interindividual PPGR differences. Glycemic responses to the oral glucose tolerance test have been linked to underlying physiology such as insulin resistance and beta cell dysfunction19. Prespecified timed blood draws may miss the peak glucose, but continuous glucose monitoring (CGM) devices enable a detailed evaluation of PPGRs and can be linked to individual metabolic traits. Indeed, using CGM, we previously showed that individuals with higher and more variable daily glucose excursions were more insulin resistant (IR) than their lower-glycemic counterparts10. Thus, CGMs have the potential to link underlying metabolic phenotypes to individual responses to food.
To further study individual PPGRs to foods and their association with metabolic subtypes and omics profiles, we conducted a rigorous investigation of responses to a wide variety of standardized carbohydrate meals and standardized preloads (‘mitigators’) to a rice meal in individuals whose metabolic traits were comprehensively profiled with gold-standard tests and whose blood and stool were profiled for metabolites, lipids, proteins and microbiome. We hypothesized that individual PPGRs are associated with the underlying metabolic physiology (for example, insulin resistance and beta cell dysfunction) as well as molecular markers.
Results
Study cohort and tests performed
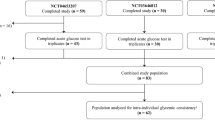
The overall study design is illustrated in Fig. 1a. A cohort of 55 participants with no prior history of T2D underwent deep metabolic phenotyping using gold-standard tests for insulin resistance by steady-state plasma glucose (SSPG), beta cell function by disposition index, hepatic insulin resistance and adipocyte insulin resistance (Methods)19,20,21,22,23,24,25,26,27,28. At baseline, we collected metabolomics, lipidomics, proteomics, microbiome and clinical data.
a, Overview of study design and data types. Left, at the baseline, the data on omics and clinical tests were collected. Mid-bottom, each individual ate seven different carbohydrate meals and three different mitigator foods with rice with CGM data and food log collected. Mid-top, an example CGM curve of a day is presented. Right, participants were then stratified into carb-response types based on which meal produced the highest spike and into metabolic traits results. b, Mean CGM curves of PPGR after different meals. c, Glucose AUC above baseline and time from baseline to peak for different meals. CGM curves are extracted into different features (and presented with mean and standard errors (error bar indicates 2 standard errors). Bars of standardized carbohydrate meals were ordered by mean value. The number of CGM curves (each dot) is as follows: rice 115, potatoes 92, bread 99, pasta 65, grapes 98, beans 46 and mixed berries 53. Extracted CGM features are compared between different carbohydrate meals. Each participant was instructed to eat each meal at least twice on two different days. d, Heatmap of extracted CGM PPGR features. Each element is the mean value of extracted CGM features. Each column (feature) is then centered and scaled. e, Association between different extracted features and nutrient contents of the standardized carbohydrate meal. For fiber, proteins and fat in study meals, it’s the Spearman correlation with statistics calculated from the R package ‘correlation’ under default settings with asymptotic t approximation with a two-sided test. The last column is treated as binary (starch versus non-starch food) with correlation (R) obtained from a simple linear model and P value obtained from a two-sided t-test of the coefficient. Asterisks indicate significance (pFDR < 0.05). P values were FDR-corrected for the whole matrix. Illustration in a created with BioRender.com.
Standardized carbohydrate meals
To quantify individual PPGRs to different carbohydrates and evaluate potential mitigation, participants underwent seven different standardized carbohydrate meal tests (50 g) and three preloaded mitigator meal tests while wearing a CGM at home and following their regular diet otherwise (Fig. 1a). The seven carbohydrate meals, each containing 50 g total carbohydrates, included five starchy meals—rice (jasmine rice), bread (buttermilk bread), potatoes (shredded), pasta (macaroni, precooked per instructions, cooled and frozen) and beans (canned black beans, high fiber)—as well as two different simple-carbohydrate meals—mixed berries (blackberries, strawberries and blueberries, high fiber) and grapes (low fiber) (Fig. 1a and Supplementary Table 6) (details in Methods). The three preloaded mitigators included pea fiber, egg white and cream, representing fiber, protein and fat, respectively (Methods), which were consumed 10 min before a standardized 50 g carbohydrate meal (for example, rice). Each meal test was performed at least twice. After processing, there were 848 CGM curves (55 participants), 267 CGM curves with mitigators (47 participants, 32 for rice + mitigators) and 33,920 glucose time points in total.
Clinical and demographic profile of the cohort
Among the 55 participants with CGMs, 24 were male, 27 were female, and 4 were unknown (Table 1). Of the participants, 26 were with healthy weight, 18 with overweight, 9 with obesity, 1 with underweight and 1 with unknown weight status. None of the participants had a prior medical history of diabetes; 27 participants were euglycemic, 26 had prediabetes, and one had T2D based on HbA1c measured during the visit (6.5%) (one was unknown). The racial and ethnic distribution was 37 European, 13 Asian, 4 Hispanic and 1 mixed Asian and European individuals. Eighteen were IR (SSPG ≥ 120 mg dl−1), 25 were insulin sensitive (IS; SSPG < 120 mg dl−1), and the mean SSPG and disposition index were 120.1 ± 72.9 mg dl−1 (mean ± s.d.) and 1.55 ± 0.82.
Individual variability and reproducibility of PPGRs
We assessed the variability between CGM curve replicates for the same meals consumed by the same individual on different days (Supplementary Fig. 1). CGM curves from most individuals were reproducible between replicates based on the area under the curve above the baseline (AUC(>baseline)) with intraindividual correlation coefficients (ICCs) of AUC(>baseline) from 0.26 for beans to 0.73 for pasta, indicating consistency between meal replicates of the same individual except for meals with low PPGRs (Supplementary Table 9)13. The correlation coefficients between replicates were also similar to previously published studies7. To reduce the effect of within-individual variation, results below were computed based on the average of each participant’s replicates.
Overall PPGRs to different standardized carbohydrates
The seven different standardized carbohydrate meals with equivalent carbohydrate loads elicited different PPGRs, emphasizing the impact of food composition (Fig. 1b, Extended Data Fig. 1 and Supplementary Table 1). Typical postprandial CGM curves of standardized carbohydrate meals slightly decreased initially after meal consumption and then increased to a single maximum and returned to baseline within three hours (Fig. 1b, Extended Data Fig. 1 and Supplementary Figs. 2 and 3). Of the five starchy carbohydrate meals, rice, bread and potatoes produced high peaks at approximately 1 hour post consumption. Grapes had high glucose peaks that reached maxima at earlier time points. Beans and pasta reached maxima at similar times as other starchy meals but exhibited significantly lower peaks, with beans eliciting the lowest (Fig. 1b,c and Supplementary Table 1). Mixed berries reached maxima at similar times as grapes but had much lower peaks. Beans and mixed berries produced the lowest peak measured by both AUC(>baseline) and delta glucose peak (the difference between the peak and baseline glucose) as compared with other meals.
From CGM curves, we extracted 11 different features of principally two types: (1) scale (for example, AUC(>baseline) and delta glucose peak) and (2) rate (for example, time baseline to peak and time return to baseline) (Fig. 1d; details in Methods). Different AUC-based features had similar patterns, which were similar to that of delta glucose peak, except for grapes (Fig. 1c,d and Extended Data Fig. 2a). Time baseline to peak, time return to baseline and delta glucose peak at 170 min show similar patterns. Using scale and rate dimensions, three exemplar meals (beans, mixed berries and rice) were clustered into partially separated groups (Extended Data Fig. 2b). Across different meals, delta glucose peak (Fig. 2a) and AUC(>baseline) (Supplementary Fig. 7a) were also positively correlated with each other. Besides single-meal features, we also defined the potato versus grape ratio (PG-ratio), which is the ratio between the delta glucose peak of potatoes and grapes, highlighting the differences between a starchy carbohydrate with a high amount of resistant starch and a simple-carbohydrate meal.
a, The Spearman correlation between PPGRs to all different meals, including both the delta glucose peak of standardized carbohydrate meals and the suppression effect of mitigator foods. The delta glucose peak was calculated as the difference between the peak and baseline. The suppression mitigator effects were defined by subtracting the delta glucose peak of rice + mitigator from that of rice and then normalizing it by that of rice. b, Examples of CGM curves of participants with different carbohydrate meals with the highest PPGR (carb-response type). c, Number of participants assigned to each carb-response type. The carb-response types are defined by both delta glucose peak and AUC(>baseline). The x axis indicates the standardized carbohydrate meal that produced the highest glucose spike, and the y axis is the number of participants for whom a given meal caused the highest spike. d, The Asian group was enriched with individuals with rice as the carbohydrate meal with the highest peak. e, Radar plot of delta glucose to different carbohydrate meals in an IS (SSPG 61 mg dl−1) and an IR (SSPG 239 mg dl−1) participant. Delta glucose values were averaged between replicates and scaled by the carbohydrate with the highest value. Insulin resistance, disposition index (DI) and HbA1c were also presented.
PPGRs to different carbohydrate meals were associated with the nutrient composition of the standardized meal. The total dietary fiber content (measured in grams) in the carbohydrate meal negatively correlated with AUC(>baseline) (−0.71, 95% confidence interval (−0.96,0.11), P = 0.088) and delta glucose peak (−0.75, 95% confidence interval (−0.96,0.04), P = 0.066) (Fig. 1e and Supplementary Table 6).
Interindividual PPGR variability and carb-response types
PPGRs varied considerably across individuals, which differed from the conventional idea of a fixed glycemic index value for a single food (Figs. 2b–e, Supplementary Figs. 2 and 18 and Extended Data Fig. 3). Although overall the PPGRs tended to correlate positively across carbohydrate meals (Fig. 2a), for different individuals, PPGRs to different meals were ranked differently; and for each individual, different meals produced the highest PPGR (Fig. 2b–e and Extended Data Fig. 3). For example, 19 participants exhibited the highest average delta glucose peak from rice (participant #18), whereas others were highest with bread, grapes, potatoes or pasta (Extended Data Fig. 3).
We aimed to type individuals based on their PPGRs to different carbohydrate meals. Participants were stratified as carb-response types (also referred to as ‘spikers’) according to the meal that produced the highest average PPGR for that individual, as measured by delta glucose peak and AUC(>baseline). Using the delta glucose peak, rice-spikers were the largest group (35%), followed by bread-spikers (24%) and grape-spikers (22%); no participants had the highest spike in response to beans or mixed berries (Fig. 2c), possibly due to higher fiber and/or protein (beans) content (Fig. 1f). Stratification based on AUC(>baseline) largely agreed with results based on delta glucose peak, except that even more individuals were classified as rice-spikers and fewer as grape-spikers (Fig. 2c). Carb-response-type classification was stable between replicates of meals, and results based on individual meal replicates largely agree with those based on the average (Extended Data Fig. 4). In addition, ICC for delta glucose peak and AUC(>baseline) was moderate to high for most meals (Supplementary Table 9). ICC values were lower for meals with smaller PPGRs (for example, beans), potentially because of a lower signal-to-noise ratio.
Metabolic physiology and PPGR to standardized carbohydrates
We investigated individuals’ PPGRs as a function of their underlying metabolic physiological traits. PPGRs, measured as both delta glucose peak and AUC(>baseline), were significantly greater in IR (measured by SSPG) compared with IS individuals for potatoes and pasta (179% and 152% for delta glucose, respectively; Figs. 2e and 3 and Supplementary Figs. 5, 13a and 17). We measured starch content and found that potatoes had significantly more resistant starch and pasta had significantly more slowly digestible starch than other starchy foods (bread and rice) (Extended Data Fig. 5). The IR group also exhibited a higher baseline (premeal) glucose and higher absolute glucose peaks compared with IS individuals (Fig. 3a and Supplementary Figs. 16 and 17). Individuals with impaired beta cell function also demonstrated higher PPGRs to potatoes (Figs. 2e and 3 and Supplementary Figs. 5 and 17). The contributions of both insulin resistance and beta cell function were still significant after adjusting for confounding factors, BMI, age and sex (Supplementary Table 2). When modeled together, only SSPG remained an independent predictor of PPGRs. Normoglycemic IR individuals also exhibited higher PPGRs to potatoes than IS individuals with prediabetes (based on HbA1c), indicating the dominance of the insulin-resistance effect (Supplementary Figs. 6c and 14). Other differences noted in potato-spikers included higher HbA1c, FBG and free fatty acids (FFA) during the SSPG test (Fig. 3c and Supplementary Fig. 8). Grape-spikers, conversely, exhibited significantly lower SSPG (indicating insulin sensitivity) and the lowest FBG. Among individuals who are IS or with normal beta cell function, grape- and rice-spikers were the most frequent groups.
a, Mean CGM curves between IR and IS groups measured by SSPG (i), and groups with normal and dysfunctional beta cells measured by disposition index (ii). b, Delta glucose peaks after eating potatoes and pasta are compared between groups with IR and IS, and delta glucose peaks after eating potatoes are compared beween groups with normal and dysfunctional (dys) beta cell functions. *, based on Holm corrected P value of the Mann–Whitney test (default, two-sided): *PH ≤ 0.05; **PH ≤ 0.01; ***PH ≤ 0.001. PH from left to right is 3.4 × 10−4, 0.02 and 9.4 × 10−3. Number of participants: IS potatoes 14, IR potatoes 16, IS pasta 12, IR pasta 16, normal beta cell potatoes 5, dysfunctional beta cell potatoes 12. Extracted CGM features for each participant are compared between different metabolic subtypes. c, Clinical and metabolic characteristics according to carb-response types. Statistical comparisons utilized Holm corrected P value of the Mann–Whitney test (default, two-sided) of the selected pairs: *PH ≤ 0.05; **PH ≤ 0.01; ***PH ≤ 0.001; ****PH ≤ 0.0001. PH: grapes versus potatoes (i) 7.8 × 10−3; grapes versus potatoes (ii) 2.5 × 10−4; rice versus potatoes (ii) 0.022, bread versus potatoes (ii) 0.016, grapes versus bread (iii) 0.019. Number of participants for (i) ((ii), (iii)): rice-spiker 18 (13, 19), grape-spiker 12 (10, 12), pasta-spiker 2 (1, 2), bread-spiker 13 (13, 13), potato-spiker 8 (6, 8). Clinical measurements were compared between carb-response-types. The boxplots in b and c show the center line as the median and the hinges as the 25th and 75th percentiles. The upper whisker extends from the hinge to the largest value not bigger than 1.5 times the distance between the hinges. Data beyond the whiskers are outliers. d, Average delta glucose peak and AUC(>baseline) between participants with IR and IS. e, Average delta glucose and AUC(>baseline) between participants with different beta cell functions (normal and dysfunctional). The solid line is the average value, and the shade is one s.d. bp, blood pressure.
We observed that whereas the PPGR to potato consumption differed between IR and IS groups and between beta cell normal and dysfunctional groups, the PPGR to grape consumption changed little (Fig. 3d and Supplementary Fig. 17). We thus calculated the PG-ratio based on delta glucose peak as a normalization of the PPGR to potatoes. The PG-ratio was consistently different between IR and IS subtypes (Supplementary Fig. 13c and Supplementary Table 2). The sex interaction with beta cell function and insulin resistance was also significant in estimating the PG-ratio, with males having less effect from insulin resistance (intersection term coefficient −0.49) and beta cell dysfunction (intersection term coefficient −0.58) (Supplementary Table 2).
Underlying metabolic physiology and mitigation effects
To test the potential suppressing effect of other foods on PPGRs to standardized carbohydrate meals, other meals containing fiber, protein and fat (mitigators) were tested. Thirty-two individuals consumed fiber (10 g, via pea fiber), protein (10 g, via egg white) or fat (15 g, via cream) 10 min before the consumption of white rice (50 g of carbohydrates), the most glucose-elevating carbohydrate meal on average. The mitigation effect was calculated as a normalized difference based on PPGRs (delta glucose peak and AUC(>baseline)) of rice + mitigator relative to rice alone. A negative mitigation effect indicated that consuming the mitigator reduced the rice PPGR.
The effects of different mitigators positively correlated with each other with a correlation of 0.79 between fat and protein and approximately 0.5 between fiber and others (Fig. 2a and Supplementary Fig. 7a). Across the entire cohort, all three mitigators, when consumed before rice, weakly but significantly decreased the delta peak glucose (Cohen’s d: fiber 0.12, protein 0.19, fat 0.05). Fat, but not protein or fiber, increased time (baseline to peak) (Fig. 4a,b and Supplementary Tables 1 and 7). However, similar to the interindividual variability in PPGRs to different carbohydrate meals (carb-response types), highly divergent patterns were observed in mitigation effects between individuals (Fig. 4e and Supplementary Fig. 3). Many individuals exhibited mild mitigation effects (for example, participants #18 and #48), one had strong mitigation effects (participant #14), and some even had higher glycemic responses with mitigators (for example, participants #33 and #69) (Fig. 4e and Supplementary Fig. 3). For most participants, (fiber 23, protein 22 and fat 20) mitigators reduced glycemic responses (Fig. 4f). The mitigation effect of protein was significantly greater in the rice-spikers versus non-rice-spikers (Fig. 4g).
a, Mean CGM curves of PPGRs to rice and rice preceded by three different mitigators (fiber, protein, fat). b: Delta glucose and time for rice and rice + mitigators. *, based on the P value of the t-test (two-sided, paired): *P ≤ 0.05; **P ≤ 0.01. The P values by comparing rice and rice + fiber, protein and fat are 0.014, 0.004 and 0.017. Each dot is a CGM curve. c, Mean CGM curves of mitigator test between individuals who are IR and IS. d, Delta glucose for rice and rice + mitigators in IS, IR, normal beta cell and dysfunctional beta cell groups. The P value is from the t-test (two-sided, paired). The number of participants for the statistic test for (i) ((ii), (iii), (iv)) is rice 13 (16, 5, 12), rice + fiber 6 (7, 2, 6), rice + protein 6 (7, 2, 6), rice + fat 6 (7, 2, 6). The plotting is based on each CGM curve (each dot) with the number of samples ((i)–(iv)): rice (24, 35, 8, 28), rice + fiber (10, 12, 3, 10), rice + protein (13, 10, 3, 8), rice + fat (12, 11, 2, 9). e, Examples of participants with different mitigation effects. f, Frequency of different directions of effects of mitigator foods on delta glucose. Positive (negative) values indicate increasing (decreasing) delta glucose in the combination meal compared with rice. g, Mitigator foods and their relative mitigation effects. The P value of the t-test (default, two-sided) between rice-spiker and non-rice-spiker is FDR-adjusted. Number of participants for rice-spiker (non-rice-spiker): rice + fiber 12 (20), rice + protein 12 (20), rice + fat 11 (20). CGM features are presented with mean and two standard errors in b and d. Boxplots in g show the center line as the median and the hinges as the 25th and 75th percentiles. The upper whisker extends from the hinge to the largest value not bigger than 1.5 times the distance between the hinges. Data beyond the whiskers are outliers.
Similar to PPGRs to standardized carbohydrate meals, mitigation effects differed according to underlying metabolic features. IR individuals showed little beneficial response to mitigators, whereas IS individuals responded more to mitigators, with fiber decreasing delta glucose peak (P = 0.031) and AUC(>baseline) (P = 0.057) (Fig. 4d, Supplementary Fig. 15 and Supplementary Table 8). Individuals with normal beta cell function also had beneficial protein mitigation effects on delta glucose peak (P = 0.028) (Fig. 4d, Supplementary Figs. 5 and 6b and Supplementary Table 8). The different mitigation responses across individuals explained the lack of strong mitigation effects in the population average (Fig. 1b,c).
Other factors correlated with PPGRs and mitigation effects
We searched for other potential factors that contributed to differential PPGRs, including demographic and clinical data. In total, 15 continuous variables and three categorical variables (ethnicity, sex and BMI groups) were examined. Asian participants were more likely than non-Asian participants to be rice-spikers (Fisher’s exact test, P < 0.05, positive false discovery rate (pFDR) = 0.21) (Fig. 2d and Supplementary Fig. 8). No other significant associations were found between PPGRs (carb-response types and mitigator effect) and ethnicity, sex or BMI groups (Supplementary Fig. 20).
Among the 15 continuous measurements, associations were uncovered between carb-response types using analysis of variance (ANOVA) with a pFDR threshold of 0.3. Bread-spikers exhibited significantly higher systolic and diastolic blood pressure compared to other carb-response types after correcting for BMI and age (Fig. 3c, Supplementary Fig. 19 and Supplementary Table 2). The analysis in this section is exploratory and has higher risks of random variation than results in other analyses.
Molecular markers linked to PPGRs and mitigation effects
To identify potential biomarkers and mechanisms of carb-response types, we performed baseline omics analyses, including untargeted metabolomics (731 annotated metabolites, nine classes), targeted lipidomics (652 lipids, 13 classes), targeted proteomics (1,470 proteins) and metagenomics microbiome analysis (Extended Data Fig. 6).
In addition to higher blood pressure levels, bread-spikers also presented significantly higher N1-Methyladenosine than other participants (Figs. 5a and 3c and Supplementary Table 3)29,30. Potato-spikers had higher levels of certain triglycerides (TAG) and fatty acids (Fig. 5a and Supplementary Fig. 21), aligning with their increased insulin resistance23,31. In addition, proteins positively associated with potato PPGRs were significantly enriched in hepatic stellate cell activation and fibroblast activation pathways (Supplementary Data 1). Black bean PPGRs were positively associated with keto acids and metabolites related to ketone-related metabolism and amino acid metabolism, including gamma-glutamylthreonine, hydroxybutyric acid, alpha-ketoisovaleric acid, 2-hydroxy-3-methylbutyric acid and beta-citryl-l-glutamic acid (Supplementary Figs. 9 and 10). In addition, metabolites enriched in the histidine pathway were both positively (1-methyl-histidine, glutamate and methylimidazole acetic acid) and negatively (histidine and imidazole acetaldehyde) associated with bean-PPGR, indicating variability in histidine catabolism (Supplementary Fig. 11 and Supplementary Data 1). PPGRs to mixed berries were positively associated with unsaturated fatty acid biosynthesis and unsaturated TAGs in plasma metabolomics and lipidomics (Supplementary Data 1).
a, Metabolites and lipids that are distinguished between carb-response types. For the carb-response types, ‘Others’ include all other participants. *, based on Holm corrected P value of the Mann–Whitney test (default, two-sided) of the selected pairs: *PH ≤ 0.05; **PH ≤ 0.01. N1-Methyladenosine was selected as it had the lowest P value when comparing bread-spikers with others. The TAGs and fatty acids were selected as lipid-related features with pFDR < 0.2. More results can be found in Supplementary Table 3 and Supplementary Fig. 21. PH: bread versus potatoes (i) 0.021, bread versus rice (i) 2.5 × 10−3, others versus grapes (ii) 0.033, grapes versus potatoes (ii) 0.035, other versus potatoes (iii) 0.016, others versus grapes (iii) 0.043, potatoes versus grapes (iii) 1.2 × 10−3, others versus potatoes (iv) 1.1 × 10−3, potatoes versus grapes (iv) 8.2 × 10−3, others versus potatoes (v) 1.9 × 10−3, potatoes versus grapes (v) 8.2 × 10−3. The boxplots show the center line as the median and the hinges as the 25th and 75th percentiles. The upper whisker extends from the hinge to the largest value not bigger than 1.5 times the distance between the hinges. Data beyond the whiskers are outliers. Number of participants in each carb-response type: rice-spiker (15), grape-spiker (6), pasta-spiker (3), bread-spiker (8), potato-spiker (7). Omics measurements were compared between carb-response types. b, Spearman correlation between T2D-related clinical features and microbiome levels (Genus). Red indicates positive correlation, blue indicates negative correlation, and asterisks indicate P value < 0.05 and pFDR < 0.2. Genera with more than one P value < 0.05 association were selected. Sample weight was controlled by partial correlation. FDR correction was implemented for the whole matrix. c, Mediation effect through the same metabolite. The mediation effect was calculated from microbiome to metabolites (mediator) and then to clinical measurements. Asterisks indicate pFDR < 0.01, and others are selected based on pFDR < 0.2. The listed microbe functions were dominated by the species Bacteroides thetaiotaomicron with the highest levels. Ca., Candidatus. Illustration in c created with BioRender.com.
Microbiome analyses also revealed taxa and functional pathways associated with PPGRs. PPGRs to most carbohydrate meals were positively associated with glycosyl hydrolase family 85 (GH85), lactate utilization ___domain and heme exporter protein B and negatively associated with d-alanyl-d-alanine carboxypeptidase (Extended Data Fig. 7). In addition, Klebsiella, Sutterella and starch degradation III were negatively or beneficially correlated with the mitigation effect (Supplementary Table 5).
Molecular features associated with metabolic physiology
To further investigate the potential mechanisms of different PPGRs, we next explored the associations between multi-omics features and metabolic traits. Correlation matrix analyses between omics and clinical measurements revealed two clusters, with some omics features associated with healthy metabolic function and others with compromised metabolic function (Fig. 5b, Extended Data Figs. 8 and 9 and Supplementary Fig. 12)18,31,32,33,34,35. For instance, tyrosine, hydroxybutyric acid, glutamic acid and TAGs were significantly positively associated with plasma glucose or insulin resistance, whereas phosphatidylcholine, lysophosphatidylcholine, lysophosphatidylethanolamine and threonic acid were significantly negatively correlated (Extended Data Figs. 8 and 9). Similarly, among microbiomes, Slackia and Tyzzerella were positively correlated with plasma glucose or insulin resistance, and Butyrivibrio, Asaccharobacter, Adlercreutzia and Fusicatenibacter were negatively correlated (Fig. 5b). In an integrated statistical model to predict SSPG, strong predictors include fasting insulin, BMI, FFA and TAGs contributing to the IR state and phosphatidylcholine contributing to the IS state (Extended Data Fig. 10).
To further trace potential causal mechanisms, we then conducted a mediation analysis with microbiomes as the input, metabolomics and lipidomics as the mediators and clinical measurements as the outcomes (Supplementary Table 4)36,37. In Bacteroides thetaiotaomicron, pathways involving protein autophosphorylation, lactose catabolism and histidine phosphotransfer kinase activity positively mediated hepatic insulin resistance and fasting insulin via a single metabolite with an inferred formula, C9H15NO3, but no annotated match in public databases (Fig. 5c).
Discussion
The results of the current study demonstrate differential PPGRs in response to a broad array of standardized carbohydrate meals and reflect underlying metabolic physiology, including insulin resistance and beta cell dysfunction. Using standardized carbohydrate meals given in replicates with CGM data captured, we showed that on average, rice was the most glucose-elevating carbohydrate, whereas some individuals exhibited higher elevations in response to consumption of potatoes, pasta, bread or grapes (Extended Data Fig. 3). Individuals with the highest PPGR to potato consumption relative to other carbohydrates (potato-spikers) were more IR with lower beta cell function, whereas grape-spikers were more IS. Furthermore, a high glucose response to potatoes relative to grapes (PG-ratio) was statistically significantly associated with insulin resistance (Fig. 3b and Supplementary Fig. 13). These results indicate that CGM-detected glycemic responses to carbohydrate meals reflect underlying physiology.
The results of this study also highlight that the mitigation effect differs between individuals according to underlying physiology, in that the mitigation effect was only beneficial in reducing delta glucose in the IS or normal beta cell function groups (Supplementary Table 8). The specific mechanisms of mitigator function were not studied in detail in this study, but a potential hypothesis is that mitigators can lower PPGRs by at least two main pathways that work in synergy. The first is decreasing carbohydrate absorption, and the second is through the process of incretin induction, insulin secretion and insulin action38. In healthy individuals, mitigators greatly reduce PPGRs because the two mechanisms work in synergy. However, with insulin resistance or beta cell dysfunction, the single effect of absorption rate reduction may be too small without the synergistic interaction with insulin function.
In addition to explaining the reduced mitigator effect in individuals with insulin resistance and beta cell dysfunction, this hypothesis also explains the elevated PPGRs to potatoes and pasta in those individuals. The potatoes provided were cooked and cooled, increasing the level of resistant starch, a type of fiber, compared with other starch meals, and so it was itself a starch food with fiber (Extended Data Fig. 5)14,39. Pasta, which was also cooked and cooled, contains some resistant starch and also more slowly digestible starch39. Grapes and rice contain lower levels of fiber and thus do not exhibit PPGR differences between individuals with different metabolic traits. Beans have higher levels of fiber and proteins but lower PPGR in general, limiting the possible effect size to be measured. We also observed that protein has the strongest mitigation effect in individuals with beta cell normal function, potentially indicating a stimulation effect on insulin secretion (Fig. 4d)40. Additional factors affecting the PPGRs and mitigation effect might come from individual microbiomes, accessibility of starch in the meal and the interaction between sex and metabolic traits.
Carb-response types are linked to glucose dysregulation subtypes and potential complications. Potato-spikers had significantly higher fasting glucose, insulin resistance and fasting levels of several TAGs and fatty acids. Enriched proteomics pathways such as hepatic stellate cell activation and fibroblast activation (Supplementary Data 1) in potato-spikers might be related to the activation of hepatic stellate cells to myofibroblast-like cells and metabolic dysfunction-associated steatohepatitis (MASH)41. In population-wide T2D studies, the severe IR diabetes group was associated with MASH17. We hypothesize that there is a similar group of individuals with higher potato PPGRs, higher insulin resistance and early signatures of MASH in the stages of prediabetes and even normoglycemia, which needs further validation in larger cohorts. In addition, Asian individuals were more likely to be rice-spikers, which aligns with higher rice glycemic responses among Asian compared to European individuals in the study of glycemic indexes15,16. We note that there may be other confounders in genetics and habits (for example, consuming rice frequently). Bread-spikers exhibited significantly higher systolic and diastolic blood pressure and N1-Methyladenosine, a metabolite associated with hypertension in mice, compared to other carb-response types, suggesting potential CVD risk (Figs. 3c, Supplementary Fig. 19 and Supplementary Table 2). The positive correlation between black bean PPGRs and keto metabolism suggests that individuals with baseline predispositions toward enhanced fatty acid oxidation or amino acid catabolism may exhibit different glucose responses to beans. The variability of histidine metabolism associated with PPGRs to beans might be shaped by differences in gut microbiota composition or habitual diet42. Nonetheless, our results demonstrate that different strategies may be useful for nutrient intervention in groups with different ethnicities and metabolic functions.
Microbiome features were also associated with PPGRs and mitigation effects. GH85 in Roseburia intestinalis was positively associated with PPGRs to all carbohydrate meals (Extended Data Fig. 7). The GH85 family is known to degrade glycosylated proteins, and Roseburia is known to degrade complex carbohydrates (for example, fiber)34,35. Elevated GH85 in Roseburia intestinalis might indicate degradation of mucins when fiber from meals was insufficient. D-alanyl-D-alanine carboxypeptidase was negatively associated with PPGRs to most carbohydrate meals and has a role in cell wall synthesis. Klebsiella (the highest level in Klebsiella pneumonia) and Sutterella (the highest level in Sutterella parvirubra) were significantly negatively (beneficially) associated with the mitigation effect (Supplementary Table 5). Klebsiella is known to reside in the small intestine, and the PPGR reduction might come from its competing usage of glucose43. Sutterella is a known factor in glucose control improvement after gastric bypass, and this might be related to its mitigation effect44. The starch degradation III in Eubacterium hallii was significantly negatively associated with the mitigation effect (Supplementary Table 5). This might indicate a competing usage of starch when the human digestive process was slowed by mitigators. Overall, a myriad of microbial biochemical pathways in different microbes could likely impact PPGRs.
Our work is built on previous PPGR studies and provides a deeper mechanistic understanding of the PPGR. Interindividual PPGR variation and correlation across foods have been discovered in both early small-scale blood-sample-based and later CGM-based studies7,13,45,46. However, early studies were limited to a small variety of standardized meals, so they did not systematically investigate the real-life heterogeneity of PPGRs7,13. Our work demonstrates that PPGRs were more consistent within individuals than between individuals, and each individual can be classified into one carb-response type with different clinical profiles. We also provided a statistical analysis of interindividual and intraindividual variability of PPGRs based on ICCs to a broad scope of carbohydrate meals. In addition, previous cohorts were not comprehensively profiled in metabolic traits. We found associations between PPGRs and blood glucose and omics features, and their relationship to the specific metabolic dysfunctional phenotype, including insulin resistance and beta cell function, enabling a deeper understanding of interindividual variation in PPGRs.
Limitations of the current study include its size of 55 well-phenotyped individuals in California. Although there was a balanced distribution of participants by sex and race, there were insufficient numbers in each race to identify more race-specific patterns. Larger studies could address these potential differences. Furthermore, for practical reasons, we cannot compare more than seven carbohydrates (for example, whole wheat bread, bananas and cooked and cooled versus hot potatoes) and include mitigator tests for all carbohydrates. Our meals also lacked variability in cooking time and solid state versus liquid state. The mitigator effects presented were systematically studied but still cannot be extrapolated to a mixed meal. Rather, we can only conclude about the effect when the mitigator was given 10 min before the rice meal. Although participants were instructed to remain sedentary for three hours post-meal, we did not track physical activity and evaluate the effects. The protocol adherence was closely monitored by the dietitian through food logs, but the meals were consumed in the home setting, so other external variables could affect the results. Our study explored the association between PPGRs and omics and clinical measurements but presented no direct links to hard clinical endpoints47,48. Further studies are needed to quantify the long-term consequences of different PPGR subtypes and intervention effects objectively and directly.
Overall, our results represent an initial step in evaluating the effect of different meals and mitigators on PPGRs and their associations with metabolic physiology, especially because individuals, both healthy and at risk, have increased access to CGMs to optimize health by modifying their diet. Our research also may be relevant for stratification-based approaches that leverage CGM data to cluster individuals based on metabolic health and potentially optimize their diet to prevent progression to T2D and CVD. Our potential approach of stratifying participants into subgroups with relatively homogeneous PPGRs could provide advantages of more precise recommendations than the broad public health recommendation and more accessible data collection than the individualized machine-learning approach. Future studies could enable routine meal recommendations through expanded PPGR profiling with an even broader scope of mitigators in bigger cohorts and novel algorithms.
Methods
Cohort design and participant recruitment
Our study aligns with the NIH definition of Basic Experimental Studies in Humans studies and aims to understand the fundamental process of postprandial responses. Male and female (based on self-report) participants of different ethnicities (based on self-report) were recruited from the San Francisco Bay Area, California. Inclusion criteria were age >18 and <75 years, BMI < 40 kg m−2 and no known diagnosis of diabetes. Exclusion criteria included major organ diseases, uncontrolled hypertension (blood pressure > 160/100 mmHg), pregnancy or lactation, use of diabetogenic medications, malabsorptive disorders (for example, celiac sprue), chronic inflammatory conditions, malignancy not at least five years in remission, alcohol use >2 standard drinks per day, use of weight loss medications or specific diets, weight change of more than 2 kg in the last three weeks and a history of bariatric surgery. Additionally, individuals were excluded if they were deemed to be unable to use the technology correctly, adhere to the study protocol or give written informed consent. Details are available in clinicaltrials.gov (NCT03919877). The first participant consented on 24 May 2018 and the last one on 15 February 2023.
Participants underwent evaluations, screening tests and metabolic tests at the Clinical and Translational Research Unit (CTRU) after an overnight fast. Metabolic tests assessed metabolic health related to T2D and included the oral glucose tolerance test, SSPG test and isoglycemic intravenous glucose infusion (IIGI) test19,20,21,22,49. There were four basic visits: a screening + OGTT visit, an SSPG visit, an IIGI visit and an omics sample collection visit. During the screening and the omics visits, stool, urine, peripheral blood mononuclear cells, plasma and serum samples were collected. Some individuals had multiple omics visits to monitor omics changes throughout the study. Participants were then instructed to complete the at-home diet cycles, during which they collected CGM and food log data.
Ethics approval and consent
The study protocol was reviewed and approved by the Institutional Review Board at Stanford University School of Medicine Human Research Protection Office (Institutional Review Board no. 43883). All participants provided written informed consent.
Standardized carbohydrate meals
Seven standardized carbohydrate meals (50 g carbohydrates each) and three mitigators were included in the study (Supplementary Table 6). We investigated individual responses to different standardized carbohydrate meals and determined whether other meals could mitigate the PPGRs. Among the seven carbohydrate meals, there were five starchy meals and two meals with mostly simple carbohydrates. The five starchy meals were jasmine rice (precooked frozen Trader Joe’s brand), potatoes (shredded, frozen precooked, Trader Joe’s brand), pasta (macaroni, cooked in the CTRU kitchen per package instructions and then frozen), white sliced bread (Trader Joe’s buttermilk bread) and beans (canned whole black beans). The two simple-carbohydrate meals were grapes (red grapes) and mixed berries (170 g raspberries and a mix of blackberries, strawberries and blueberries, about 85 g each). The three mitigators were fiber (pea fiber powder, 14 g providing 10 g of fiber), protein (boiled egg white, 100 g, providing 10 g of protein) and fat (Bellwether Farms Creme Fraiche, 38 g, providing 15 g total fat). The weight of the mitigators was chosen to approximate regular meal sizes.
Participants were instructed to use microwaves to heat the meal at home and consume each on separate days without adding any condiments or other meals. They were to consume the standardized meals as the first meal in the morning, after a 10–12 hour fast, not consume any other food or drink and remain sedentary for 3 hours after consumption of the provided meals. For mitigator tests, participants were instructed to consume the mitigator first, followed by the carbohydrate meal (rice) 10 min later. They were also advised to avoid exercise the afternoon and evening before the test. The starting time point (zero minutes) was the recorded time of consuming the standardized carbohydrate meals in the food log.
Diet cycles
During the diet cycle, participants were instructed to consume the standardized meals as described in the last section while otherwise following their regular diet. Participants were instructed to keep detailed logs of food intake, including the time and portions of all foods and drinks consumed using the ‘Cronometer’ app. A dietitian assisted participants in adhering to the cook and eat instructions. Food logs with specific food and time were collected to evaluate adherence to the assigned instructions.
Two versions of CGM were used during the study period: Dexcom G4 and G6 Pro. For the G4, calibration was needed, requiring participants to measure blood glucose levels by fingerstick with provided glucose meters, glucose-reading strips and a lancet device. Participants were instructed to calibrate before the standardized meals. The G6 Pro required no calibration. Glucose values were blinded to participants until after the study. Participants were instructed to wear the CGM devices continuously throughout the study. The G4 sensor lasted seven days, and the G6 Pro lasted ten days. Trained study personnel inserted the sensor and educated participants on its use and care. Multiple CGM sensors were provided as needed.
Demographic and clinical information
Metadata were downloaded from REDCap and cleaned manually. The data included demographic information, clinical blood test results and responses from health and physical activity surveys19,26. The data were homogenized and manually checked with unreasonable values labeled and removed. Missing clinical values (for example, BMI) were estimated if the corresponding independent variables for calculation existed. If multiple values existed, they were averaged across visits.
Metabolic tests
Participants were instructed to fast for 12 hours before each metabolic test. The tests were conducted on three different days. Details of the metabolic tests are available in previous publications19,26.
The OGTT measured postprandial glycemic response to a standard glucose drink (75 g glucose)19. Blood samples were collected through an antecubital intravenous catheter before and after the glucose drink at −10, 0, 15, 30, 60, 90, 120, 150 and 180 min. Blood glucose was checked at the aforementioned time points and at 10, 20, 40, 50, 75, 105, 135 and 150 min. Baseline levels of insulin, C-peptide, glucagon-like peptide-1, glucose-dependent insulinotropic polypeptide and glucagon were measured.
The two-stage insulin suppression test (called the SSPG test in the paper) quantified insulin suppression of lipolysis (adipose tissue insulin resistance) and insulin-mediated glucose uptake (muscle insulin resistance), as previously described20,21,22,23,49. During the test, participants received an infusion of octreotide (0.27 μg m−2 min−1) for 240 min to suppress endogenous insulin secretion. During the first stage of the test, insulin was infused at 6 mU m−2 min−1 and glucose at 50 mg m−2 min−1, and blood samples were collected at 100, 110 and 120 min for measurement of glucose, insulin and FFA concentrations23. The glucose, insulin and FFA values collected during the last 20 min of the first stage were averaged to calculate SSPG, steady-state plasma insulin and steady-state plasma FFA concentrations. During the second stage of the test, insulin was infused at 32 mU m−2 min−1 and glucose at 267 mg m−2 min−1. Blood samples were drawn at 210, 220, 230 and 240 min to measure the second-stage steady-state plasma insulin and SSPG concentrations.
The IIGI test measured the incretin effect, which is the ability of oral nutrients to stimulate insulin secretion beyond that stimulated by comparable plasma glucose administered intravenously27. It quantified the incretin effect by comparison with OGTT measurements. During the IIGI test, an intravenous catheter was placed in the antecubital vein for administration of continuous dextrose infusion at a rate needed to obtain the desired glucose level to match the curve generated by the OGTT. Blood samples were collected from a second intravenous catheter at the same time points as the OGTT, with immediate results generated by a bedside YSI plasma glucose analyzer and the rate of dextrose infusion adjusted as needed to maintain the desired glucose curve that matched values obtained during the OGTT. Plasma from this test as well as the OGTT was collected for measurement of insulin and C-peptide to calculate the incretin effect.
Although all participants were free of diabetes by self-report, they were stratified for analysis purposes according to HbA1c drawn at their baseline visit and American Diabetes Association criteria as follows: normoglycemia (HbA1c < 5.7%), prediabetes (HbA1c 5.7%–6.4%) and T2D (HbA1c ≥ 6.5%).
SSPG (mg dl−1) measured the level of insulin resistance, with higher values indicating greater insulin resistance20,21,22,23,49. A cutoff at 120 mg dl−1 was used to classify participants into IR and IS groups19,24. This threshold was based on physiology, prospective studies and a natural separation point between clustering observed in our cohort24. An alternative cutoff excluded intermediate results (IR, SSPG > 120; IS, SSPG < 100). Results followed the default cutoff if not otherwise specified.
Beta cell function was expressed as disposition index, reflecting insulin secretion adjusted for insulin resistance. The calculation was by insulin SECretion (ISEC) software, based on C-peptide concentrations measured during OGTT at time points 0, 15 and 30 min and with adjustment for age, sex and BMI19,50. We calculated the disposition index (pmol dl) per (kg ml), defined as the area under the insulin secretion rate divided by the SSPG (as measured during the insulin suppression test). Participants were stratified into normal (disposition index > 2.2), intermediate (1.2 ≤ disposition index ≤ 2.2) and dysfunctional (disposition index < 1.2) beta cell function17.
Incretin effect was calculated based on the difference in C-peptide concentrations between OGTT and IIGI at seven time points (0, 15, 30, 60, 90, 120 and 180 min)17,24,28. A higher incretin effect may indicate either increased secretion of incretin (for example, glucagon-like peptide-1, glucose-dependent insulinotropic polypeptide) and/or increased beta cell responsiveness to incretin. Participants were stratified into normal (incretin effect% > 64), intermediate (39 ≤ incretin effect% ≤ 64) and dysfunctional (incretin effect% < 39) incretin effects. An alternative cutoff excluded intermediate results (dysfunctional, incretin effect% < 40; normal, incretin effect% > 60). Results followed the default cutoff if not otherwise specified.
Hepatic IR was estimated using a surrogate index based on insulin, HDL, BMI, age and sex. Participants were stratified into hepatic IS (index < 3.95), intermediate (3.95 ≤ index ≤ 4.8) and IR (index > 4.8) (refs. 26,28,51). Participants were stratified into adipose IS (FFA ≤ 0.15), intermediate (0.15 < FFA < 0.5) and IR (FFA ≥ 0.5) (ref. 23). We followed previous papers in the calculation of homeostatic model assessment values52. Details of calculations are available in the previous publications19,26.
There were two groups of participants for metabolic tests, including groups 1 and 2. Group 1 had the most metabolic test measurements on most participants and is the one we focused on for metabolic traits. Group-wise comparisons across metabolic subtypes (different insulin resistances, beta cell function and incretin function) were based on group 1 unless otherwise specified, and other correlation analyses are based on all available data. Group 2 had a distribution shift, was mostly IS and had SSPG only among subtype measurements. In group 1 and among those with standardized meal CGM data, we quantified SSPG (n = 30), disposition index (n = 28), incretin effect (n = 28), hepatic IR (n = 28), FFA (n = 22) and homeostatic model assessment insulin-resistance surrogate (n = 28). Group 2 had basic clinical measurements and SSPG (n = 13) and was mostly IS (SSPG > 120, n = 2; SSPG > 130, n = 1).
CGM data processing and quality control
CGM data were analyzed over a period from 25 min before to 170 min after the consumption of standardized carbohydrate meals. The reference time point (0 min) corresponds to the consumption time of the standardized carbohydrate meals, as recorded in the food logs. The CGM data were downloaded from Dexcom devices, processed and integrated with food log data. Standardized meals were identified and selected from the Cronometer food logs. Food names were cleaned and unified for the standardized meals. Duplicate records of identical meals were merged. We only kept those mitigator records correctly following the procedure: (1) rice must be consumed within 30 min after the mitigators; (2) rice must be consumed after the mitigators. Other food intake before and during (2 hours) the standardized meal tests was also recorded. Time shifts from daylight saving were automatically recorded and corrected. CGM data were then merged with food log data.
We kept CGM records with relatively complete data in the time range of interest ([−25,170] minutes) and removed data with more than 30-minute gaps. We then applied a smoothing spline and resampled the data into a uniform time grid of 40 time points.
We manually checked each CGM curve and corrected mistakes in dates and times, including traveling between time zones. CGM curves with mistakes, such as discontinuities after device calibration or an indication of noncompliance, were removed. Traveling between time zones often produced an integer-hour time discrepancy between CGM and food log records, which was corrected by shifting integer hours.
The reproducibility and consistency of PPGRs were assessed based on AUC(>baseline). The glucose response was analyzed from the time of carbohydrate meal consumption until either 180 or 120 min post consumption. Replicate 1 and replicate 2 data for each carbohydrate meal type were extracted, and Pearson correlation coefficients were calculated. ICC was also calculated to evaluate consistency within replicates of the same individuals.
CGM feature extraction and definition of PPGRs
We projected each CGM time series ([0, 170] minutes) into features19. The AUC (mg min dl−1) was calculated using the composite trapezoidal rule. AUC above a certain value (for example, AUC(>baseline) and AUC(>140)) was calculated by shifting the curves to these thresholds and integrating the positive parts. The baseline glucose level was defined as the glucose value with the minimal absolute distance to the zero time point. The peak value was defined as the maximal glucose value in the time range. Delta glucose peak (mg dl−1) was defined as the difference between the peak glucose value and the baseline glucose value. Delta glucose peak at a specific time point (for example, delta glucose peak at 120 min) was defined as the difference between the glucose value at the chosen time point and the baseline glucose value. Time (baseline to peak) (min) was defined as the time difference between the baseline time point and peak value. Time (return to baseline) was defined as the minimal time for the glucose level to decrease to or below the baseline glucose level. If the glucose level had not returned to baseline within 170 min, the time was set as 170 min. Slope (baseline to peak) (mg (dL min)−1) was defined as the division between delta glucose peak value and time from baseline to peak.
Mitigation effects were measured by subtracting the delta glucose peak of rice + mitigator from the delta glucose peak of rice alone and then normalizing by the rice delta glucose peak. A similar quantification of the mitigation effect was also calculated based on AUC(>baseline). The mitigation effect measured the relative suppression effect on glycemic response.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data are shared through supplementary tables, public repositories and an integrated web interface. This Article focused on participants with CGM and, particularly, with complete metabolic testing, a subset of the whole cohort. Only patient data with consent to data sharing were shared. The participant IDs were also reassigned to prevent identification. Metabolomics and lipidomics data were deposited into Metabolomics Workbench with IDs ST003630 and ST003636 (ref. 53). Microbiome data were deposited into NCBI SRA under PRJNA1189566. The affinity proteomics Olink data have been deposited into the PRIDE repository with the dataset identifier PAD000001 (ref. 54), which will be open after publication. Clinical data, CGM data and omics data were also shared in integration through a web interface for the whole cohort (NCT03919877) when consent for data sharing was obtained from the participants. The link is https://cgmdb.stanford.edu/. New types of data will be shared through the web interface when finalized. This manuscript also used datasets/databases including human genome GRCh38 (https://benlangmead.github.io/aws-indexes/bowtie) and metid based database-mapping55. Source data are provided with this paper.
Code availability
The analysis is based on R, Python and MATLAB, and detailed information is provided in the supplementary files. The used packages include Rcpm v.1.0.4, masscleaner v.1.0.6, lipidr v.2.12.0, SummarizedExperiment v.1.28.0, GenomicRanges v.1.50.1, GenomeInfoDb v.1.34.2, MatrixGenerics v.1.10.0, matrixStats v.0.62.0, pathview v.1.38.0, ggfortify v.0.4.15, ComplexUpset v.1.3.3, Rodin v.0.1.43, visNetwork v.2.1.2, gridExtra v.2.3, mediation v.4.5.0, sandwich v.3.0-2, mvtnorm v.1.1-3, Matrix v.1.5-3, doSNOW v.1.0.20, snow v.0.4-4, iterators v.1.0.14, KEGG.db v.2.7.1, MSnbase v.2.24.0, ProtGenerics v.1.30.0, mzR v.2.32.0, Rcpp v.1.0.9, massdataset v.1.0.19, masstools v.1.0.8, metid v.1.2.25, metid v.0.2.0, metpath v.1.0.5, ReactomePA v.1.42.0, org.Hs.eg.db v.3.16.0, AnnotationDbi v.1.60.0, IRanges v.2.32.0, S4Vectors v.0.36.0, Biobase v.2.58.0, BiocGenerics v.0.44.0, clusterProfiler v.4.6.0, ppcor v.1.1, MASS v.7.3-57, igraph v.1.3.5, psych v.2.2.9, reshape2 v.1.4.4, lsr v.0.5.2, circlize v.0.4.15, ggpubr v.0.6.0, ComplexHeatmap v.2.14.0, data.table v.1.14.4, dplyr v.1.0.10, readxl v.1.4.1, npreg v.1.0-9, Hmisc v.5.1-0, tidyr v.1.3.0, ggplot2 v.3.4.0, magrittr v.2.0.3, foreach v.1.5.2, stringr v.1.5.0, caret v.4.4.0, glmnet v.4.4.0, pandas 2.0.3, matplotlib 3.5.1, seaborn 0.11.2, numpy 1.22.3 and scipy 1.7.3. The codes of this study are shared through open-source GitHub (https://github.com/mikeaalv/cgm_meal_manuscript).
References
Bansal, N. Prediabetes diagnosis and treatment: a review. World J. Diab. 6, 296–303 (2015).
Klil-Drori, A. J., Azoulay, L. & Pollak, M. N. Cancer, obesity, diabetes, and antidiabetic drugs: is the fog clearing? Nat. Rev. Clin. Oncol. 14, 85–99 (2017).
Coles, B. et al. Cardiovascular events and mortality in people with and without type 2 diabetes: an observational study in a contemporary multi-ethnic population. J. Diab. Investig. 12, 1175–1182 (2021).
Retnakaran, R. et al. Risk factors for renal dysfunction in type 2 diabetes: U.K. Prospective Diabetes Study 74. Diabetes 55, 1832–1839 (2006).
Klein, R., Klein, B. E., Moss, S. E., Davis, M. D. & DeMets, D. L. The Wisconsin epidemiologic study of diabetic retinopathy. III. Prevalence and risk of diabetic retinopathy when age at diagnosis is 30 or more years. Arch. Ophthalmol. 102, 527–532 (1984).
Callaghan, B. C. et al. Role of neurologists and diagnostic tests on the management of distal symmetric polyneuropathy. JAMA Neurol. 71, 1143–1149 (2014).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Cavalot, F. et al. Postprandial blood glucose predicts cardiovascular events and all-cause mortality in type 2 diabetes in a 14-year follow-up: lessons from the San Luigi Gonzaga diabetes study. Diabetes Care 34, 2237–2243 (2011).
Gallwitz, B. Implications of postprandial glucose and weight control in people with type 2 diabetes: understanding and implementing the International Diabetes Federation guidelines. Diabetes Care 32, S322 (2009).
Hall, H. et al. Glucotypes reveal new patterns of glucose dysregulation. PLoS Biol. 16, e2005143 (2018).
Foster-Powell, K., Holt, S. H. & Brand-Miller, J. C. International table of glycemic index and glycemic load values: 2002. Am. J. Clin. Nutr. 76, 5–56 (2002).
Gannon, M. C. & Nuttall, F. Q. Factors affecting interpretation of postprandial glucose and insulin areas. Diabetes Care 10, 759–763 (1987).
Berry, S. E. et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 26, 964–973 (2020).
Perera, A., Meda, V. & Tyler, R. T. Resistant starch: a review of analytical protocols for determining resistant starch and of factors affecting the resistant starch content of foods. Food Res. Int. 43, 1959–1974 (2010).
Wolever, T. M., Giddens, J. L. & Sievenpiper, J. L. Effect of ethnicity on glycaemic index: a systematic review and meta-analysis. Nutr. Diab. 5, e170 (2015).
Kataoka, M. et al. Glycaemic responses to glucose and rice in people of Chinese and European ethnicity. Diabet. Med. 30, e101–e107 (2013).
Ahlqvist, E. et al. Novel subgroups of adult-onset diabetes and their association with outcomes: a data-driven cluster analysis of six variables. Lancet Diabetes Endocrinol. 6, 361–369 (2018).
Zaghlool, S. B. et al. Metabolic and proteomic signatures of type 2 diabetes subtypes in an Arab population. Nat. Commun. 13, 7121 (2022).
Metwally, A. A. et al. Prediction of metabolic subphenotypes of type 2 diabetes via continuous glucose monitoring and machine learning. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-024-01311-6 (2024).
Shen, S. W., Reaven, G. M. & Farquhar, J. W. Comparison of impedance to insulin-mediated glucose uptake in normal subjects and in subjects with latent diabetes. J. Clin. Invest. 49, 2151–2160 (1970).
Greenfield, M. S., Doberne, L., Kraemer, F., Tobey, T. & Reaven, G. Assessment of insulin resistance with the insulin suppression test and the euglycemic clamp. Diabetes 30, 387–392 (1981).
Pei, D., Jones, C. N., Bhargava, R., Chen, Y. D. & Reaven, G. M. Evaluation of octreotide to assess insulin-mediated glucose disposal by the insulin suppression test. Diabetologia 37, 843–845 (1994).
McLaughlin, T., Yee, G., Glassford, A., Lamendola, C. & Reaven, G. Use of a two-stage insulin infusion study to assess the relationship between insulin suppression of lipolysis and insulin-mediated glucose uptake in overweight/obese, nondiabetic women. Metabolism 60, 1741–1747 (2011).
Yeni-Komshian, H., Carantoni, M., Abbasi, F. & Reaven, G. M. Relationship between several surrogate estimates of insulin resistance and quantification of insulin-mediated glucose disposal in 490 healthy nondiabetic volunteers. Diabetes Care 23, 171–175 (2000).
Bergman, R. N., Ader, M., Huecking, K. & Van Citters, G. Accurate assessment of beta-cell function: the hyperbolic correction. Diabetes 51, S212–S220 (2002).
Park, H. et al. Lifestyle profiling using wearables and prediction of glucose metabolism in individuals with normoglycemia or prediabetes. Preprint at medRxiv https://doi.org/10.1101/2024.09.05.24312545 (2024).
Nauck, M. A., Vardarli, I., Deacon, C. F., Holst, J. J. & Meier, J. J. Secretion of glucagon-like peptide-1 (GLP-1) in type 2 diabetes: what is up, what is down. Diabetologia 54, 10–18 (2011).
Vangipurapu, J. et al. A novel surrogate index for hepatic insulin resistance. Diabetologia 54, 540–543 (2011).
Furman, D. et al. Expression of specific inflammasome gene modules stratifies older individuals into two extreme clinical and immunological states. Nat. Med. 23, 174–184 (2017).
Xiong, W. et al. N1-methyladenosine formation, gene regulation, biological functions, and clinical relevance. Mol. Ther. 31, 308–330 (2023).
Raju, S. C. et al. Microbial-derived imidazole propionate links the heart failure-associated microbiome alterations to disease severity. Genome Med. 16, 27 (2024).
Miao, G. et al. Longitudinal plasma lipidome and risk of type 2 diabetes in a large sample of American Indians with normal fasting glucose: the strong heart family study. Diabetes Care 44, 2664–2672 (2021).
Morze, J. et al. Metabolomics and type 2 diabetes risk: an updated systematic review and meta-analysis of prospective cohort studies. Diabetes Care 45, 1013–1024 (2022).
Wang, Z. et al. Gut microbiota and blood metabolites related to fiber intake and type 2 diabetes. Circ. Res. 134, 842–854 (2024).
Qin, J. et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60 (2012).
Zhou, X. et al. Longitudinal profiling of the microbiome at four body sites reveals core stability and individualized dynamics during health and disease. Cell Host Microbe 32, 506–526.e509 (2024).
Tingley, D., Yamamoto, T., Hirose, K., Keele, L. & Imai, K. mediation: R package for causal mediation analysis. J. Stat. Softw. 59, 1–38 (2014).
Goff, H. D., Repin, N., Fabek, H., El Khoury, D. & Gidley, M. J. Dietary fibre for glycaemia control: towards a mechanistic understanding. Bioact. Carbohydr. Diet. Fibre 14, 39–53 (2018).
Hodges, C. et al. Method of food preparation influences blood glucose response to a high-carbohydrate meal: a randomised cross-over trial. Foods 9, 23 (2020).
Clemmensen, C. et al. Oral L-arginine stimulates GLP-1 secretion to improve glucose tolerance in male mice. Endocrinology 154, 3978–3983 (2013).
Tang, Y., Zheng, S. & Chen, A. Curcumin eliminates leptin’s effects on hepatic stellate cell activation via interrupting leptin signaling. Endocrinology 150, 3011–3020 (2009).
Cuparencu, C. et al. Biomarkers of meat and seafood intake: an extensive literature review. Genes Nutr. 14, 35 (2019).
Kastl, A. J. Jr., Terry, N. A., Wu, G. D. & Albenberg, L. G. The structure and function of the human small intestinal microbiota: current understanding and future directions. Cell Mol. Gastroenterol. Hepatol. 9, 33–45 (2020).
Wang, C. et al. The genus Sutterella is a potential contributor to glucose metabolism improvement after Roux-en-Y gastric bypass surgery in T2D. Diabetes Res. Clin. Pract. 162, 108116 (2020).
Vrolix, R. & Mensink, R. P. Variability of the glycemic response to single food products in healthy subjects. Contemp. Clin. Trials 31, 5–11 (2010).
Vega-Lopez, S., Ausman, L. M., Griffith, J. L. & Lichtenstein, A. H. Interindividual variability and intra-individual reproducibility of glycemic index values for commercial white bread. Diabetes Care 30, 1412–1417 (2007).
Martínez-González, M. A., Gea, A. & Ruiz-Canela, M. The Mediterranean diet and cardiovascular health. Circ. Res. 124, 779–798 (2019).
Brand-Miller, J., Hayne, S., Petocz, P. & Colagiuri, S. Low-glycemic index diets in the management of diabetes: a meta-analysis of randomized controlled trials. Diabetes Care 26, 2261–2267 (2003).
Reaven, G. M. Banting lecture 1988. Role of insulin resistance in human disease. Diabetes 37, 1595–1607 (1988).
Hovorka, R., Soons, P. A. & Young, M. A. ISEC: a program to calculate insulin secretion. Comput. Methods Programs Biomed. 50, 253–264 (1996).
Petersen, M. C., Vatner, D. F. & Shulman, G. I. Regulation of hepatic glucose metabolism in health and disease. Nat. Rev. Endocrinol. 13, 572–587 (2017).
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–D470 (2016).
Perez-Riverol, Y. et al. The PRIDE database at 20 years: 2025 update. Nucleic Acids Res. 53, D543–D553 (2024).
Shen, X. et al. metID: an R package for automatable compound annotation for LC–MS-based data. Bioinformatics 38, 568–569 (2022).
Acknowledgements
We thank all participants, investigators and CTRU nurses and staffs of the CGM1.0 study. We thank A. Chen and L. Stainton for administrative support. This work was supported by NIH/NIDDK grant no. R01 DK110186-01 and a Stanford PHIND award (M.P.S and T.M.). We also acknowledge the support of the Stanford Diabetes Research Center (P30DK116074). This research is also supported by the American Diabetes Association Grant 11-23-PDF-76 (Y.W.). It is also supported by National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health grant no. F32DK126287 to A.W.B. and National Library of Medicine (grant no. 2T15LM007033) for B.E. H.P. was supported by the NIH institutional research training grant no. NIH 2T32HL09804911 and the Stanford Lifestyle Medicine grant. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under grant no. UM1TR004921. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was partially supported by proteomic profiling conducted through NIH/NIDDK grant no. R01 DK114183 (Principal Investigator: Themistocles L. Assimes). This study was also supported by Stanford Center for Clinical and Translational Research and Education: MedTech Pilot Grant. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Y.W., B.E. and A.A.M. conducted the main data analysis. D.P., C.B., A.A.M., Y.W. and H.P. contributed to clinical data curation. D.P., A.A.M., Y.W. and B.E. conducted CGM data curation and quality control. A.A.M., H.P., A.W.B., F.A., B.M. and Y.W. contributed to the sample arrangement for omics collection and assays. A.W.B., D. Bogumil, S.P., Z.S. and D.L. contributed to microbiome data collection. Y.W. and A.W.B. contributed to microbiome data preprocessing. A.A.M. and F.A. collected Olink data. B.M., F.W. and D. Bradley collected metabolomics and lipidomics data. B.M., Y.W., F.W. and D. Bradley processed metabolomics and lipidomics data. Y.W. and D.H. ran the lipidomics analysis. D.P., A.C., C.B., E.A., F.A. and T.M. recruited patients and conducted clinical tests. L.R., Y.W. and D.P. conducted biochemical experiments. Y.W., B.E., A.A.M. and Y.L. conducted omics analysis. Y.W., D.P., A.W.B. and S.M.L. interpreted omics results. T.M. and H.R. provided clinical interpretations. B.E. and Y.W. built machine-learning models. Y.W., B.E., M.P.S., T.M. and D.P. wrote the initial draft with edits and comments from the whole group. Y.W., L.M. and B.E. contributed to visualization. A.A.M., D.P., T.M. and M.P.S. conceptualized and designed the study and coordinated the cohort collection. M.P.S. and T.M. funded the study. G.G., E.S.R. and P.V.R. contributed to the website development implementation. G.G., E.S.R., P.V.R., T.W., L.M. and Y.W. contributed to the data sharing and visualization.
Corresponding authors
Ethics declarations
Competing interests
M.P.S. is a cofounder, scientific advisor and shareholder of Filtricine, Iollo, January AI, Marble Therapeutics, Next Thought AI, Personalis, Protos Biologics, Qbio, RTHM, SensOmics. M.P.S. is a scientific advisor and equity holder of Abbratech, Applied Cognition, Enovone, M3 Helium, Onza. M.P.S. is a scientific advisor and stock option holder of Jupiter Therapeutics, Mitrix, Neuvivo, Sigil Biosciences, WndrHLTH, Yuvan Research. M.P.S. is a cofounder and stock option holder of Crosshair Therapeutics. M.P.S. is an investor in and scientific advisor of R42 and Swaza. M.P.S. is an investor in Repair Biotechnologies. M.P.S. is a cofounder, shareholder and director of Exposomics, Fodsel, InVu Health. M.P.S. is a cofounder and equity holder of Mirvie, NiMo Therapeutics, Orange Street Ventures. A.A.M. is currently an employee of Google. D.P. and T.M. are members of the scientific advisory board of January AI. The other authors declare no competing interests.
Peer review
Peer review information
Nature Medicine thanks Ellen Blaak, Clemens Wittenbecher and Tao Zuo for their contribution to the peer review of this work. Primary Handling Editor: Ming Yang, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Mean curve with confidence interval for each meal.
The x axis is time (minute), and the y axis is glucose level (mg/dL). The blue line is the mean curve of the meal, computed from all participants and replicates. The pink area is the confidence interval calculated by 2 standard errors.
Extended Data Fig. 2 Separation of PPGRs to meals by engineered features.
a: Delta glucose peak (relative peak value to baseline) (mg/dL) and return time (min) for different food combinations. CGM curves are extracted into different features and presented with mean and standard errors (the error bar indicates 2 standard errors). The x axis is different food combinations, and the y axis is the feature value. Bars are ordered by mean value. The number of CGM curves (each dot) is: rice 115, potatoes 92, bread 99, pasta 65, grapes 98, beans 46, mixed berries 53, rice+fiber 64, rice+protein 61, and rice+fat 60. Extracted CGM features are compared between different carbohydrate meals. Each participant was instructed to eat each meal at least twice on two different days. b: the scatter plot for three exemplar meals visualized with 2 relatively independent features. The x axis is time to peak (the slope from baseline to peak, mg/(dL*min)), and the y axis is delta glucose peak. Different colors indicate different meals. Each point is one CGM curve.
Extended Data Fig. 3 Delta glucose of each meal in each participant.
The value was averaged among replicates and normalized by the maximum for that participant. Insulin resistance (insulin resistant IR vs insulin sensitive IS), SSPG, disposition index (DI), and HbA1c were also presented.
Extended Data Fig. 4 Reproducibility of carb-response-type classification for each replicate.
Carb-response-type in this paper was classified based on the average of PPGRs (delta glucose peak) of replicates. Here, we also classified each individual into different carb-response-type based on each single meal replicate (1 and 2) and compared it with the classification based on averaged PPGRs. The x axis is different carb-response-types from the averaged PPGR classification. The y axis indicates the percent of average-based classification to be assigned to the same group based on single replicates. Green indicates that the meal with the highest PPGR corresponding to the carb-response-type is still classified as the highest and red indicates that the original highest meal is classified as the second highest. For example, in participants classified to be bread-spiker by the averaged PPGR, most meal replicates still hold the same classification (bread as the highest PPGR meal) (62%), and a small chunk of meal replicates (15%) classified bread as the second highest meal.
Extended Data Fig. 5 Starch composition of standardized carbohydrate meals.
The starch composition was measured, and the amount of rapidly digestible starch, slowly digestible starch, and resistant starch were quantified (detail in Methods). The boxplot presents the ratio between resistant starch and total starch and the ratio between slow digestible starch and total starch. Asterisks indicate Holm corrected p-value of the t-test (default, two-sided) of the selected pairs, *: pFDR < =0.05, **: pFDR < =0.01. The number of replicates is 3 for each food. The boxplots show the center line as the median and the hinges as the 25th and 75th percentiles. The upper whisker extends from the hinge to the largest value not bigger than 1.5 times the distance between the hinges. Data beyond the whiskers are outliers. pFDR is: potatoes vs rice (left) 0.0014, potatoes vs bread (left) 0.019, pasta vs rice (right) 0.047, and pasta vs bread (right) 0.021.
Extended Data Fig. 6 PCA scatter plot of metabolomics and lipidomics.
Omics samples are colored according to sex (row 1) and hepatic insulin resistant (hepatic IR) states (row 2). The x axis is principal component 1 (PC1), and the y axis is PC2. Color distinguishes different groups, and each point is one sample. Each column is a different omics.
Extended Data Fig. 7 Spearman correlation of microbiome functional unit with PPGRs to standardized meals (delta glucose peak).
Strong microbiome features are selected (pFDR<0.1) from features of GO, KEGG, metacyc pathway, pfam, and reactions. Red indicates positive and blue indicates negative correlation. Asterisks indicate correlation original p-value < 0.05. The partial Spearman correlation was run with additional covariates, sample weight, and two-sided t-tests were run.
Extended Data Fig. 8 Spearman correlation between clinical measurement and metabolomics.
Metabolic features that are significantly associated (pFDR<0.05) with selected T2D-related clinical measurements (A1C, FBG, OGTT @120 mins, SSPG, disposition index (DI), Incretin effect, hepatic insulin resistance (hepatic IR), HOMA IR, adipose IR) are listed in the row. All clinical features are listed in columns. Red indicates positive and blue indicates negative correlation. Asterisks indicate p-value < 0.05 and double asterisks indicate pFDR<0.05. FDR corrections were implemented for each clinical feature. P-values were calculated with asymptotic t approximation with a two-sided test.
Extended Data Fig. 9 Spearman correlation between clinical measurement and lipidomics.
Lipidomics features that are significantly associated (pFDR<0.05) with selected T2D-related clinical measurements (A1C, FBG, OGTT @120 mins, SSPG, DI, Incretin effect, hepatic IR, HOMA IR, adipose IR) are listed in the row. All clinical features are listed in columns. Red indicates positive and blue indicates negative correlation. Asterisks indicate p-value < 0.05 and double asterisks indicate pFDR<0.05. FDR corrections were implemented for each clinical feature. P-values were calculated with asymptotic t approximation with a two-sided test.
Extended Data Fig. 10 Model estimation of SSPG and PG ratio.
a: SSPG estimation model. Left: Root mean squared error (RMSE) of SSPG estimation. The x axis indicates different tested models. Right: Coefficient values of the top predictive features of the cooperative model for SSPG estimation. Coefficients with more than 30% of the absolute value of the maximum features are presented. Positive value contributed to higher SSPG. The Null model used mean as the estimation. b: Model performance and coefficient of the PG (Potatoes/Grapes) ratio estimation model. Each of these models was built 100 times with different train/test splits. The model performance is presented with mean and standard errors (the error bar indicates 2 standard errors).
Supplementary information
Supplementary Information
Supplementary Methods 1–11 and Figs. 1–21.
Supplementary Tables
Supplementary Tables 1–10 with captions and titles in the file.
Supplementary Data 1
Pathway enrichment result for metabolomics, lipidomics and proteomics associated with meal-specific postprandial response. Omics features significantly (P < 0.05) associated with PPGRs (delta glucose peak) to each meal were selected and enriched with GO, KEGG, Reactome and HMDB. The P value was adjusted by FDR for each cluster, within each omic and for each database or ontology (for example, GO pathways enriched from proteins positively correlated with rice PPGRs). Details can be found in the Methods.
Source data
Source Data of Figs. 1–5 and Extended Data Figs. 1–2 and 4–10
Statistical source data.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, Y., Ehlert, B., Metwally, A.A. et al. Individual variations in glycemic responses to carbohydrates and underlying metabolic physiology. Nat Med (2025). https://doi.org/10.1038/s41591-025-03719-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41591-025-03719-2