Abstract
Recent research highlights the significant impact of methionine metabolism on glioma progression. An increasing amount of compelling evidence bridges long non-coding RNAs to abnormal metabolism in gliomas. However, the specific role of long non-coding RNAs in methionine metabolism regulating glioma progression remains unclear. This study reveals that methionine deprivation inhibits the proliferation, migration, and invasion capabilities of gliomas. Interestingly, the expression of TP53TG1, a long non-coding RNA, is also suppressed. TP53TG1 is highly expressed in gliomas and associated with poor patient outcomes. Subsequently, our data proves that inhibition of TP53TG1 suppresses glioma cell proliferation and the epithelial-mesenchymal transition process both in vitro and in vivo. Ultimately, we found that the underlying mechanism involves a competing endogenous RNA regulating network, in which TP53TG1 modulates the target protein STK17B by competitively binding to miR-96-5p, thus regulating glioma progression. These findings suggest that targeting methionine deprivation could be a promising approach for the clinical treatment of glioma.
Similar content being viewed by others
Background
Gliomas, common malignant tumors in the central nervous system, pose a grim prognosis for patients, with current treatments showing limited efficacy. Consequently, there is a growing acknowledgment of the potential in targeting glioma metabolism as an innovative therapeutic approach1,2. Characterized by rapid proliferation and dysregulated metastasis, tumor cells demand a substantial energy supply, compelling them to modify their metabolic patterns, notably in glycolysis, the pentose phosphate pathway, and amino acid metabolism3. Studies indicate that abnormal metabolism of glutamine, asparagine, lysine, and other amino acids hold significant importance in driving glioma progression4,5,6. For example, glutamine is absorbed and metabolized in significant quantities, subsequently providing energy and essential materials for the proliferation of glioma cells7,8. Moreover, asparagine has been reported to confer resistance to apoptosis in tumor cells4. Within the glioma microenvironment, the abnormal metabolism of arginine and tryptophan contributes to the formation of an immunosuppressive environment9,10,11. Methionine, an essential amino acid, plays a crucial role in various tumors, including gliomas. Substituting external methionine with homocysteine in the environment inhibits tumor cell proliferation, causing cell cycle blockage and apoptosis, a phenomenon known as the Hoffman effect12,13. This observation hints at the possibility that limiting methionine uptake could impede glioma advancement, although the precise mechanisms remain unclear.
Long non-coding RNAs (lncRNAs), RNA transcripts exceeding 200 nucleotides in length, have recently emerged as crucial players in intricate regulating network, influencing both transcriptional and post-transcriptional regulation processes14,15,16. Moreover, they are closely associated with glioma progression, impacting proliferation, migration, invasion, or metabolic reprogramming of glioma cells. LncRNAs exert their effects in glioma through various mechanisms, including the induction of mRNA instability, participation in competing endogenous RNA networks, interaction with proteins to regulate their functions, and facilitating intercellular communication via exosome release17. For instance, LINC00606 regulates glioblastoma progression by sponging miR-486-3p and targeting TCF1218. Furthermore, lncRNAs can also influence related signaling pathways in glioma, such as the PI3K/Akt/mTOR pathway, MAPK pathway, Wnt/β-catenin pathway, and others19. LncRNA NCK1-AS1 has been reported to activate the Wnt/β-catenin pathway, promoting tumorigenesis in glioma20. Recent studies have confirmed that lncRNAs are involved in methionine metabolism in a variety of tumors, but most studies concentrate on investigating the role of lncRNAs as upstream regulators by influencing methionine or its metabolites, including S-adenosylmethionine (SAM), S-adenosylhomocysteine (SAH), or genome-wide methylation levels. For example, LINC00662 and lncRNA SNHG6 have been shown to regulate the progression of hepatocellular carcinoma by modulating methionine metabolism21,22. However, there is limited research on whether methionine metabolism can regulate tumor progression, particularly gliomas, through its effects on lncRNAs which serve as downstream regulators.
Recently, Salmena et al. proposed a novel hypothesis regarding competing endogenous RNA (ceRNAs), which encompasses circRNAs, lncRNAs, and mRNAs capable of binding to miRNAs via microRNA response elements (MREs) possessing identical or similar complementary sequences23. This interaction facilitates the communication between previously unrelated transcriptional samples to communicate, using the “language of RNAs” MREs24. LncRNAs can function as ceRNAs by competitively absorbing miRNAs in gliomas. Consequently, the inhibitory effect of miRNAs on downstream target mRNAs is rescued, affecting glioma progression25,26,27. However, further investigation is required to ascertain whether lncRNAs functioning as ceRNAs are involved in methionine metabolism and influence glioma progression.
This study aims to explore the impacts of methionine deprivation on glioma progression and the associated molecular mechanisms. Here, our findings demonstrate a substantial inhibitory impact on glioma progression both in vitro and in vivo following methionine deprivation. Further analysis unveils the regulatory mechanism of methionine metabolism in glioma progression, implicating lncRNA TP53TG1 acting as a ceRNA by sponging miR-96-5p, thereby modulating the expression of STK17B, which in turn affects glioma proliferation and epithelial-mesenchymal transition (EMT) process. These results suggest that a methionine-deprived diet holds promise as a potential therapeutic strategy for glioma treatment. Additionally, lncRNA TP53TG1 may serve as a promising prognostic marker and a novel therapeutic target for patients with glioma.
Methods
Tissue samples
This study used 20 pairs of low-grade gliomas, glioblastoma multiforme, and their corresponding adjacent tissue samples obtained from patients undergoing surgical resection at the First Affiliated Hospital of Harbin Medical University in Harbin, China. All tissue samples were promptly frozen in liquid nitrogen upon collection and stored at -80 °C. Ethical approval was acquired from the ethics committee of the First Affiliated Hospital of Harbin Medical University and adhered to the principles of the Declaration of Helsinki, and informed consent was obtained.
Cell lines and cell culture
The human glioblastoma cell lines U251 and LN229 and normal human astrocyte cell lines were purchased from the China Infrastructure of Cell Line Resource (National Science & Technology Infrastructure). The normal group cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (BOSTER, China), whereas the methionine-deprived group cells were cultured in customized DMEM without methionine (BOSTER, China), all containing 10% fetal bovine serum (Sigma, USA) and 1% penicillin–streptomycin solution (Cat#SC118-01, Seven, Beijing, China). Cells were maintained in an incubator (Thermo, USA) with 5% CO2 and a temperature of 37 °C.
Dual-luciferase assay
The binding sites for TP53TG1 and STK17B with miR-96-5p were predicted using RNAhybrid and starBase 3.0. The Wild type and Mutant 3′UTR sequences of TP53TG1 and STK17B were designed and inserted into the luciferase reporter GV272 vector (Shanghai Genechem Co., Ltd). These plasmids were then co-transfected with miR-Nc or miR-96-5p Mimics into 293 T cells. The luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, USA) on an enzyme meter (Tecan Infinite).
Gene set enrichment analysis (GSEA)
GSEA was used to explore the relationship between TP53TG1 and STK17B with the EMT process. The cluster profile package obtained from https://www.gsea-msigdb.org was used for this analysis.
Cell transfection
LN229 and U251 cells were transfected with Lipofectamine 8000 reagent (Cat#C0533, Beyotime, China). The small interfering RNAs (siRNAs) (si-TP53TG1#1,2, si-STK17B#1,2), overexpression plasmids (oe-TP53TG1, oe-STK17B), miR-96-5p inhibitor, mimics, and negative controls, were purchased from China General BIOL (Anhui) Co., Ltd. Lentiviruses shNC (negative control) and shTP53TG1 (TP53TG1 knockdown) were used to establish stable glioma cell lines obtained from Shanghai Genechem Co., Ltd. Transfections were conducted in accordance with the instructions provided in the manual. The sequences mentioned above are displayed in Supplementary Table 1.
qRT-PCR
Glioma tissue samples and cells’ total RNA were extracted by TRIzol reagent (Cat#T9424, Sigma, USA). cDNA synthesis was conducted with the Transcriptor cDNA Synthesis Kit (Cat#4897030001, Roche, Switzerland). Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed by applying an ABI Prism 7500 rapid thermocycler (Applied Biosystems, USA) in conjunction with SYBR Green Master (Cat#4913914001, Roche, Switzerland) as per the standard protocol. GAPDH and U6 SnRNA (small nuclear RNA) served as control genes. Primers used were customized by China General BIOL (Anhui) Co., Ltd., and their sequences are provided in Supplementary Table 2.
MTT assay and Colony formation assay
Cell viability was evaluated using MTT reagent (Cat#HY-15924, MCE, USA). Approximately 2000 glioma cells were seeded in each well of a 96-well plate containing 100 µL of culture solution. After 24 h, the cells were treated with relative siRNAs or overexpression plasmids for 48 h. Subsequently, an MTT solution was added following standard protocols. After a 4 h incubation, 150 µL of DMSO was added to each well after removing the supernatant. Finally, the absorbance was detected at 490 nm using an enzyme meter (TeCan Sunrise).
For colony formation assay, transfected glioma cells were cultured at a density of 500 cells per well in 6-well plates with sufficient culture medium and incubated to form colonies. After 14 days, washed the cells with phosphate buffer saline (Cat#SC106-01, Seven, Beijing, China), fixed with 4% paraformaldehyde (Cat#SI101-01, Seven, Beijing, China), and stained with 0.1% crystal violet (Cat#C0121, Beyotime, China). The number of colonies was calculated using ImageJ software (version 1.54 d, National Institutes of Health).
EdU assay
The EdU Kits (Cat#C0071S, Beyotime, China) were utilized to evaluate glioma cell proliferation. LN229 and U251 cells were cultured on a cover glass in 6‐well plates and transfected with the respective siRNAs or overexpression plasmids for 48 h. Subsequently, EdU reagent was added per well follow the instruction. Then fixed the cells with 4% paraformaldehyde and permeabilized with Triton X-100 (Cat#P0096, Beyotime, China). After staining with the EdU reaction mixture, the cell nuclei were labeled using mounting medium with DAPI (Cat#AB104139, Abcam, UK). EdU-positive cells were observed by the fluorescence microscope (Leica, Germany).
Transwell assay
The migration and invasion abilities of glioma cells were evaluated by using Transwell chambers (Cat#TCS003024, JET BIOFIL, China) coated with or without Matrigel (Cat#356234, Corning, USA). Approximately 5 × 104 cells/200 µL of serum-free DMEM were added to the upper chamber, whereas 500 µL DMEM mixed with 10% fetal bovine serum was added to the lower chamber. After 48 h, cells that moved to the other side of the membrane were fixed and stained. The cells were observed by microscope, and images were captured.
Western blot analysis
Glioma tissue and cell proteins were extracted using RIPA Lysis Buffer (Cat#SW104-02, Seven, Beijing, China), and the protein concentration was determined with Enhanced BCA Protein Assay Kit (Cat#P0010, Beyotime, China). Protein extraction and western blot assays were conducted according to the protocols outlined in our previous study28. Protein bands were visualized and recorded using the GeneGnome XRQ Imaging System (Syngene, UK), and protein expression intensity was quantified employing ImageJ software (version 1.54 d, National Institutes of Health). Primary antibodies used were anti-GAPDH (Cat#BM1623, BOSTER, China), anti-E-cadherin (Cat#20874-1-AP, proteintech, China), anti-N-cadherin (Cat#22018-1-AP, proteintech, China), anti-vimentin (Cat#10366-1-AP, proteintech, China), anti-STK17B (Cat#2294, Cell Signaling TECHNOLOGY, USA), and secondary antibodies used were HRP Goat Anti-Rabbit IgG (Cat#AS014, ABclonal, China) and HRP Goat Anti-Mouse IgG (Cat#AS003, ABclonal, China).
Animal model
Female nude mice (4–6 weeks) with weight of 15–20 g were purchased from Liaoning Changsheng Experimental Animal. Technology Co. Ltd (China) and housed in a specific pathogen-free environment at 23–25 °C. Subcutaneous and orthotopic xenograft nude mouse models were established to evaluate glioma cell growth in vivo. For the orthotopic xenograft model, the mice were randomly divided into two groups for LV-shNC (n = 6) and LV-shTP53TG1 (n = 6). The mice were anesthetized via intraperitoneal injection. Then, the lentivirus transfected stable LN229 cells (1 × 106) in serum-free DMEM (6 µL) were implanted into the mice brain. The injection site was 2.5 mm to the right of the sagittal suture, 0.5 mm posterior to the coronal suture, and 3.5 mm below the skull. Tumor size was assessed using magnetic resonance imaging after 3 weeks, and the mice were then monitored until death, with survival curves plotted accordingly. Tumor volumes in magnetic resonance imaging were measured by ImageJ software and MicroDicom. For the subcutaneous xenograft model, 1 × 106 LN229 cells were subcutaneously injected into the right armpit of the mice. After 2 weeks, the mice were randomly divided into two groups for normal (n = 4) or methionine deprivation (n = 4) feeding. Tumors were removed and measured after an additional 2 weeks. Mice were euthanized by cervical dislocation following anesthesia. The selection of our sample size was consistent with previous report from our group which was mentioned in Western blot section. No animals or data points were excluded from this study, and data from three of the most representative MRI images were selected for statistical analysis. All animal experiments were conducted in a blinded manner and were approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University. These experiments complied with relevant laws and regulations and adhered to the ARRIVE guidelines.
Immunohistochemical (IHC) staining
Glioma tissues were obtained from orthotopic xenograft nude mouse models, and tissue sections were stained for Ki-67 (Cat#A16919, ABclonal, China), E-cadherin (Cat#20874-1-AP, Proteintech, China), and vimentin (Cat#10366-1-AP, Proteintech, China). All procedures were performed according to the standard IHC staining protocols29.
Microarray analysis
Differentially expressed lncRNAs in glioma cells following methionine deprivation were detected and analyzed using the Arraystar Human LncRNA microarray (KangChen Bio-tech, Shanghai. China). The data were subsequently normalized.
Statistical analysis
Data were obtained from at least three independent replications. Students’ t-tests or one-way analyses of variance (Prism software version 9.5.1) were used to analyze differences among groups. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, in the graphs. All experiments were repeated more than three times.
Results
Methionine deprivation exerts inhibitory effects on glioma in vitro and in vivo
We cultured LN229 and U251 glioma cells in regular and methionine-deprived mediums for 48 h to investigate the effect of methionine deprivation on glioma cells. Compared with the group cultured in a normal medium, the viability of glioma cells cultured in a methionine-deprived medium for 48 h showed a significant reduction (Fig. 1A). Methionine deprivation’s impact on glioma cell proliferation was confirmed using a colony formation assay. A significant decrease in colony counts of LN229 and U251 cells cultured under methionine-deprived conditions was observed (Fig. 1B). Additional confirmation was attained using the EdU assay, which exhibited a notably lower proportion of green fluorescence (indicating cell proliferation activity) in the methionine-deprived group than in the normal group (Fig. 1C). The transwell assay demonstrated that the malignant migration and invasion abilities of glioma cells were weakened after methionine deprivation (Fig. 1D). To validate these findings in vivo, we established an ectopic planting model in nude mice by inoculating them with LN229 glioma cells. The glioma tissues of the methionine-deprived diet group exhibited evidently smaller volume and lower weight than those of the normal feeding group, confirming the earlier in vitro results (Fig. 1E).
A Cell viability of glioma cells is assessed using the MTT assay after culturing with methionine-deprived medium for 48 h. B The size and number of colonies are observed using colony formation experiments after culturing glioma cells in methionine-deprived medium. C EdU assay is used to examine cell proliferation after culturing glioma cells in methionine-deprived medium for 48 h. The scale bar is 100 μm. D Transwell assay is used to assess the migration and invasion ability after culturing LN229 and U251 cells in methionine-deprived medium. The scale bar is 100 μm. E Tumor volume and weight of normal or methionine deprivation feeding group nude mice. Error bars represent mean ± standard deviation (SD). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Methionine deprivation affects the lncRNA TP53TG1/miR-96-5p/STK17B ceRNA network in glioma
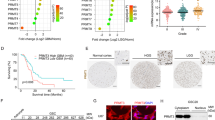
We investigated the underlying mechanisms involved based on the experimental results. LncRNAs is a series of RNA molecules involved in the glioma metabolic pathways. We hypothesized that lncRNAs may contribute significantly to the effects of methionine deprivation on glioma; thus, we analyzed the expression levels of lncRNAs through RNA sequencing after culturing U251 glioma cells in normal or methionine-deprived medium for 48 h. A total of 99 lncRNAs exhibited reduced expression levels following methionine deprivation (p < 0.05, |log2FC |≥1). We identified two intersecting genes, TP53TG1 and LINC00461, by overlapping the 99 downregulated lncRNAs with the 132 upregulated lncRNAs observed in glioma tissues from The Cancer Genome Atlas (TCGA) database (Fig. 2A). Next, the overall survival times associated with these two genes were analyzed using the GEPIA2 database30. Patients with high expression of TP53TG1 exhibited relatively short survival times, whereas the opposite result was observed for LINC00461 (Fig. 2B). Additionally, TP53TG1 showed increased expression levels in glioma tissues and cell lines (Supplementary Figure 1A).
A Two intersecting genes (TP53TG1 and LINC00461) are obtained. B Kaplan–Meier survival curves are generated in patients with high and low TP53TG1 or LINC00461 expression in glioblastoma multiforme and low-grade gliomas obtained from the TCGA databases. C A network diagram representing associations was constructed. D, E The differential expression and correlation analyses of the 68 mRNAs. F The protein levels of STK17B in normal and high-grade glioma patient tissues. G GSEA reveals high TP53TG1 and STK17B expression levels associated with epithelial mesenchymal transition (EMT). H qRT-PCR employed to assess the relative expression levels of TP53TG1 and miR-96-5p after 48 h of methionine deprivation. I The STK17B protein level after 48 h of methionine deprivation in LN229 and U251 cells. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
As lncRNAs exhibit different regulatory modes based on their subcellular localization, we predicted the localization of TP53TG1 using several websites, including lncATLAS, iLoc-lncRNA, lncLocator, and RNALocate v2.0. All analyses consistently indicated that TP53TG1 was predominantly localized in the cytoplasm rather than in the nucleus31,32,33,34 (Supplementary Figure 1B). Given that cytoplasmic lncRNAs typically exert their regulatory roles through the ceRNA network, we hypothesized that TP53TG1 may function similarly. We searched for mRNAs and miRNAs associated with TP53TG1 in the ceRNA network to validate this hypothesis. We used miRcode, starBase, miRDB, MiRTarBase, and TargetScan websites to predict 2 miRNAs and 68 mRNAs associated with TP53TG135,36,37,38,39 (Fig. 2C). Subsequently, we performed a differential expression analysis for these 68 mRNAs and identified STK17B as the mRNA exhibiting upregulated expression in glioma tissues with the largest differential value. This finding was corroborated by western blot analysis results. Correlation analysis also revealed a robust association between STK17B and TP53TG1 (Fig. 2D–F, Supplementary Fig. 1C, D). To further confirm TP53TG1 and STK17B as target genes, we performed a GSEA, which indicated a positively correlated between TP53TG1 and STK17B with EMT process (Fig. 2G). EMT refers to a process in which cells derived from epithelial tissue transition to a mesenchymal phenotype, resulting in enhanced migratory and invasive capabilities of the cells40. This finding aligns with earlier observations of altered migration and invasion abilities of glioma cells following methionine deprivation. After identifying the lncRNAs and mRNA in the relevant ceRNA network, we focused on the sole miRNA, miR-96-5p, which TP53TG1 and STK17B simultaneously adsorb.
Subsequently, we identified the expression levels of these three genes in glioma cells under methionine deprivation. qRT-PCR results revealed decreased TP53TG1 and increased miR-96-5p expression levels, while western blot experiments validated a reduction in STK17B protein levels, aligning with our previous predictions (Fig. 2H, I, Supplementary Fig. 1E).
Validation of the lncRNA TP53TG1/miR-96-5p/STK17B ceRNA network mechanism
To further confirm the establishment of the ceRNA network, we designed the overexpression plasmid and two siRNAs to upregulate or downregulate TP53TG1’s expression in glioma cells (Supplementary Fig. 1F, G). We observed that the protein level of STK17B was increased while the expression level of miR-96-5p decreased when TP53TG1 was overexpressed. Contrasting results were observed when TP53TG1 expression was downregulated (Fig. 3A–D, Supplementary Fig. 2A, B). Furthermore, following the addition of miR-96-5p inhibitor or mimics (Supplementary Fig. 1H), the protein level of STK17B was upregulated or downregulated (Fig. 3E, F, Supplementary Fig. 2C, D). Moreover, after knocking down TP53TG1, adding a miR-96-5p inhibitor resulted in a rescue effect on reducing STK17B protein levels (Fig. 3G, Supplementary Fig. 2E). Dual luciferase reporter assays’ results revealed that miR-96-5p overexpression inhibited luciferase expression levels of the TP53TG1-WT and STK17B-WT groups (Fig. 3H). These data indicate that lncRNA TP53TG1 competitively absorbs miR-96-5p to regulate STK17B as part of a ceRNA network in gliomas.
A, B TP53TG1 is either overexpressed or knocked down in glioma cells, and protein level of STK17B is examined. C, D The expression level of miR-96-5p determined following TP53TG1 overexpression or knockdown. E, F Protein level of STK17B is assessed after adding miR-96-5p inhibitor or mimics. G The protein level of STK17B is assessed using western blot. H Binding sites are predicted, and wild-type and mutant luciferase vectors constructed. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
TP53TG1 promotes glioma proliferation and EMT process in vitro and in vivo
To investigate the specific impact of TP53TG1 on glioma progression, we altered its expression by either overexpressing or knocking down TP53TG1 in glioma cells. The MTT assay results indicated that the viability of glioma cells, with overexpressed or knocked down TP53TG1, was significantly enhanced or reduced, respectively (Fig. 4A, B). The colony formation and EdU assays revealed that TP53TG1 knockdown decreased colony counts and reduced the proportion of green fluorescence, while TP53TG1 overexpression increased them (Fig. 4C–F, Supplementary Fig. 3A–D). Moreover, glioma cells’ migration and invasion abilities were similarly enhanced when TP53TG1 was overexpressed but decreased when TP53TG1 was knocked down (Fig. 4G–H, Supplementary Fig. 3E–H). To further investigate the association between TP53TG1 and EMT, we analyzed the protein levels of relevant EMT markers, including N-cadherin, E-cadherin, and vimentin. As shown in Fig. 4I–J and Supplementary Fig. 3I–N, TP53TG1 overexpression resulted in upregulated expression of N-cadherin and vimentin but downregulated expression of E-cadherin. Conversely, the results were opposite when TP53TG1 was knocked down.
A, B Cell viability of LN229 and U251 cells is assessed using the MTT assay. C–F Glioma cell proliferation is evaluated using the colony formation and EdU assays. The scale bar represents 100 μm. G, H Glioma cell migration and invasion abilities are determined using the transwell assay. The scale bar represents 100 μm. I, J The protein levels of EMT markers (N-cadherin, E-cadherin, and vimentin) in LN229 glioma cells when TP53TG1 is either overexpressed or knocked down. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To further investigate its effects in vivo, we established nude mice xenograft models by injecting LN229 cells transfected with lentivirus LV-shNC or LV-shTP53TG1. Tumor growth in the LV-shTP53TG1 group was significantly inhibited compared to that in the LV-shNC group, resulting in prolonged survival time (Fig. 5A–D). IHC analysis of intracranial tumor tissues in nude mice revealed that the LV-shTP53TG1 group exhibited lower expression levels of Ki67 and vimentin but higher expression levels of E-cadherin than the LV-shNC group (Fig. 5E). These results suggest that TP53TG1 facilitates the proliferation of glioma and promotes the EMT process in vitro and in vivo.
A LN229 cells transfected with lentivirus LV-shNC and LV-shTP53TG1, which carry the GFP fragment, are observed under a fluorescence microscope. B qRT-PCR validates TP53TG1 expression level in LN229 cells after transfection with lentivirus LV-shNC and LV-shTP53TG1. C, D Representative MRI scans, quantitative data and survival curves are presented. Scale bar represents 2 mm. The nude mice are divided into two groups: those injected with LN229 cells transfected with lentivirus LV-shNC (median survival time: 25.5 days) and those with LV-shTP53TG1 (median survival time: 41.5 days) (p = 0.0005; data from six animals/group). E IHC staining results for Ki67, vimentin, and E-cadherin from mice glioma tissues. Scale bar represents 100 μm. Error bars indicate mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
miR-96-5p restrains glioma proliferation and inhibits the EMT process
To assess the effect of miR-96-5p on glioma, LN229 and U251 cells were added with miR-96-5p mimics or inhibitor. Cell viability notably increased in the inhibitor group while significantly decreasing in the mimics group, as demonstrated by the MTT assay (Fig. 6A, B). Moreover, colony formation and EdU assays demonstrated that cell proliferation was increased when miR-96-5p levels were reduced but inhibited when miR-96-5p levels were elevated (Fig. 6C–F, Supplementary Fig. 4A–D). Furthermore, glioma cell migration and invasion abilities were enhanced when miR-96-5p expression was decreased, whereas the opposite results were observed when miR-96-5p expression was increased (Fig. 6G–H, Supplementary Fig. 4E–H). Additionally, western blot experiments demonstrated that the EMT process, which can be promoted using a miR-96-5p-inhibitor and suppressed by miR-96-5p-mimics, was evident (Fig. 6I–J, Supplementary Fig. 4I–N). These data suggest that miR-96-5p has the potential to inhibit glioma progression.
A, B Cell viability of glioma cells assessed using MTT assay after transfection with miR-96-5p inhibitor or mimics. C–F The colony formation and EdU assays used to evaluate the cell proliferation of LN229 and U251 cells transfected with either miR-96-5p-inhibitor or miR-96-5p-mimics. The scale bar represents 100 μm. G-H The migration and invasion abilities of LN229 and U251 cells are assessed using the transwell assay. The scale bar represents 100 μm. I, J. Western blot is used to analyze the expression levels of EMT proteins following the addition of miR-96-5p-inhibitor or miR-96-5p-mimics in LN229 cells. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
STK17B promotes glioma proliferation and the EMT process
STK17B overexpression led to a significant increase in cell activity and proliferative capacity, whereas its knockdown resulted in a notable decrease (Fig. 7A–F, Supplementary Fig. 1I, Supplementary Fig. 5A–D). Furthermore, data from the transwell assay revealed that STK17B positively regulated glioma cells’ migratory and invasive abilities (Fig. 7G–H, Supplementary Fig. 5E–H). Based on this phenomenon, we investigated the changes in EMT-associated proteins following the overexpression or knockdown of STK17B. Enhanced STK17B expression was correlated with elevated N-cadherin and vimentin and decreased E-cadherin protein levels, thus contributing to the EMT (Fig. 7I, Supplementary Fig. 5I, J, M). Conversely, reducing STK17B expression led to opposite outcomes (Fig. 7J, Supplementary Fig. 5K, L, N). These findings demonstrate that STK17B promotes glioma proliferation and enhances EMT.
A, B LN229 and U251 cells are transfected with either oe-STK17B (overexpression plasmid) or si-STK17B (siRNAs), and cell viability assessed using the MTT assay. C–F The cell proliferation of LN229 and U251 transfected with either oe-STK17B or si-STK17B evaluated using colony formation and EdU assays. The scale bar represents 100 μm. G, H The migration and invasion abilities of LN229 and U251 cells transfected with either oe-STK17B or si-STK17B evaluated using the transwell assay. The scale bar represents 100 μm. I, J The levels of EMT protein after STK17B overexpression or knockdown were assessed. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
TP53TG1 regulates glioma proliferation and EMT by sponging miR-96-5p targeted STK17B
We overexpressed STK17B or added a miR-96-5p inhibitor after TP53TG1 knockdown to address whether lncRNA TP53TG1, miRNA miR-96-5p, and mRNA STK17B influence glioma progression through the ceRNA network. Compared with the TP53TG1 siRNA alone group, the proliferation, migration, and invasion abilities were significantly restored in the group overexpressing STK17B or with the addition of a miR-96-5p-inhibitor (Fig. 8A–D, Supplementary Fig. 6A–L). The protein outcomes also showed that STK17B overexpression or the addition of a miR-96-5p inhibitor prevented the inhibitory effect on EMT following TP53TG1 knockdown (Fig. 8E–F, Supplementary Fig. 6M–P). These results confirm our hypothesis and provide evidence that TP53TG1 regulates glioma proliferation and EMT by sponging miR-96-5p and targeting STK17B.
A EdU assay used to assess cell proliferation of LN229 cells transfected with NC, si-TP53TG1, and si-TP53TG1+ oe-STK17B. The scale bar represents 100 μm. B The proliferation of LN229 cells transfected with NC, si-TP53TG1, and si-TP53TG1+ miR-96-5p-inhibitor is evaluated. The scale bar represents 100 μm. C The migration abilities of LN229 and U251 cells transfected with NC, si-TP53TG1, and si-TP53TG1+ oe-STK17B are assessed using the transwell assay. The scale bar represents 100 μm. D The migration abilities of LN229 and U251 cells transfected with NC, si-TP53TG1, and si-TP53TG1+ miR-96-5p-inhibitor are evaluated. The scale bar represents 100 μm. E EMT protein levels analyzed after transfection with NC, si-TP53TG1, and si-TP53TG1+ oe-STK17B. F EMT protein levels are assessed after transfection with NC, si-TP53TG1, and si-TP53TG1+ miR-96-5p-inhibitor. Error bars represent mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
Gliomas exhibit resistance to conventional treatments due to their complex tumor microenvironment and inherent heterogeneity. Given that gliomas depend on reprogrammed and aberrant metabolic patterns for survival, targeting glioma metabolic pathways has emerged as a promising therapeutic strategy. Various inhibitors have been developed to target glucose metabolism, glycolytic enzymes, and mitochondrial oxidative phosphorylation (OXPHOS), yielding encouraging results in clinical trials41,42. Moreover, tryptophan metabolism related IDO inhibitors, and arginine deprivation therapy also have been shown to remodel the immunosuppressive microenvironment and effectively inhibited glioma progression8,10,11. Methionine is closely associated with tumor progression. Professor Hoffman proposed that most tumor cells depend on external methionine sources43. Subsequently, numerous studies have explored the effects of limiting the intake of exogenous methionine by tumor cells. Gao et al. revealed the therapeutic effects of a methionine-deprived dietary pattern on mouse colon cancer tumor models44. Our study showed a notable reduction in glioma cell proliferation, migration, and invasion following methionine deprivation. Additionally, we observed that the methionine-deprived diet efficiently restrained tumor growth in the nude mouse xenogeneic subcutaneous implant model. These effects are attributed to the ceRNA (TP53TG1/miR-96-5p/STK17B) regulatory network. These findings provide strong evidence supporting glioma’s high dependency on methionine (Fig. 9).
The ceRNA network is a competitive endogenous RNA network where lncRNAs competitively bind to miRNAs, thereby alleviating miRNAs’ inhibitory effect on target mRNA23. Multiple studies have demonstrated the involvement of ceRNAs in regulating glioma progression20,25,45. In this study, we identified lncRNA TP53TG1, which exhibited decreased expression following methionine deprivation. Our findings indicated higher expression of TP53TG1 in glioma tissues and cells, suggesting its potential role in glioma progression. The functions of lncRNA differ depending on its subcellular localization. In the nucleus, it controls the epigenetic state of specific genes, regulates transcription, and engages in alternative splicing. In the cytoplasm, it is crucial as a ceRNA regulating gene expression46,47. Therefore, we predicted the subcellular localization of TP53TG1 using multiple websites, confirming its presence in the cytoplasm. We performed several verification experiments to investigate the competitive endogenous regulatory relationships among TP53TG1, miR-96-5p, and STK17B. The dual-luciferase assay results suggesting the binding of miR-96-5p to either TP53TG1 or STK17B. Additionally, we validated that this ceRNA network was modulated by methionine metabolism and could affect the EMT process of glioma.
In this study, we discovered TP53TG1 was linked to methionine metabolism and promoted glioma progression, including cell proliferation and EMT. This result aligns with previous studies identifying TP53TG1 as an oncogene in nasopharyngeal carcinoma and pancreatic ductal adenocarcinoma48,49. The potential mechanism by which methionine deprivation inhibits TP53TG1 expression in gliomas may be related to findings in gastric cancer, where the mRNA stability of TP53TG1 is positively regulated by m6A modification50. During methionine deprivation cultures, the methionine metabolite SAM, which serves as a methyl donor, is markedly decreased. Generally, a reduction in SAM correlates with a substantial decrease in overall m6A modification levels51,52,53,54. Therefore, it is reasonable to speculate that the decreased level of m6A modification of TP53TG1 following methionine deprivation in this experiment led to a reduction in its mRNA stability and, consequently, an inhibition of its expression. The specific mechanisms involved need to be addressed in future research.
MiR-96-5p has been reported to demonstrate tumor suppressor effects in various malignant tumors. For instance, Yu et al.55 demonstrated that miR-96 downregulates KRAS expression, thereby reducing cancer cell invasion, migration, and tumor growth in pancreatic cancer. Similarly, Yao et al.56 discovered that upregulation of miR-96 expression inhibits proliferation, migration, invasion, and tumor formation in human osteosarcoma cells. Consistent with these findings, our experimental results confirmed the tumor suppressor effect of miR-96-5p in glioma.
STK17B, a serine/threonine protein kinase, belongs to the death-associated protein kinase (DAPK) family57. Studies have demonstrated that STK17B is implicated in the progression of various diseases. For instance, STK17B exacerbates the progression of nonalcoholic fatty liver disease58. Furthermore, STK17B is also involved in the regulation of breast cancer and colorectal cancer59,60. However, the role and mechanism of action of STK17B in glioma is unclear. Our study indicated a significant upregulation of STK17B expression in glioma and identified its positive regulation of glioma proliferation and EMT. Furthermore, STK17B is regulated by TP53TG1 through endogenous competition.
EMT is a vital process wherein cells originating from epithelial tissues lose polarity and transform into a mesenchymal phenotype. This transition empowers cells with migratory and invasive capabilities, significantly contributing to tumor advancement. EMT is governed by multiple signaling pathways, such as TGF-β/Smad, Wnt/β-catenin, MAPK, Notch, and PI3K/Akt/GSK-3β40,61,62,63,64. Previous studies by Chen et al. and Lan et al. also observed the participation of TP53TG1 and STK17B, respectively, in EMT regulation in hepatocellular carcinoma65,66. These findings indirectly support our results regarding the connection between the TP53TG1/miR-96-5p/STK17B ceRNA network and the EMT process in glioma.
In recent years, the significant role of lncRNAs in tumor progression has been increasingly recognized, positioning them as potential diagnostic and therapeutic targets. TP53TG1 has been reported to be involved in the progression of hepatocellular carcinoma, pancreatic ductal adenocarcinoma, and nasopharyngeal carcinoma48,49,65. In this study, we identified the oncogenic role of TP53TG1 as a competing endogenous RNA in gliomas, suggesting that targeting the TP53TG1-related ceRNA pathway may represent a novel therapeutic approach for gliomas. Most tumors exhibit methionine-dependent characteristics; for instance, methionine deprivation in gastric cancer results in cell death67. Additionally, the combined deprivation of cysteine and methionine can enhance ferroptosis in gliomas68. In our experiments, we also observed that methionine deprivation inhibited glioma progression. Given the aforementioned therapeutic role of methionine deprivation in tumors, we propose that a methionine deprivation diet may serve as a potential therapeutic strategy for glioma.
In this experiment, we still had several shortcomings. We did not directly validate the cellular localization of TP53TG1, which requires resolution in future studies. Additionally, in the dual luciferase experiment, the fluorescence intensity in the MUT group did not fully rebound, suggesting the potential existence of multiple binding sites during competitive binding. Furthermore, the specific mechanisms underlying TP53TG1/miR-96-5p/STK17B ceRNA network’s influence on EMT warrant further exploration. Given that the methionine deprivation diet is a long-term treatment, its long-term safety and potential side effects require additional investigation.
In conclusion, our findings revealed that methionine deficiency downregulated the levels of lncRNA TP53TG1, consequently reducing the expression of STK17B by sponging miR-96-5p. As a result, we observed inhibition of glioma cell proliferation and the EMT in vitro and in vivo. Furthermore, we found a correlation between high TP53TG1 expression and short survival time in patients with glioma, indicating the potential of TP53TG1 as a novel prognostic marker for glioma. Our study shed light on a potential mechanism underlying glioma inhibition by methionine deprivation, suggesting that a methionine-deprived diet could represent a promising therapeutic strategy for glioma treatment.
Data availability
The datasets used and analyzed during the current study are available from Huailei Liu on reasonable request.
Code availability
Detailed code is available from the corresponding author upon request.
References
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro Oncol. 16, iv1–iv63 (2014).
Lee, J. H. et al. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature 5607717, 243–247 (2018).
Martinez-Reyes, I. & Chandel, N. S. Cancer metabolism: looking forward. Nat. Rev. Cancer 2110, 669–680 (2021).
Landis, C. J., Tran, A. N., Scott, S. E., Griguer, C. & Hjelmeland, A. B. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochim Biophys. Acta Rev. Cancer 18692, 175–188 (2018).
Deshmukh, R., Allega, M. F. & Tardito, S. A map of the altered glioma metabolism. Trends Mol. Med 2711, 1045–1059 (2021).
An, S. et al. Amino Acid Metabolism Abnormity and Microenvironment Variation Mediated Targeting and Controlled Glioma Chemotherapy. Small 1240, 5633–5645 (2016).
Caniglia, J. L. et al. Beyond glucose: alternative sources of energy in glioblastoma. Theranostics 115, 2048–2057 (2021).
Bernstock, J. D. et al. Targeting oncometabolism to maximize immunotherapy in malignant brain tumors. Oncogene 4119, 2663–2671 (2022).
Platten, M., Friedrich, M., Wainwright, D. A., Panitz, V. & Opitz, C. A. Tryptophan metabolism in brain tumors - IDO and beyond. Curr. Opin. Immunol. 70, 57–66 (2021).
Kesarwani, P., Kant, S., Prabhu, A. & Chinnaiyan, P. The interplay between metabolic remodeling and immune regulation in glioblastoma. Neuro Oncol. 1910, 1308–1315 (2017).
Hajji, N. Arginine deprivation alters microglial polarity and synergizes with radiation to eradicate non-arginine-auxotrophic glioblastoma tumors. J. Clin. Invest. 1326, e142137 (2022).
Kaiser, P. Methionine Dependence of Cancer. Biomolecules 104, 568 (2020).
Sowers, ML. & Sowers, LC. Glioblastoma and Methionine Addiction. Int J Mol Sci 2313, 7156 (2022).
Zhang, Y. et al. Noncoding RNAs link metabolic reprogramming to immune microenvironment in cancers. J. Hematol. Oncol. 141, 169 (2021).
Liu, B., Qiang, L., Wang, G. D., Duan, Q. & Liu, J. LncRNA MALAT1 facilities high glucose induced endothelial to mesenchymal transition and fibrosis via targeting miR-145/ZEB2 axis. Eur. Rev. Med Pharm. Sci. 238, 3478–3486 (2019).
Wan, D. et al. miR-203 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting oncogene ADAM9 and oncogenic long non-coding RNA HULC. Anticancer Agents Med Chem. 164, 414–423 (2016).
Peng, Z., Liu, C. & Wu, M. New insights into long noncoding RNAs and their roles in glioma. Mol. Cancer 171, 61 (2018).
Dong, N. et al. LINC00606 promotes glioblastoma progression through sponge miR-486-3p and interaction with ATP11B. J. Exp. Clin. Cancer Res 431, 139 (2024).
Kim, S. H., Lim, K. H., Yang, S. & Joo, J. Y. Long non-coding RNAs in brain tumors: roles and potential as therapeutic targets. J. Hematol. Oncol. 141, 77 (2021).
Huang, L. et al. Long non-coding RNA NCK1-AS1 promotes the tumorigenesis of glioma through sponging microRNA-138-2-3p and activating the TRIM24/Wnt/beta-catenin axis. J. Exp. Clin. Cancer Res 391, 63 (2020).
Guo, T. et al. LINC00662 promotes hepatocellular carcinoma progression via altering genomic methylation profiles. Cell Death Differ. 277, 2191–2205 (2020).
Guo, T. et al. SNHG6 Acts as a Genome-Wide Hypomethylation Trigger via Coupling of miR-1297-Mediated S-Adenosylmethionine-Dependent Positive Feedback Loops. Cancer Res 7814, 3849–3864 (2018).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 1463, 353–358 (2011).
Zhong, Y. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 171, 79 (2018).
Zhu, J., Gu, W. & Yu, C. MATN1-AS1 promotes glioma progression by functioning as ceRNA of miR-200b/c/429 to regulate CHD1 expression. Cell Prolif. 531, e12700 (2020).
Wei, C. et al. LncRNA HOXA11-AS promotes glioma malignant phenotypes and reduces its sensitivity to ROS via Tpl2-MEK1/2-ERK1/2 pathway. Cell Death Dis. 1311, 942 (2022).
Wang, S. et al. Hypoxia-induced lncRNA PDIA3P1 promotes mesenchymal transition via sponging of miR-124-3p in glioma. Cell Death Dis. 113, 168 (2020).
Yan, T. et al. Suppression of the hyaluronic acid pathway induces M1 macrophages polarization via STAT1 in glioblastoma. Cell Death Discov. 81, 193 (2022).
Yan, T. et al. Interfering with hyaluronic acid metabolism suppresses glioma cell proliferation by regulating autophagy. Cell Death Dis. 125, 486 (2021).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47W1, W556–W560 (2019).
Mas-Ponte, D. et al. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 237, 1080–1087 (2017).
Su, Z. D. et al. iLoc-lncRNA: predict the subcellular ___location of lncRNAs by incorporating octamer composition into general PseKNC. Bioinformatics 3424, 4196–4204 (2018).
Cao, Z., Pan, X., Yang, Y., Huang, Y. & Shen, H. B. The lncLocator: a subcellular localization predictor for long non-coding RNAs based on a stacked ensemble classifier. Bioinformatics 3413, 2185–2194 (2018).
Cui, T. et al. RNALocate v2.0: an updated resource for RNA subcellular localization with increased coverage and annotation. Nucleic Acids Res. 50D1, D333–D339 (2022).
Jeggari, A., Marks, D. S. & Larsson, E. miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 2815, 2062–2063 (2012).
Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42 Database issue:D92-D97 (2014).
Chen, Y. & Wang, X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48D1, D127–D131 (2020).
Huang, H. Y. et al. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res 48D1, D148–D154 (2020).
Mcgeary, S.E. et al. The biochemical basis of microRNA targeting efficacy. Science. 3666472 (2019).
Brabletz, S., Schuhwerk, H., Brabletz, T. & Stemmler, M. P. Dynamic EMT: a multi-tool for tumor progression. Embo J. 4018, e108647 (2021).
Zhao, J. et al. Advancing glioblastoma treatment by targeting metabolism. Neoplasia 51, 100985 (2024).
Bi, J. et al. Altered cellular metabolism in gliomas - an emerging landscape of actionable co-dependency targets. Nat. Rev. Cancer 201, 57–70 (2020).
Hoffman, R. M. & Erbe, R. W. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc. Natl Acad. Sci. USA 735, 1523–1527 (1976).
Gao, X. et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 5727769, 397–401 (2019).
Mu, M., Niu, W., Zhang, X., Hu, S. & Niu, C. LncRNA BCYRN1 inhibits glioma tumorigenesis by competitively binding with miR-619-5p to regulate CUEDC2 expression and the PTEN/AKT/p21 pathway. Oncogene 3945, 6879–6892 (2020).
Rashid, F., Shah, A. & Shan, G. Long Non-coding RNAs in the Cytoplasm. Genomics Proteom. Bioinforma. 142, 73–80 (2016).
Carlevaro-Fita, J. & Johnson, R. Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol. Cell 735, 869–883 (2019).
Zhang, Y. et al. Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA-96. Cancer Sci. 1109, 2760–2772 (2019).
Yuan, J. et al. Super-Enhancers Promote Transcriptional Dysregulation in Nasopharyngeal Carcinoma. Cancer Res. 7723, 6614–6626 (2017).
Fang, D. et al. m6A modification-mediated lncRNA TP53TG1 inhibits gastric cancer progression by regulating CIP2A stability. Cancer Sci. 11312, 4135–4150 (2022).
Ramalingam, H. et al. A methionine-Mettl3-N(6)-methyladenosine axis promotes polycystic kidney disease. Cell Metab. 336, 1234–1247 (2021).
Li, T. et al. Methionine deficiency facilitates antitumour immunity by altering m (6)A methylation of immune checkpoint transcripts. Gut 723, 501–511 (2023).
Jin, X. et al. Unveiling the methionine cycle: a key metabolic signature and NR4A2 as a methionine-responsive oncogene in esophageal squamous cell carcinoma. Cell Death Differ. 315, 558–573 (2024).
Boulias, K. & Greer, E. L. Biological roles of adenine methylation in RNA. Nat. Rev. Genet 243, 143–160 (2023).
Yu, S. et al. miRNA-96 suppresses KRAS and functions as a tumor suppressor gene in pancreatic cancer. Cancer Res. 7014, 6015–6025 (2010).
Yao, Q., Pei, Y., Zhang, X. & Xie, B. microRNA-96 acts as a tumor suppressor gene in human osteosarcoma via target regulation of EZRIN. Life Sci. 203, 1–11 (2018).
Gozuacik, D. & Kimchi, A. DAPk protein family and cancer. Autophagy 22, 74–79 (2006).
Li, Y. et al. DRAK2 aggravates nonalcoholic fatty liver disease progression through SRSF6-associated RNA alternative splicing. Cell Metab. 3310, 2004–2020 (2021).
Doherty, G. A. et al. Regulation of the apoptosis-inducing kinase DRAK2 by cyclooxygenase-2 in colorectal cancer. Br. J. Cancer 1013, 483–491 (2009).
Yang, K. M. et al. DRAK2 participates in a negative feedback loop to control TGF-beta/Smads signaling by binding to type I TGF-beta receptor. Cell Rep. 25, 1286–1299 (2012).
Peng, D., Fu, M., Wang, M., Wei, Y. & Wei, X. Targeting TGF-beta signal transduction for fibrosis and cancer therapy. Mol. Cancer 211, 104 (2022).
Ding, D. et al. Timosaponin BII inhibits TGF-beta mediated epithelial-mesenchymal transition through Smad-dependent pathway during pulmonary fibrosis. Phytother. Res 377, 2787–2799 (2023).
Jiang, W. et al. miRNA-101 Suppresses Epithelial-to-Mesenchymal Transition by Targeting HMGA2 in Pancreatic Cancer Cells. Anticancer Agents Med. Chem. 164, 432–439 (2016).
Yu, Z. et al. Polysaccharide from Rhizopus nigricans inhibits the invasion and metastasis of colorectal cancer. Biomed. Pharmacother. 103, 738–745 (2018).
Chen, B. et al. Long noncoding RNA TP53TG1 suppresses the growth and metastasis of hepatocellular carcinoma by regulating the PRDX4/beta-catenin pathway. Cancer Lett. 513, 75–89 (2021).
Lan, Y. et al. STK17B promotes carcinogenesis and metastasis via AKT/GSK-3beta/Snail signaling in hepatocellular carcinoma. Cell Death Dis. 92, 236 (2018).
Xin, L. et al. Methionine deficiency promoted mitophagy via lncRNA PVT1-mediated promoter demethylation of BNIP3 in gastric cancer. Int J. Biochem Cell Biol. 141, 106100 (2021).
Upadhyayula, P. S. et al. Dietary restriction of cysteine and methionine sensitizes gliomas to ferroptosis and induces alterations in energetic metabolism. Nat. Commun. 141, 1187 (2023).
Acknowledgements
This study was funded by the First Affiliated Hospital of Harbin Medical University Fund for Distinguished Young Scholars (HYD2020JQ0015), Natural Science Foundation of Heilongjiang Province (YQ2023H009), China Postdoctoral Science Foundation (2023M732121), the National Natural Science Foundation of China (No. 82002634) and the National Natural Science Foundation of China (No. 81672486).
Author information
Authors and Affiliations
Contributions
J.L., R.L., H.H., Y.H.: Acquisition, analysis, bioinformatics analysis, writing, draft the figure. H.L. (Honglei Li), H.C., N.W., M.C.: Data interpretation, investigation. Y.S.: magnetic resonance imaging scan and recording. H.L. (Huailei Liu)., K.W., T.Y.: Conceptualization, supervision, design, funding acquisition. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All experimental ethics and animal research were approved by the ethics committee of the First Affiliated Hospital of Harbin Medical University and adhered to the principles of the Declaration of Helsinki.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, J., Liu, R., Hu, H. et al. Methionine deprivation inhibits glioma proliferation and EMT via the TP53TG1/miR-96-5p/STK17B ceRNA pathway. npj Precis. Onc. 8, 270 (2024). https://doi.org/10.1038/s41698-024-00763-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-024-00763-y