Abstract
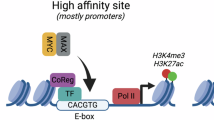
Dysregulation and enhanced expression of MYC transcription factors (TFs) including MYC and MYCN contribute to the majority of human cancers. For example, MYCN is amplified up to several hundredfold in high-risk neuroblastoma. The resulting overexpression of N-myc aberrantly activates genes that are not activated at low N-myc levels and drives cell proliferation. Whether increasing N-myc levels simply mediates binding to lower-affinity binding sites in the genome or fundamentally changes the activation process remains unclear. One such activation mechanism that could become important above threshold levels of N-myc is the formation of aberrant transcriptional condensates through phase separation. Phase separation has recently been linked to transcriptional regulation, but the extent to which it contributes to gene activation remains an open question. Here we characterized the phase behavior of N-myc and showed that it can form dynamic condensates that have transcriptional hallmarks. We tested the role of phase separation in N-myc-regulated transcription by using a chemogenetic tool that allowed us to compare non-phase-separated and phase-separated conditions at equivalent N-myc levels, both of which showed a strong impact on gene expression compared to no N-myc expression. Interestingly, we discovered that only a small percentage (<3%) of N-myc-regulated genes is further modulated by phase separation but that these events include the activation of key oncogenes and the repression of tumor suppressors. Indeed, phase separation increases cell proliferation, corroborating the biological effects of the transcriptional changes. However, our results also show that >97% of N-myc-regulated genes are not affected by N-myc phase separation, demonstrating that soluble complexes of TFs with the transcriptional machinery are sufficient to activate transcription.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The RNA-seq raw data were uploaded to Gene Expression Omnibus database (accession number GSE259300). All other data are available within the text, Supplementary Information and source data section. Source data are provided with this paper.
Change history
12 June 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41594-024-01351-1
References
Baluapuri, A., Wolf, E. & Eilers, M. Target gene-independent functions of MYC oncoproteins. Nat. Rev. Mol. Cell Biol. 21, 255–267 (2020).
Dang, C. V. MYC on the path to cancer. Cell 149, 22–35 (2012).
Meyer, N. & Penn, L. Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 (2008).
Carroll, P. A., Freie, B. W., Mathsyaraja, H. & Eisenman, R. N. The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis. Front. Med. 12, 412–425 (2018).
Cheng, J. M. et al. Preferential amplification of the paternal allele of the N-myc gene in human neuroblastomas. Nat. Genet. 4, 191–194 (1993).
Brodeur, G. M., Seeger, R. C., Schwab, M., Varmus, H. E. & Bishop, J. M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124 (1984).
Kohl, N. E., Gee, C. E. & Alt, F. W. Activated expression of the N-myc gene in human neuroblastomas and related tumors. Science 226, 1335–1337 (1984).
Pugh, T. J. et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet. 45, 279–284 (2013).
Chanthery, Y. H. et al. Paracrine signaling through MYCN enhances tumor-vascular interactions in neuroblastoma. Sci. Transl. Med. 4, 115ra3 (2012).
Murphy, D. J. et al. Distinct thresholds govern Myc’s biological output in vivo. Cancer Cell 14, 447–457 (2008).
Soucek, L. et al. Modelling Myc inhibition as a cancer therapy. Nature 455, 679–683 (2008).
Smith, D. P., Bath, M. L., Metcalf, D., Harris, A. W. & Cory, S. MYC levels govern hematopoietic tumor type and latency in transgenic mice. Blood 108, 653–661 (2006).
Felsher, D. W. & Bishop, J. M. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4, 199–207 (1999).
Zeid, R. et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet. 50, 515–523 (2018).
Choi, J. M., Holehouse, A. S. & Pappu, R. V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 49, 107–133 (2020).
Wheeler, R. J. & Hyman, A. A. Controlling compartmentalization by non-membrane-bound organelles. Philos. Trans. R. Soc. B 373, 20170193–20170199 (2018).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell. Biol. 18, 285–298 (2017).
Hyman, A. A., Weber, C. A. & Jülicher, F. Liquid–liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 30, 39–58 (2014).
Hyman, A. A. & Simons, K. Cell biology. Beyond oil and water-phase transitions in cells. Science 337, 1047–1049 (2012).
Zamudio, A. V. et al. Mediator condensates localize signaling factors to key cell identity genes. Mol. Cell. 76, 753–766.e6 (2019).
Boija, A. et al. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell 175, 1842–1855.e16 (2018).
Chong, S. et al. Imaging dynamic and selective low-complexity ___domain interactions that control gene transcription. Science 361, eaar2555–11 (2018).
Hnisz, D., Shrinivas, K., Young, R. A., Chakraborty, A. K. & Sharp, P. A. Perspective. Cell 169, 13–23 (2017).
Dekker, J. et al. Spatial and temporal organization of the genome: current state and future aims of the 4D nucleome project. Mol. Cell 83, 2624–2640 (2023).
Lyon, A. S., Peebles, W. B. & Rosen, M. K. A framework for understanding the functions of biomolecular condensates across scales. Nat. Rev. Mol. Cell Biol. 22, 215–235 (2021).
Alberti, S. & Hyman, A. A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 22, 196–213 (2021).
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Li, C. H. et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature 586, 440–444 (2020).
Narlikar, G. J. et al. Is transcriptional regulation just going through a phase? Mol. Cell 81, 1579–1585 (2021).
Cai, D. et al. Phase separation of YAP reorganizes genome topology for long-term YAP target gene expression. Nat. Cell Biol. 21, 1578–1589 (2019).
Ahn, J. H. et al. Phase separation drives aberrant chromatin looping and cancer development. Nature 595, 591–595 (2021).
Tuttle, L. M. et al. Gcn4-Mediator specificity is mediated by a large and dynamic fuzzy protein–protein complex. Cell Rep. 22, 3251–3264 (2018).
Kar, M. et al. Phase-separating RNA-binding proteins form heterogeneous distributions of clusters in subsaturated solutions. Proc. Natl Acad. Sci. USA 119, e2202222119 (2022).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Trojanowski, J. et al. Transcription activation is enhanced by multivalent interactions independent of phase separation. Mol. Cell 82, 1878–1893.e10 (2022).
Thody, S. A. et al. Small molecule properties define partitioning into biomolecular condensates. Preprint at bioRxiv https://doi.org/10.1101/2022.12.19.521099 (2022).
Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020).
Kilgore, H. R. et al. Distinct chemical environments in biomolecular condensates. Nat. Chem. Biol. 20, 291–301 (2024).
Chung, C.-I., Yang, J. & Shu, X. Chemogenetic minitool for dissecting the roles of protein phase separation. ACS Cent. Sci. 9, 1466–1479 (2023).
Han, H. et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 36, 483–497.e15 (2019).
Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Brangwynne, C. P., Mitchison, T. J. & Hyman, A. A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc. Natl Acad. Sci. USA 108, 4334–4339 (2011).
Malhis, N., Jacobson, M. & Gsponer, J. MoRFchibi SYSTEM: software tools for the identification of MoRFs in protein sequences. Nucleic Acids Res. 44, W488–W493 (2016).
Andresen, C. et al. Transient structure and dynamics in the disordered c-Myc transactivation ___domain affect Bin1 binding. Nucleic Acids Res. 40, 6353–6366 (2012).
McEwan, I. J., Dahlman-Wright, K., Ford, J. & Wright, A. P. Functional interaction of the c-Myc transactivation ___domain with the TATA binding protein: evidence for an induced fit model of transactivation ___domain folding. Biochemistry 35, 9584–9593 (1996).
Chen, L. et al. p53 is a direct transcriptional target of MYCN in neuroblastoma. Cancer Res. 70, 1377–1388 (2010).
Reisman, D., Elkind, N. B., Roy, B., Beamon, J. & Rotter, V. c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth Differ. 4, 57–65 (1993).
Sabari, B. R. et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361, eaar3958–13 (2018).
Rai, A. K., Chen, J. X., Selbach, M. & Pelkmans, L. Kinase-controlled phase transition of membraneless organelles in mitosis. Nature 559, 211–216 (2018).
Spencer, C. A., Kruhlak, M. J., Jenkins, H. L., Sun, X. & Bazett-Jones, D. P. Mitotic transcription repression in vivo in the absence of nucleosomal chromatin condensation. J. Cell Biol. 150, 13–26 (2000).
Gottesfeld, J. M. & Forbes, D. J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22, 197–202 (1997).
Yu, D. et al. A naturally monomeric infrared fluorescent protein for protein labeling. Nat. Methods 12, 763–765 (2015).
Shu, X. Imaging dynamic cell signaling in vivo with new classes of fluorescent reporters. Curr. Opin. Chem. Biol. 54, 1–9 (2020).
Vagnarelli, P. Mitotic chromosome condensation in vertebrates. Exp. Cell. Res. 318, 1435–1441 (2012).
Nair, S. K. & Burley, S. K. X-ray structures of Myc–Max and Mad–Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 112, 193–205 (2003).
Morin, J. A. et al. Sequence-dependent surface condensation of a pioneer transcription factor on DNA. Nat. Phys. 18, 271–276 (2022).
Zhang, Q. et al. Visualizing dynamics of cell signaling in vivo with a phase separation-based kinase reporter. Mol. Cell 69, 334–345.e5 (2018).
Chung, C. I., Zhang, Q. & Shu, X. Dynamic imaging of small molecule induced protein–protein interactions in living cells with a fluorophore phase transition based approach. Anal. Chem. 90, 14287–14293 (2018).
Jiang, X. et al. Annexin A8 (ANXA8) regulates proliferation of porcine endometrial cells via Akt signalling pathway. Reprod. Domest. Anim. 54, 3–10 (2019).
Zeng, Y. et al. SERINC2-knockdown inhibits proliferation, migration and invasion in lung adenocarcinoma. Oncol. Lett. 16, 5916–5922 (2018).
Zeller, K. I., Jegga, A. G., Aronow, B. J., O’Donnell, K. A. & Dang, C. V. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 4, R69 (2003).
Sabò, A. et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature 511, 488–492 (2014).
Walz, S. et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature 511, 483–487 (2014).
Zenk, F. et al. HP1 drives de novo 3D genome reorganization in early Drosophila embryos. Nature 593, 289–293 (2021).
Chandra, B. et al. Phase separation mediates NUP98 fusion oncoprotein leukemic transformation. Cancer Discov. 12, 1152–1169 (2022).
Shi, B. et al. UTX condensation underlies its tumour-suppressive activity. Nature 597, 726–731 (2021).
Peeples, W. & Rosen, M. K. Mechanistic dissection of increased enzymatic rate in a phase-separated compartment. Nat. Chem. Biol. 17, 693–702 (2021).
Case, L. B., Zhang, X., Ditlev, J. A. & Rosen, M. K. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097 (2019).
Huang, W. Y. C. et al. A molecular assembly phase transition and kinetic proofreading modulate Ras activation by SOS. Science 363, 1098 (2019).
Riback, J. A. et al. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168, 1028–1040.e19 (2017).
Mittag, T. & Pappu, R. V. A conceptual framework for understanding phase separation and addressing open questions and challenges. Mol. Cell 82, 2201–2214 (2022).
Chung, C.-I. et al. Phase separation of YAP-MAML2 differentially regulates the transcriptome. Proc. Natl Acad. Sci. USA 121, e2310430121 (2024).
Mastop, M. et al. Characterization of a spectrally diverse set of fluorescent proteins as FRET acceptors for mTurquoise2. Sci. Rep. 7, 11999 (2017).
Tsutsui, H., Karasawa, S., Okamura, Y. & Miyawaki, A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat. Methods 5, 683–685 (2008).
Jao, C. Y. & Salic, A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA 105, 15779–15784 (2008).
Pillai-Kastoori, L., Schutz-Geschwender, A. R. & Harford, J. A. A systematic approach to quantitative western blot analysis. Anal. Biochem. 593, 113608 (2020).
Farina, A., Faiola, F. & Martinez, E. Reconstitution of an E box-binding Myc:Max complex with recombinant full-length proteins expressed in Escherichia coli. Protein Expr. Purif. 34, 215–222 (2004).
Bremer, A. et al. Quantifying coexistence concentrations in multi-component phase-separating systems using analytical HPLC. Biomolecules 12, 1480 (2022).
Roehrl, M. H. A., Wang, J. Y. & Wagner, G. A general framework for development and data analysis of competitive high-throughput screens for small-molecule inhibitors of protein–protein interactions by fluorescence polarization. Biochemistry 43, 16056–16066 (2004).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Acknowledgements
We thank H. Madhani, B. Huang, V. Ramani and E. Holland for their critical suggestions. This work was supported by National Institutes of Health (NIH) grants R01CA258237, U01DK127421 and R35GM131766 and a Benioff Initiative for Prostate Cancer Research Award (to X.S.); NIH grants P01CA217959, P30CA082103 and U01CA217864, and grants from the Alex Lemonade Stand, St. Baldrick and Samuel Waxman Cancer Research Foundations (to W.A.W.); U01DA052713 and R21DA056293 (to Y.S.); and ALSAC and the St. Jude Research Collaborative on the Biology and Biophysics of RNP granules (to T.M.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
X.S. conceived the project. J.Y., C.-I.C. and L.H. made the constructs. J.Y. performed N-myc phase separation and colocalization with other proteins in cells. C.-I.C. conducted imaging of small-molecule-induced N-myc phase separation and analyzed colocalization with other proteins. C.-I.C. performed and analyzed nascent RNA labeling, RT–qPCR and RNA-seq. J.Y., J.K., X.S. and W.A.W. planned and performed experiments to analyze expression of endogenous N-myc protein in the neuroblastoma cells. H.L. processed RNA-seq data. H.L., H.H., Z.M., C.-I.C, J.Y., Q.Z., X.Y., X.S. and Y.S. analyzed RNA-seq data. A.N. and T.M. designed and analyzed the in vitro experiments. A.N. conducted the in vitro experiments. J.Y., C.-I.C, T.M. and X.S. wrote the paper. All authors contributed to the final draft.
Corresponding author
Ethics declarations
Competing interests
X.S. and W.A.W. are co-founders of Granule Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Structural & Molecular Biology thanks Robert Eisenman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Dimitris Typas, in collaboration with the Nature Structural & Molecular Biology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Immunofluorescence images of Kelly cells treated with DMSO or MYC/MAX dimerization inhibitor MYCi975.
(Left) Kelly cells were incubated with DMSO or MYCi975 (30 μM) for 20 hours, followed by immunostaining with N-myc antibody. Stars (*) indicate representative cells. Triangle (∆) indicates few (that is un-representative) cells with high MYCN expression. Scale bar, 5 μm. (Right) Normalized average N-myc immunofluorescence intensity in DMSO or MYCi975 treated cells. Data are shown as mean ± SD. N = 3 biological replicates.
Extended Data Fig. 2 Neuroblastoma cell lines with various degrees of MYC (N-myc, c-MYC) protein expression.
Western blot analysis of MYC proteins in various neuroblastoma cells.
Extended Data Fig. 3 Estimation of N-myc concentration in the Kelly cells.
The overall procedure is (a) calculation of an mEGFP concentration-brightness standard curve using purified mEGFP protein. (b) Western blot analysis to determine relative amount of N-myc in Kelly vs SH-EP cells. (c) Fluorescence imaging (Hoechst)-based measurement of the nuclear volume of SH-EP and Kelly cells. The N-myc-mEGFP concentration in SH-EP cells is determined by comparing the fluorescence intensity of N-myc-mEGFP versus that of the purified mEGFP. (d) Estimation of the N-myc concentration in the nucleus of Kelly cells. Data are mean + SD (n = 20 cells). Scale bar, 10 μm.
Extended Data Fig. 4 Relationship of N-myc condensate number and size to the N-myc protein levels.
(a) Number of N-myc condensates was plotted against the N-myc protein level per nucleus. SH-EP cells expressing N-myc-mEGFP were imaged under the spinning disc confocal microscope. (b) Size of N-myc condensates was plotted against the N-myc protein level in single nucleus. (c) SH-EP cells with different expressing levels of N-myc-mEGFP. Images are max-projected to show all condensates. Dim cells are shown with brightness adjustment as listed. Scale bar: 5 μm.
Extended Data Fig. 5 Endogenous N-myc condensates contain DNA-binding and dimerization partner, transcriptional machinery and nascent RNA but do not co-localize with c-Jun.
Immunostained N-myc condensates colocalized with MED1 (a), Pol II S5p (b), MAX (c), nascent RNA (d) and c-Jun (e). Kelly cells were stained with antibody against N-myc, and antibodies against MED1, Pol II S5p, or c-Jun, or co-expressed with MAX-mKO3, or labeled by 5-ethynyluridine for labeling nascent RNA. The arrows point to representative condensates. The fluorescence intensity profile (right panel) is extracted from the position shown by the dashed line. Co-localization assessment by Pearson’s correlation coefficient is shown in right panels. Center lines show the median values. Green boxes contain the 25th to 75th percentiles of dataset. Black whiskers mark the 10th and 90th percentiles. Outliers are marked with grey dots. NMED1 = 76, NPol II-S5P = 67, NMAX = 61, Nnascent RNA = 82 cells, Nc-Jun = 67 cells. Scale bars, 5 μm.
Extended Data Fig. 6 Condensate number and size related to protein levels of the truncated N-myc lacking the DNA-binding ___domain.
(a) Number of N-myc1–365 condensates was plotted against the N-myc protein level per nucleus. (b) Size of N-myc1–365 condensates was plotted against the N-myc protein level in single nucleus. (c) SH-EP cells with different expressing levels of N-myc1–365-mEGFP. Images are max-projected to show all condensates. Dim cells are shown with brightness adjustment as listed. Scale bar: 5 μm.
Extended Data Fig. 7 MAX contributes to N-myc phase separation.
(a) MAX was knocked out by sgRNA in SH-EP cells expressing MYCN-mEGFP. Top left: SPARK value against N-myc concentration in wild type SH-EP cells expressing MYCN-mEGFP. Top middle: SPARK value against N-myc concentration in MAX-KO SH-EP cells expressing MYCN-mEGFP. Lower left: western blot showing MAX protein level in wild type (WT) and MAX-KO (sgMAX) SH-EP cells. Lower middle: SPARK value in MAX-KO SH-EP cells expressing MYCN-mEGFP and MAX-mKO3. Right panel: N-myc saturation concentration in WT, MAX-KO and rescued SH-EP cells. The MAX-KO SH-EP cells were rescued by expressing MAX-mKO3. Center lines show the median values. Boxes contain the 25th to 75th percentiles of dataset. Whiskers mark the minimum and maximum values. NWT = 40, NsgMAX = 49, NsgMAX+MAX-mKO3 = 54. P value, two-sided non-paired t-test. (b) Left: SPARK value against N-myc concentration in SH-EP cells expressing MYCN-mEGFP. Right: SPARK value against N-myc concentration in SH-EP cells expressing MYCN-mEGFP and MAX-mKO3.
Extended Data Fig. 8 Phase separation of full-length N-myc protein is enhanced by DNA oligonucleotides containing non-canonical Myc E-box sequences.
(a) Fluorescence anisotropy assay showing binding of the N-Myc / MAX heterodimer to non-canonical E-box DNA in 20 mM HEPES, 150 mM KCl, 10 mM MgCl2, 1 mM DTT, and 0.01% NP-40 containing buffer. The concentration of fluorophore-labeled oligonucleotide DNA containing 1 NCE-box sequence (CATCTG or CATATG) was fixed at 50 nM and a ratio of Myc / Max of 3:1 was used. The dissociation constant (Kd) of the interaction between proteins and DNA was determined by fitting the experimental data in Origin Pro 8.0 {Roehrl, 2004 #1}. MAX/CEbox and N-myc/MAX/Cebox data are the same as in Fig. 4b. (b) Brightfield microscopy images of solutions containing the N-myc / Max complex (10 µM Myc / 3 µM Max) in the presence of 1NCEbox (26-mer), and 7NCEbox (182 mer) DNA in 20 mM HEPES, 150 mM KCl, 10 mM MgCl2, 1 mM DTT. Purple boxes highlights the images with N-myc phase separation observed. The images show similar phase separation as with canonical E-box sequences. The scale bar is 5 μm.
Extended Data Fig. 9 Saturation concentration of SparkDrop-tagged N-myc and the stable N-myc/SparkDrop SH-EP cells used for the RNAseq experiments.
(a) Saturation concentration of SparkDrop-tagged N-myc with and without lenalidomide. Each green dot represents data from a single cell (~200 cells for each group). The concentration was calculated based on the green fluorescence of mEGFP in SparkDrop. (b) Representative images of stable SparkDrop-MYCN cells before and 30 minutes after 1 µM lenalidomide (lena) incubation. Scale bar: 20 µm. (c) N-myc concentration is 0.20 ± 0.01 µM (Mean ± SD). N = 200 cells.
Extended Data Fig. 10 RT-qPCR of SERINC2 mRNA in SH-EP cells expressing N-myc/SparkDrop treated with 1 µM lenalidomide for different duration.
Data are shown as mean ± SD. N = 3 biological independent replicates. P value, two-sided non-paired t-test.
Supplementary information
Supplementary Information
Supplementary Figs. 1–13 and Supplementary Table 1.
Supplementary Table 2
Supplementary Table 2: List of DEGs from RNA-seq.
Supplementary Data 1
Source data for Supplementary Figs. 1–13.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6i
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3b
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7a
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, J., Chung, CI., Koach, J. et al. MYC phase separation selectively modulates the transcriptome. Nat Struct Mol Biol 31, 1567–1579 (2024). https://doi.org/10.1038/s41594-024-01322-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41594-024-01322-6
This article is cited by
-
Super-enhancers in tumors: unraveling recent advances in their role in Oncogenesis and the emergence of targeted therapies
Journal of Translational Medicine (2025)
-
Emerging roles of transcriptional condensates as temporal signal integrators
Nature Reviews Genetics (2025)
-
Biomolecular condensates in immune cell fate
Nature Reviews Immunology (2025)
-
Transcription regulation by biomolecular condensates
Nature Reviews Molecular Cell Biology (2025)
-
Targeting a key disulfide linkage to regulate RIG-I condensation and cytosolic RNA-sensing
Nature Cell Biology (2025)