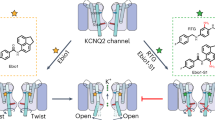
Abstract
Voltage-gated ion channels (VGICs) are crucial targets for neuropsychiatric therapeutics owing to their role in controlling neuronal excitability and the established link between their dysfunction and neurological diseases, highlighting the importance of identifying modulators with distinct mechanisms. Here we report two small-molecule modulators with the same chemical scaffold, Ebio2 and Ebio3, targeting a potassium channel KCNQ2, with opposite effects: Ebio2 acts as a potent activator, whereas Ebio3 serves as a potent and selective inhibitor. Guided by cryogenic electron microscopy, patch-clamp recordings and molecular dynamics simulations, we reveal that Ebio3 attaches to the outside of the inner gate, employing a unique non-blocking inhibitory mechanism that directly squeezes the S6 pore helix to inactivate the KCNQ2 channel. Ebio3 also showed efficacy in inhibiting currents of KCNQ2 pathogenic gain-of-function mutations, presenting an avenue for VGIC-targeted therapies. Overall, these findings contribute to the understanding of KCNQ2 inhibition and provide insights into developing selective, non-blocking VGIC inhibitors.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
269,00 € per year
only 22,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All data are available from the corresponding authors upon reasonable request. The 3D cryo-EM density maps and structure coordinates have been deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) with accession codes EMD-60981 and PDB 9IXY for KCNQ2–Ebio2 and EMD-60982 and PDB 9IXZ for KCNQ2–Ebio3, respectively. Source data are provided with this paper.
References
Hodgkin, A. L. & Huxley, A. F. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117, 500–544 (1952).
Bean, B. P., Nowycky, M. C. & Tsien, R. W. β-Adrenergic modulation of calcium channels in frog ventricular heart cells. Nature 307, 371–375 (1984).
Liao, P., Yong, T. F., Liang, M. C., Yue, D. T. & Soong, T. W. Splicing for alternative structures of CaV1.2 Ca2+ channels in cardiac and smooth muscles. Cardiovasc. Res. 68, 197–203 (2005).
Hetzenauer, A., Sinnegger-Brauns, M. J., Striessnig, J. & Singewald, N. Brain activation pattern induced by stimulation of L-type Ca2+-channels: contribution of CaV1.3 and CaV1.2 isoforms. Neuroscience 139, 1005–1015 (2006).
Hemmerlein, B. et al. Overexpression of Eag1 potassium channels in clinical tumours. Mol. Cancer 5, 41 (2006).
Fertleman, C. R. et al. SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron 52, 767–774 (2006).
Nappi, M. et al. Gain of function due to increased opening probability by two KCNQ5 pore variants causing developmental and epileptic encephalopathy. Proc. Natl Acad. Sci. USA 119, e2116887119 (2022).
Barcia, G. et al. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat. Genet. 44, 1255–1259 (2012).
Wulff, H., Christophersen, P., Colussi, P., Chandy, K. G. & Yarov-Yarovoy, V. Antibodies and venom peptides: new modalities for ion channels. Nat. Rev. Drug Discov. 18, 339–357 (2019).
Santos, R. et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 16, 19–34 (2017).
Bezanilla, F. The voltage sensor in voltage-dependent ion channels. Physiol. Rev. 80, 555–592 (2000).
Jensen, M. Ø. et al. Mechanism of voltage gating in potassium channels. Science 336, 229–233 (2012).
Labro, A. J., Lacroix, J. J., Villalba-Galea, C. A., Snyders, D. J. & Bezanilla, F. Molecular mechanism for depolarization-induced modulation of Kv channel closure. J. Gen. Physiol. 140, 481–493 (2012).
Gao, S. et al. Structural basis for human CaV1.2 inhibition by multiple drugs and the neurotoxin calciseptine. Cell 186, 5363–5374.e16 (2023).
Lenaeus, M., Gamal El-Din, T. M., Tonggu, L., Zheng, N. & Catterall, W. A. Structural basis for inhibition of the cardiac sodium channel by the atypical antiarrhythmic drug ranolazine. Nat. Cardiovasc. Res. 2, 587–594 (2023).
Li, X. et al. Structural basis for modulation of human NaV1.3 by clinical drug and selective antagonist. Nat. Commun. 13, 1286 (2022).
Vitali Serdoz, L., Rittger, H., Furlanello, F. & Bastian, D. Quinidine—a legacy within the modern era of antiarrhythmic therapy. Pharmacol. Res. 144, 257–263 (2019).
Grace Andrew, A. & Camm, A. J. Quinidine. N. Engl. J. Med. 338, 35–45 (1998).
Wang, H.-S. et al. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science 282, 1890–1893 (1998).
Jentsch, T. J. Neuronal KCNQ potassium channels: physiology and role in disease. Nat. Neurosci. 1, 21–30 (2000).
Brown, D. A. & Adams, P. R. Muscarinic suppression of a novel voltage-sensitive K+ current in a vertebrate neurone. Nature 283, 673–676 (1980).
Mulkey, S. B. et al. Neonatal nonepileptic myoclonus is a prominent clinical feature of KCNQ2 gain-of-function variants R201C and R201H. Epilepsia 58, 436–445 (2017).
Millichap, J. J. et al. Infantile spasms and encephalopathy without preceding neonatal seizures caused by KCNQ2 R198Q, a gain-of-function variant. Epilepsia 58, e10–e15 (2017).
Miceli, F. et al. Early-onset epileptic encephalopathy caused by gain-of-function mutations in the voltage sensor of Kv7.2 and Kv7.3 potassium channel subunits. J. Neurosci. 35, 3782 (2015).
Miceli, F. et al. KCNQ2 R144 variants cause neurodevelopmental disability with language impairment and autistic features without neonatal seizures through a gain-of-function mechanism. eBioMedicine 81, 104130 (2022).
Fontán-Lozano, Á., Suárez-Pereira, I., Delgado-García, J. M. & Carrión, Á. M. The M-current inhibitor XE991 decreases the stimulation threshold for long-term synaptic plasticity in healthy mice and in models of cognitive disease. Hippocampus 21, 22–32 (2011).
Liu, H., Jia, L., Chen, X., Shi, L. & Xie, J. The Kv7/KCNQ channel blocker XE991 protects nigral dopaminergic neurons in the 6-hydroxydopamine rat model of Parkinson’s disease. Brain Res. Bull. 137, 132–139 (2018).
Li, X. et al. Molecular basis for ligand activation of the human KCNQ2 channel. Cell Res. 31, 52–61 (2021).
Zhang, Y.-M. et al. Discovery of HN37 as a potent and chemically stable antiepileptic drug candidate. J. Med. Chem. 64, 5816–5837 (2021).
Zhang, S. et al. A small-molecule activation mechanism that directly opens the KCNQ2 channel. Nat. Chem. Biol. 20, 847–856 (2024).
Smart, O. S., Neduvelil, J. G., Wang, X., Wallace, B. A. & Sansom, M. S. P. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996).
Sun, J. & MacKinnon, R. Cryo-EM structure of a KCNQ1/CaM complex reveals insights into congenital long QT syndrome. Cell 169, 1042–1050 (2017).
Li, T. et al. Structural basis for the modulation of human KCNQ4 by small-molecule drugs. Mol. Cell 81, 25–37 (2021).
Moldenhauer, H., Díaz-Franulic, I., González-Nilo, F. & Naranjo, D. Effective pore size and radius of capture for K+ ions in K-channels. Sci. Rep. 6, 19893 (2016).
Zhou, Y., Morais-Cabral, J. H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Singh, N. A. et al. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat. Genet. 18, 25–29 (1998).
Dedek, K. et al. Myokymia and neonatal epilepsy caused by a mutation in the voltage sensor of the KCNQ2 K+ channel. Proc. Natl Acad. Sci. USA 98, 12272–12277 (2001).
Rundfeldt, C. The new anticonvulsant retigabine (D-23129) acts as an opener of K+ channels in neuronal cells. Eur. J. Pharmacol. 336, 243–249 (1997).
French, J. et al. XEN1101, a novel potassium channel modulator: interim data from an ongoing, long-term, open-label extension of a phase 2b study (X-TOLE) in adults with focal epilepsy (S44.008). Neurology 100, 4207 (2023).
Zaczek, R. et al. Two new potent neurotransmitter release enhancers, 10,10-bis(4-pyridinylmethyl)-9(10h)-anthracenone and 10,10-bis(2-fluoro-4-pyridinylmethyl)-9(10h)-anthracenone: comparison to linopirdine. J. Pharmacol. Exp. Ther. 285, 724 (1998).
Lamas, J. A., Selyanko, A. A. & Brown, D. A. Effects of a cognition-enhancer, linopirdine (DuP 996), on M-type potassium currents (IK(M)) some other voltage- and ligand-gated membrane currents in rat sympathetic neurons. Eur. J. Neurosci. 9, 605–616 (1997).
Rivas-Ramírez, P., Reboreda, A., Rueda-Ruzafa, L., Herrera-Pérez, S. & Lamas, J. A. Contribution of KCNQ and TREK channels to the resting membrane potential in sympathetic neurons at physiological temperature. Int. J. Mol. Sci. 21, 2020 (2020).
Zhao, Y. et al. Molecular basis for ligand modulation of a mammalian voltage-gated Ca2+ channel. Cell 177, 1495–1506 (2019).
Lenaeus, M. J., Vamvouka, M., Focia, P. J. & Gross, A. Structural basis of TEA blockade in a model potassium channel. Nat. Struct. Mol. Biol. 12, 454–459 (2005).
Huang, J. et al. Cannabidiol inhibits NaV channels through two distinct binding sites. Nat. Commun. 14, 3613 (2023).
Huang, J., Fan, X., Jin, X., Teng, L. & Yan, N. Dual-pocket inhibition of NaV channels by the antiepileptic drug lamotrigine. Proc. Natl Acad. Sci. USA 120, e2309773120 (2023).
Zhao, Y. et al. Cryo-EM structures of apo and antagonist-bound human CaV3.1. Nature 576, 492–497 (2019).
Fernández-Mariño, A. I. et al. Inactivation of the Kv2.1 channel through electromechanical coupling. Nature 622, 410–417 (2023).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021).
Webb, B. & Sali, A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016).
Jo, S., Kim, T., Iyer, V. G. & Im, W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008).
Berendsen, H. J. C., van der Spoel, D. & van Drunen, R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91, 43–56 (1995).
Vanommeslaeghe, K. & MacKerell, A. D. Jr Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012).
Acknowledgements
This work is funded by grants from the National Key Research and Development Program of China (2022YFE0205600 to H.Y. and P.H. and 2022YFC3400504 to J.S.); the National Natural Science Foundation of China (82273857 to Q.Z., 82373792 to H.Y. and 32371204 to J.G.); the Fundamental Research Funds for the Central Universities, Joint Funding of the Macau Science and Technology Development Fund and the Ministry of Science and Technology of the People’s Republic of China (grant no. 0006/2021/AMJ to P.H.); and the East China Normal University Medicine and Health Joint Fund (2022JKXYD07001 to H.Y. and Z.C.). J.G. is supported by the Ministry of Education Frontier Science Center for Brain Science & Brain–Machine Integration, Zhejiang University. Single-particle cryo-EM data were collected at the Center of Cryo-Electron Microscopy at Zhejiang University. We thank X. Zhang and S. Chang for support with facility access and data acquisition. We are also thankful for the support of the East China Normal University Multifunctional Platform for Innovation (001).
Author information
Authors and Affiliations
Contributions
H.Y., Q.Z. and J.G. conceived the project. Q.Z. and Y.Y. performed the molecular modification. J. Li, J.H., H.H., W.K., J. Liu and Y.M. performed electrophysiology tests. Z.Y. and Y.Z. collected the EM data and calculated the EM map. Z.Y. and J.G. built and refined the atomic model and analyzed the structure. S.Z., Q.Z., J.S. and J. Li performed MD simulations. Y.Y., Q.Z. and L.M. assisted in compound preparation and synthesis. Z.C. and P.H. provided intellectual expertise and shared key methodologies. J. Li and Q.Z. prepared the draft of the paper, with input from all authors. H.Y., Q.Z. and J.G. wrote the paper. All authors read and commented on the paper.
Corresponding authors
Ethics declarations
Competing interests
J. Li, Y.Y., L.M., J.S., J.G., Q.Z. and H.Y. are inventors of patent application 202411521894.9 that covers the potential usage of Ebio2, Ebio3 and their derivatives. The other authors declare no competing interests.
Peer review
Peer review information
Nature Chemical Biology thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 General structural modification strategy of Ebio1.
Upper panel showing chemical structures and binding modes of Ebio1, HN37, and XEN1101. Red dashed lines represent hydrogen bonding interactions between ligands and the KCNQ channel. Burgundy and yellow dashed lines denote two regions that can be exploited in the molecular modification. Lower panel showing structures of Ebio1 derivatives and their effects on the KCNQ2 channel at a concentration of 1 μM. The holding potential was −80 mV. The KCNQ2 current was elicited by a 2,000-ms voltage step to +50 mV. Data are presented as mean ± s.e.m. n = 8, 4, 3, 3, 4, 15, 3, and 7 biological replicates, respectively.
Extended Data Fig. 2 The effects of Ebio2 on KCNQ2.
a, Current-voltage relationship of the KCNQ2 channel in response to 10 nM Ebio2. b, Normalized activation (left) and deactivation (right) phase from full traces in the absence and presence of 10 nM Ebio2. The test voltages were +50 mV and –120 mV, respectively. c, Analysis of activation (left) and deactivation (right) time constants for the KCNQ2 channel in the absence and presence of 10 nM Ebio2. n = 11 biological replicates. d, Representative single-channel recordings from inside-out patches of KCNQ2 at +50 mV in the absence and presence of 1 μM Ebio2 (left), and corresponding all-point amplitude histograms fitted by Gaussian distributions (red solid line, right). e,f, Histogram showing the single-channel conductance (e) and the open probability (f) of KCNQ2 in the absence and presence of 1 μM Ebio2. n = 5 biological replicates. g, Concentration-dependent curves of Ebio2 on KCNQ members, obtained from the +50 mV traces at steady state. h, Concentration-dependent curves of Ebio2 effects on the half-activation voltage shift (ΔV1/2) for KCNQ members. The holding potential was −80 mV. The KCNQ current was elicited by a series of voltage stepping from −90 mV to +60 mV in 10 mV increments (a,g,h). The error bars represent the mean ± s.e.m. of each data point calculated from 3–14 biological replicates. Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
Extended Data Fig. 3 The effects of Ebio3 on KCNQ2.
a, Current-voltage relationship of the KCNQ2 channel in response to 10 nM Ebio3. The holding potential was −80 mV. The KCNQ2 current was elicited by a series of voltage stepping from −90 mV to +60 mV in 10 mV increments. b, The normalized activation (left) and deactivation (right) phase from full traces in the absence and presence of 10 nM Ebio3. c, Analysis of activation (left) and deactivation (right) time constants for the KCNQ2 channel in the absence and presence of 10 nM Ebio3. n = 5 biological replicates. d, Representative current traces of KCNQ2 obtained from whole-cell patch-clamp recordings before (black) and after (blue) the application of 1 nM Ebio3. The holding potential was −80 mV. The KCNQ2 current was elicited by a 2,000-ms voltage step to +50 mV. e, Effect of 1 nM Ebio3 on the extent of inactivation assessed using a three-pulse (P1–P3) protocol with a holding voltage of −80 mV. P1 was to +50 mV, P2 was from −90 to +50 mV in 20 mV increments for 1,500 ms, and P3 was to +50 mV to assess the fraction of inactivated channels (insert). f, Fraction of non-inactivated channels during each P2 voltage step before (black) and after (blue) the application of 1 nM Ebio3, obtained by measuring the peak current at P3 normalized to P1. The error bars represent the mean ± s.e.m. n = 10 biological replicates. Statistical analysis was performed using an unpaired two-tailed Student’s t-test.
Extended Data Fig. 4 Structure determination of KCNQ2-Ebio2 complex.
a, Flowchart of image processing for KCNQ2-Ebio2 particles, and representative cryo-EM micrograph of KCNQ2-Ebio2 (inset). n = 3 independent experiments. b, The gold-standard FSC curves of the final 3D reconstruction of KCNQ2-Ebio2, and the FSC curve for cross-validation between the map and the model of KCNQ2-Ebio2. c, The angular distribution of particles that gave rise to the KCNQ2-Ebio2 cryo-EM map reconstruction. d, The density map of KCNQ2-Ebio2 is colored by local resolution. The local resolution is estimated with RELION 3.1 and generated in Chimera. e, The isolated densities of PIP2 are observed between VSD and PD in the map of KCNQ2-Ebio2 complex. The EM densities are contoured at the level of 0.0085 in UCSF ChimeraX.
Extended Data Fig. 6 Structure determination of KCNQ2-Ebio3 complex.
a, Flowchart of image processing for KCNQ2-Ebio3 particles, and representative cryo-EM micrograph of KCNQ2-Ebio3 (inset). n = 3 independent experiments. b, Size-exclusion chromatography of KCNQ2-CaM on Superose 6 and SDS-PAGE analysis of the final sample. n = 3 independent experiments. c, The gold-standard FSC curves of the final 3D reconstruction of KCNQ2-Ebio3, and the FSC curve for cross-validation between the map and the model of KCNQ2-Ebio3. d, The angular distribution of particles that gave rise to the KCNQ2-Ebio3 cryo-EM map reconstruction. e, The density map of KCNQ2-Ebio3 is colored by local resolution. The local resolution is estimated with RELION 3.1 and generated in Chimera.
Extended Data Fig. 7 The structural comparison of inner gates among KCNQ2-Ebio3, KCNQ2-Ebio2, and apo-state KCNQ2 structures.
a, Structural comparison between Ebio3- and Ebio2-bound KCNQ2. b-d, The inner gate of KCNQ2-Ebio3 (b), KCNQ2-Ebio2 (c), and KCNQ2-apo (d, PDB ID: 7CR3) structures. The dashed lines show diagonal atom-to-atom distances (in Å) at the constriction-lining residues G310, S314, and L318.
Extended Data Fig. 8 Ebio3-dependent dynamic rearrangement of pore domains for WT KCNQ2 and the L307A mutant channel.
a-d, Molecular dynamics simulations for system I (a), II (b), III (c), and IV (d). Four panels on the left showing the ensemble plot of Ebio3 molecules in the binding pocket during MD simulations, and RMSD of Ebio3 molecules against simulation time across the three independent repeats. The two rightmost panels showing Ebio3-induced conformational changes of the pore ___domain before (left) and after (right) MD simulation. Ion-conduction pores of the initial and final snapshots are highlighted and shown as gray surfaces. Key gating residues are shown in sticks and colored orange for S314 and L318.
Extended Data Fig. 9 Dynamic rearrangement of pore domains for WT KCNQ2 and the L307A mutant channel without Ebio3.
a-d, Representation of the channel pore diameter for MD simulation systems I’ to IV’, without the addition of Ebio3. Three independent repetitions were performed for each system.
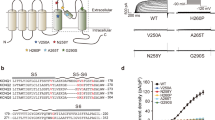
Extended Data Fig. 10 Ebio3 inhibits KCNQ2 GOF mutants with increased voltage-sensitivity.
a, Current density of WT KCNQ2 and GOF mutant channels with increased voltage sensitivity at +60 mV. n = 16, 9, 8, and 5 biological replicates. b, V1/2 of WT KCNQ2 and GOF mutant channels. Due to the drastically leftward shift of the G-V curve, the exact V1/2 value of the R201C mutant was not obtained in our recordings. n = 20, 8, 9, and 8 biological replicates. c, Summary of the percent of inhibition by 10 nM Ebio3 on outward current of WT KCNQ2 and GOF mutant channels, analyzed at +50 mV. n = 10, 9, 10, and 10 biological replicates. d-f, Current density-voltage curves of 10 nM Ebio3 effects on the KCNQ2 GOF mutant channels. The holding potential was −80 mV. The KCNQ2 current was elicited by a series of voltage stepping from −90 mV to +60 mV in 10 mV increments. The error bars represent the mean ± s.e.m. of each data point calculated from 5–20 biological replicates (a-f). An unpaired two-tailed Student’s t-test was performed to compare WT KCNQ2 and each mutant (a,b). NA, not available.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2, Supplementary Figs. 1–14 and Supplementary Note.
Supplementary Data
Statistical source data for Supplementary Figs. 1, 12 and 14.
Source data
Source Data Fig. 1
Statistical source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed gel.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, J., Yang, Z., Zhang, S. et al. Small molecule inhibits KCNQ channels with a non-blocking mechanism. Nat Chem Biol 21, 1100–1109 (2025). https://doi.org/10.1038/s41589-024-01834-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41589-024-01834-8