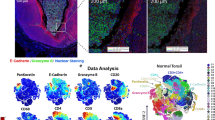
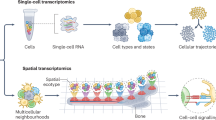
Abstract
Delineating cell populations is crucial for understanding immune function in health and disease. Spatial omics technologies offer insights by capturing three complementary domains: single-cell molecular biomarker expression, cellular spatial relationships and tissue architecture. However, current computational methods often fail to fully integrate these multidimensional data, particularly for immune cell populations and intrinsic functional states. We introduce Cell Local Environment and Neighborhood Scan (CellLENS), a self-supervised computational method that learns cellular representations by fusing information across three spatial omics domains (expression, neighborhood and image). CellLENS markedly enhances de novo discovery of biologically relevant immune cell populations at fine granularity by integrating individual cells’ molecular profiles with their neighborhood context and tissue localization. By applying CellLENS to diverse spatial proteomic and transcriptomic datasets across multiple tissue types and disease settings, we uncover unique immune cell populations functionally stratified according to their spatial contexts. Our work demonstrates the power of multi-___domain data integration in spatial omics to reveal insights into immune cell heterogeneity and tissue-specific functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
This study did not generate any new experimental data. The CODEX spleen dataset was generated from ref. 2 and the original data can be accessed at https://data.mendeley.com/datasets/zjnpwh8m5b/1. The Xenium tonsil dataset was generated from ref. 35, and the original data can be accessed at https://www.10xgenomics.com/datasets/human-tonsil-data-xenium-human-multi-tissue-and-cancer-panel-1-standard. The Xenium LN dataset was generated from ref. 38, and the original data can be accessed at https://www.10xgenomics.com/datasets/preview-data-xenium-prime-gene-expression. The CODEX tonsil dataset was generated from ref. 34, and the data can be found in our Zenodo link https://doi.org/10.5281/zenodo.14617085. The CODEX cHL dataset was generated from ref. 39, and the original data is available via Zenodo at https://doi.org/10.5281/zenodo.10067010 (ref. 66). The CosMx liver dataset was generated from ref. 42, and the original data can be accessed at https://nanostring.com/products/cosmx-spatial-molecular-imager/ffpe-dataset/human-liver-rna-ffpe-dataset/. For convenient access to this study’s data, the files are available via Zenodo at https://doi.org/10.5281/zenodo.14617085 (ref. 65).
Code availability
The CellLENS Python package, along with the code used in this study, is available via GitHub at https://github.com/sggao/celllens/.
References
Giesen, C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014).
Goltsev, Y. et al. Deep profiling of mouse splenic architecture with codex multiplexed imaging. Cell 174, 968–981 (2018).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Wang, X. et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Samusik, N., Good, Z., Spitzer, M. H., Davis, K. L. & Nolan, G. P. Automated mapping of phenotype space with single-cell data. Nat. Methods 13, 493–496 (2016).
Levine, J. H. et al. Data-driven phenotypic dissection of AML reveals progenitor-like cells that correlate with prognosis. Cell 162, 184–197 (2015).
Traag, V. A., Waltman, L. & Van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. https://doi.org/10.1038/s41598-019-41695-z (2019).
Bhate, S. S., Barlow, G. L., Schürch, C. M. & Nolan, G. P. Tissue schematics map the specialization of immune tissue motifs and their appropriation by tumors. Cell Syst. 13, 109–130 (2022).
Schürch, C. M. et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359 (2020).
Kumar, V., Abbas, A. & Aster, J. Robbins & Cotran Pathologic Basis of Disease (Robbins Pathology) (Saunders, 2014).
Eroschenko, V. P. DiFiore's Atlas of Histology with Functional Correlations (Lippincott, Williams & Wilkins, 2013).
Phillips, D. et al. Immune cell topography predicts response to PD-1 blockade in cutaneous T cell lymphoma. Nat. Commun. 12, 6726 (2021).
Yeo, Y. Y. et al. Epstein–Barr virus orchestrates spatial reorganization and immunomodulation within the classic Hodgkin lymphoma tumor microenvironment. Preprint at bioRxiv https://doi.org/10.1101/2024.03.05.583586 (2024).
Chen, S. et al. Integration of spatial and single-cell data across modalities with weakly linked features. Nat. Biotechnol. 42, 1096–1106 (2024).
Zhu, B. et al. Robust single-cell matching and multimodal analysis using shared and distinct features. Nat. Methods 20, 304–315 (2023).
Brbić, M. et al. Annotation of spatially resolved single-cell data with STELLAR. Nat. Methods 19, 1411–1418 (2022).
Amitay, Y. et al. CellSighter: a neural network to classify cells in highly multiplexed images. Nat. Commun. 14, 4302 (2023).
Bao, F. et al. Integrative spatial analysis of cell morphologies and transcriptional states with MUSE. Nat. Biotechnol. 40, 1200–1209 (2022).
Chidester, B., Zhou, T., Alam, S. & Ma, J. SPICEMIX enables integrative single-cell spatial modeling of cell identity. Nat. Genet. 55, 78–88 (2023).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Janesick, A. et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 14, 8353 (2023).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. Preprint at https://arxiv.org/abs/1609.02907 (2016).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT, 2016).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems 25 https://proceedings.neurips.cc/paper_files/paper/2012/file/c399862d3b9d6b76c8436e924a68c45b-Paper.pdf (NeurIPS, 2012).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Commun. ACM 60, 84–90 (2017).
Rousseeuw, P. J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987).
Davies, D. L. & Bouldin, D. W. A cluster separation measure. IEEE Trans. Pattern Anal. Mach. Intell. 2, 224–227 (2009).
Caliński, T. & Harabasz, J. A dendrite method for cluster analysis. Commun. Stat. Theory Methods 3, 1–27 (1974).
Newman, M. E. J. Modularity and community structure in networks. Proc. Natl Acad. Sci. USA 103, 8577–8582 (2006).
Argelaguet, R. et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. https://doi.org/10.1186/s13059-020-02015-1 (2020).
Cumpelik, A. et al. Dynamic regulation of B cell complement signaling is integral to germinal center responses. Nat. Immunol. 22, 757–768 (2021).
Steiniger, B. S. Human spleen microanatomy: why mice do not suffice. Immunology 145, 334–346 (2015).
Kennedy-Darling, J. et al. Highly multiplexed tissue imaging using repeated oligonucleotide exchange reaction. Eur. J. Immunol. 51, 1262–1277 (2021).
Human tonsil data: Xenium human multi-tissue and cancer panel 1 standard. 10x Genomics www.10xgenomics.com/datasets/human-tonsil-data-xenium-human-multi-tissue-and-cancer-panel-1-standard (2024).
Herling, M. et al. High TCL1 levels are a marker of B-cell receptor pathway responsiveness and adverse outcome in chronic lymphocytic leukemia. Blood 114, 4675–4686 (2009).
Papa, I. & Vinuesa, C. G. Synaptic interactions in germinal centers. Front. Immunol. 9, 1858 (2018).
FFPE human lymph node with 5k pan tissue and pathways panel. 10x Genomics www.10xgenomics.com/datasets/preview-data-xenium-prime-gene-expression (2024).
Shaban, M. et al. MAPS: pathologist-level cell type annotation from tissue images through machine learning. Nat. Commun. 15, 28 (2024).
Veldman, J. et al. CD4+ T cells in classical Hodgkin lymphoma express exhaustion associated transcription factors TOX and TOX2: characterizing CD4+ T cells in Hodgkin lymphoma. Oncoimmunology 11, 2033433 (2022).
Nagasaki, J. et al. The critical role of CD4+ T cells in PD-1 blockade against MHC-II–expressing tumors such as classic Hodgkin lymphoma. Blood Adv. 4, 4069–4082 (2020).
CosMx SMI human liver FFPE dataset. NanoString https://nanostring.com/products/cosmx-spatial-molecular-imager/ffpe-dataset/human-liver-rna-ffpe-dataset/ (2025).
Cheng, S. et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792–809 (2021).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Li, Z., Wang, T., Liu, P. & Huang, Y. SpatialDM for rapid identification of spatially co-expressed ligand–receptor and revealing cell–cell communication patterns. Nat. Commun. 14, 3995 (2023).
Kotliar, D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-seq. eLife 8, e43803 (2019).
Yuan, Y. et al. Mechanisms of tumor-associated macrophages affecting the progression of hepatocellular carcinoma. Front. Pharmacol. 14, 1217400 (2023).
Tian, Z., Hou, X., Liu, W., Han, Z. & Wei, L. Macrophages and hepatocellular carcinoma. Cell Biosci. https://doi.org/10.1186/s13578-019-0342-7 (2019).
Mages, S. et al. TACCO unifies annotation transfer and decomposition of cell identities for single-cell and spatial omics. Nat. Biotechnol. 41, 1465–1473 (2023).
Aran, D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Lee, J. L. & Schaffenrath, L. Benchmarking_tissue_preparation_pilotdata. Zenodo https://doi.org/10.5281/zenodo.11410274 (2024).
Hu, J. et al. SpaGCN: integrating gene expression, spatial ___location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat. Methods 18, 1342–1351 (2021).
Pham, D. et al. Robust mapping of spatiotemporal trajectories and cell–cell interactions in healthy and diseased tissues. Nat. Commun. 14, 7739 (2023).
Xu, H. et al. Unsupervised spatially embedded deep representation of spatial transcriptomics. Genome Med. 16, 12 (2024).
Singhal, V. et al. BANKSY unifies cell typing and tissue ___domain segmentation for scalable spatial omics data analysis. Nat. Genet. 56, 431–441 (2024).
Varrone, M., Tavernari, D., Santamaria-Martínez, A., Walsh, L. A. & Ciriello, G. CellCharter reveals spatial cell niches associated with tissue remodeling and cell plasticity. Nat. Genet. 56, 74–84 (2024).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Dosovitskiy, A. An image is worth 16×16 words: transformers for image recognition at scale. Preprint at https://arxiv.org/abs/2010.11929 (2020).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Chen, A. et al. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell 185, 1777–1792 (2022).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Kim, J. et al. Unsupervised discovery of tissue architecture in multiplexed imaging. Nat. Methods 19, 1653–1661 (2022).
Hu, Y. et al. Unsupervised and supervised discovery of tissue cellular neighborhoods from cell phenotypes. Nat. Methods 21, 267–278 (2024).
Zhu, B. Cross-___domain information fusion for enhanced cell population delineation in single-cell spatial-omics data. Zenodo https://doi.org/10.5281/zenodo.14617085 (2025).
Shaban, M. et al. Data for MAPS: pathologist-level cell type annotation from tissue images through machine learning. Zenodo https://doi.org/10.5281/zenodo.10067010 (2023).
Acknowledgements
We thank the insightful discussion with lab members from the G.P.N., A.K.S., S.J. and Z.M. labs. G.P.N. is supported in part by the Rachford and Carlota A. Harris Endowed Professorship. A.K.S. is supported in part by a National Institutes of Health (NIH) grant no. P01AI177687, the Bill & Melinda Gates Foundation (INV-055706), an NIH grant no. 75N93019C00071, the Koch Institute Frontier Research Program, the Virginia and D.K. Ludwig Fund for Cancer Research and the Koch Institute Support (core) grant no. P30-CA014051 from the National Cancer Institute. S.J. is supported in part by NIH grant nos. DP2AI171139, P01AI177687, R01GM152585, R01AI149672 and U24CA224331, a Gilead’s Research Scholars Program in Hematologic Malignancies, a Sanofi Award, the Bill & Melinda Gates Foundation, the Dye Family Foundation and previously by the Leukemia Lymphoma Society Career Development Program. Z.M. is supported by National Science Foundation grant nos. 2345215 and 2245575. S.J. and Z.G.J. acknowledge support by a Beth Israel Deaconess Medical Center Spark grant. This article reflects the views of the authors and should not be construed as representing the views or policies of the institutions that provided funding.
Author information
Authors and Affiliations
Contributions
S.G., Z.M., S.C. and B.Z. conceptualized the study. S.G., Z.M., B.Z. and Y.W. developed and implemented the algorithm. B.Z., J.Y., Y.B., A.Y.H., Y.Y.Y., G.L., S.M. and Y.W. carried out the analysis. Z.G.J., S.J.R., K.W., G.P.N., A.K.S., S.J. and Z.M. provided the resources. G.P.N., A.K.S., S.J. and Z.M. supervised the study. B.Z., S.G. and S.C. contributed equally and have the right to list their names first in their CVs.
Corresponding authors
Ethics declarations
Competing interests
S.J. is a cofounder of Elucidate Bio, has received speaking honoraria from Cell Signaling Technology and has received research support from Roche, Novartis and Sanofi unrelated to this work. G.P.N. received research grants from Pfizer, Vaxart, Celgene and Juno Therapeutics during the time of and unrelated to this work. G.P.N. is a cofounder of Akoya Biosciences and Ionpath; an inventor on patent US9909167; and a scientific advisory board member for Akoya Biosciences. A.K.S. reports compensation for consulting or scientific advisory board membership from Honeycomb Biotechnologies, Cellarity, Ochre Bio, Relation Therapeutics, IntrECate Biotherapeutics, Bio-Rad Laboratories, Fog Pharma, Passkey Therapeutics and Dahlia Biosciences unrelated to this work. S.J.R. receives research support from Bristol Myers Squibb and KITE/Gilead. S.J.R. is a member of the scientific advisory board of Immunitas Therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Raphael Gottardo, Ahmed Mahfouz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available. Primary Handling Editor: Nick Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Detailed illustration of LENS-CNN and LENS-GNN-duo model architectures in the CellLENS pipeline.
The simplified versions are presented in Fig. 1. Additional details on CellLENS model architectures are described in the Methods section. Related code is deposited in the GitHub repository with documentation (see the Code Availability section for details).
Extended Data Fig. 2 Refined B cell subpopulations discovered by CellLENS in a tonsil CODEX dataset.
(A) Metrics evaluation of clustering performances on CODEX human tonsil tissue. Embeddings/representations of cells, from 7 different methods, were used as input: CellSNP representation, feature (protein expression table), concact (protein expression + neighborhood composition table), CCA representation, MOFA+ representation, SpiceMix representation, and MUSE representation. Clusters (for the calculation of Silhouette score, CH, and DB index) were generated with Leiden clustering using the same parameters. (B) UMAP visualization of the embedding and Leiden clustering result (cc: CellLENS clusters; fc; feature-only clusters). Left panel: CellLENS embedding; Right panel: feature expression table. (C) Zoom-in of the UMAP visualization of B cells (green), germinal center B cells (purple), and replicating non-GC cells (red) on CellLENS embedding (left) and feature-only embedding (right). Cells colored according to cell populations identified using CellLENS clusters in both UMAPs. (D) Zoom-in of the UMAP visualization of the same cell types shown in C, but colored by cluster numbers from CellLENS embedding (left) and feature-only embedding (right). CellLENS successfully separated replicating non-GC cells from GC B cells (c10 and c8), where feature-only failed (c8 partial). (E) B cell and replication-related marker expression (z-normed) heatmap. Left panel: clusters from CellLENS; Right panel: clusters from feature-only. (F) Visualization of cell type locations: B cells (green), GC B cells (purple), and replicating non-GC cells (red). Based on the ___location, replicating non-GC cells should not be mixed with GC B cells, as shown in the clustering result from feature-only.
Extended Data Fig. 3 Refined rare immune subpopulations discovered by CellLENS in a lymph node Xenium dataset (5k gene panel).
(A) UMAP visualization of the embedding and Leiden clustering result (cc: CellLENS clusters; fc; feature-only clusters). Left panel: CellLENS embedding; Right panel: feature expression table. (B) Heatmaps of top 10 mRNA marker genes for each cluster. Clusters from feature-only representation with Leiden clustering. Genes were identified via function ‘FindALLMarker’ in R package ‘Seurat’. (C) Same heatmap setup but clusters from CellLENS representation with Leiden clustering.
Extended Data Fig. 4 Supplementary plots illustrating the application of CellLENS on the human HCC CosMx-SMI data in Fig. 5.
(A) UMAP visualization of the embedding and Leiden clustering result (cc: CellLENS clusters; fc; feature-only clusters). Left panel: CellLENS embedding; Right panel: feature expression table. (B) Heatmaps of top 10 mRNA marker genes for each cluster. Genes were identified via function ‘FindALLMarker’ in R package ‘Seurat’. Left panel: clusters from CellLENS representation; Right panel: clusters from feature-only representation. (C) Visualization of spatial locations in the HCC tissue of different Macrophage subpopulations identified by feature-only representation. In each plot, cells in a target cluster are colored in red, HCC tumor cells are colored in grey, and other cells and empty spaces are colored in black.
Extended Data Fig. 5 Evaluation of CellLENS performance stability on adjacent CODEX tonsil sections: correspondence of CellLENS clusters identified individually on two sections.
CODEX tonsil tissue from two adjacent sections52 were used to evaluate the robustness of CellLENS performance. Shown here are Leiden clustering results (resolution = 1) based on CellLENS embeddings. In each of the three columns, left are clusters identified from CellLENS embedding of slide 1, and right are corresponding clusters in slide 2 aligned to their counterparts in slide 1.
Extended Data Fig. 6 Comprehensive benchmarking with additional methods.
A total of 12 methods are benchmarked here: CellLENS (our method). SpaGCN: Designed for spot-level spatial modalities (to identify spatial domains). StLearn: Designed for spatial modalities (to identify cell populations). SEDR: Designed for spatial modalities (to identify cell populations). MUSE: Designed for single-cell spatial modalities (to identify cell populations). SpiceMix: Designed for spatial modalities (to identify cell populations). BANKSY: Designed for spatial modalities (to identify cell populations or spatial domains). CellCharter: Designed for spatial modalities (to identify spatial domains). MOFA+: Designed for general modalities. CCA: General statistical procedure with canonical correlation analysis. Concatenation: Direct concatenation between feature and ___location matrix. Feature-only profile: Conventional way of cell type identification. We applied all 12 methods to five datasets presented in our manuscript (CODEX spleen, Xenium tonsil, CODEX tonsil, CODEX cHL, and CosMx HCC) and evaluated them using four different metrics (See Methods for details). Here we aggregated all the results across metrics and datasets into one summary figure. Each subpanel represents a specific metric (for example, Modularity score). The Y-axis indicates the average ranking of a method across benchmarking conditions (for example, K clusters or resolution numbers). On the X-axis, methods are arranged by their average ranking across all four metrics, such that methods on the left perform the best overall. In the summary figure, the rankings were averaged across all five datasets.
Extended Data Fig. 7 Comparison between embeddings generated by CellLENS and SpaGCN on CODEX mouse spleen data.
While initially designed for spot-level spatial-omics data, SpaGCN could run on the whole CODEX mouse spleen data ( ~ 50k cells). We compared the cell type delineation ability between the embeddings from CellLENS and SpaGCN. The figure setup is the same as described in Fig. 2. SpaGCN failed to identify the various B cell subpopulations captured by CellLENS.
Extended Data Fig. 8 Comparison between embeddings generated by CellLENS and SpaGCN on CosMx HCC data.
While initially designed for spot-level spatial-omics data, SpaGCN could run on the whole CosMx-SMI human HCC data ( ~ 50k cells). We compared the cell type delineation ability between the embeddings from CellLENS and SpaGCN. The figure setup is the same as described in Fig. 5. SpaGCN failed to identify the various Macrophage subpopulations captured by CellLENS.
Extended Data Fig. 9 Loss quantification of the CellLENS model when using different image feature extraction processes.
(A) We compared the CellLENS model training losses, across three CellLENS variations: 1) Default CellLENS, where the imaging feature extraction part is done by training an Alex-Net like CNN encoder (supervised by local cell type neighborhood composition vector). 2) CellLENS with a pre-trained ResNet50, where the image features were directly extracted with the pre-trained ResNet 50 model, flattened, and reduced to a vector with 128 dimensions. This vector is swapped with the original image feature vector obtained from the retrained Alex-Net model, and the rest of the CellLENS training process remains the same; 3) CellLENS with a pre-trained ViT (transformer), using a similar process as the pre-trained ResNet50 in (2). (B) We compared the losses from three CellLENS variations: 1) CellLENS default with Alex-Net as described above. 2) CellLENS but swapping out the Alex-Net architecture with a ResNet50 architecture, and retraining its weights (initialized at pretrained weights). 3) CellLENS but swapping out the Alex-Net architecture with a ViT architecture, and retraining its weights (initialized at pretrained weight). The model loss was calculated the same as described in the Methods section paragraph ‘Information retrieval efficacy evaluation of the LENS-GNN duo module’. In these cases, we implemented a 80/20 train test data split. Retraining was only done on the train data, and loss values were calculated on test data.
Extended Data Fig. 10 Memory and run-time benchmarking for training CellLENS on datasets of different sizes.
We ran CellLENS on increasing numbers of cells: ~10k, ~50k, ~150k, ~500k, ~1.5 mil, and recorded the memory and run-time statistics. The benchmarking was performed on an NVIDIA A5000 GPU with an SSD disk (PCIe 4.0) for I/O. Left panel: GPU memory usage during CNN and GNN training stages of CellLENS, across variable cell numbers. Middle panel: Run-times of CellLENS CNN training stage. Recorded run-time for both default LENS-CNN or LENS-CNN with a ViT architecture. Right panel: Run-time of CellLENS GNN training stage.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, B., Gao, S., Chen, S. et al. CellLENS enables cross-___domain information fusion for enhanced cell population delineation in single-cell spatial omics data. Nat Immunol 26, 963–974 (2025). https://doi.org/10.1038/s41590-025-02163-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41590-025-02163-1