Abstract
Gastric cancer (GC) is a prevalent malignant tumor of the digestive system that is often diagnosed at advanced stages owing to inconspicuous early symptoms and a lack of specific examination methods. Effective treatment of advanced stages remains challenging, emphasizing the need for new therapeutic targets. Metabolic reprogramming, a hallmark of tumors, plays a pivotal role in tumor progression, immune evasion, and immune surveillance. DNA damage-regulated autophagy modulator 1 (DRAM1) encodes a hexameric transmembrane protein that is predominantly located in lysosomes and induces autophagy; however, its mechanism of action in gastric cancer remains unclear. Our study found that elevated DRAM1 expression in patients with GC correlated with survival and prognosis. DRAM1 knockdown suppressed energy metabolism in GC cells through the PI3K/AKT/mTOR signaling pathway, thereby mitigating GC progression. Atorvastatin, a focus of recent tumor research, significantly enhanced apoptosis levels in DRAM1 knockdown GC cells compared to the control group. Therefore, through metabolic reprogramming, DRAM1 may serve as a potential therapeutic target for GC prevention.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is a very common type of cancer globally. The 2020 Global Cancer Morbidity and Mortality Survey revealed that GC ranks fifth in occurrence and fourth as the leading cause of death1. Additionally, evidence linking metabolism and tumor cells has accumulated over the years. However, tumor cells can reprogram their metabolism by altering glucose transport, glutamine metabolism, the electron transport chain, and the pentose phosphate pathway (PPP) to adapt and acquire the necessary nutrients and energy for survival2.
DRAM1, located in the lysosomes, functions as a hexatransmembrane protein that initiates autophagy3. It directs newly synthesized amino acid transporters to the lysosomes and facilitates the movement of amino acids within the lysosomes, promoting mTORC1 activation and regulating amino acid metabolism4. In alcoholic fatty liver disease, DRAM1 is highly expressed and interacts with PKM2 to enhance macrophage activation, thereby exacerbating the progression of adrenoleukodystrophy5. Additionally, DRAM1 is implicated in the occurrence and development of various conditions, including tumors, inflammatory bowel disease, myocardial ischemia, and leukemia6,7,8,9,10. DRAM1 recruits Bax to lysosomes, leading to the release of lysosomal cathepsin B, which triggers tumor cell apoptosis via the truncated BH3-interacting ___domain death agonist pathway11. Furthermore, DRAM1 promotes the lysosomal transport of marine mycobacteria in macrophages in models of Mycobacterium tuberculosis, as well as the phagocytosis of Gigi bacteria to defend against Gigi bacterium infection12.
Currently, the molecular functions and mechanisms of action of DRAM1 in GC are poorly understood. This study aimed to elucidate the molecular functions and underlying mechanisms of DRAM1 in GC.
Results
DRAM1 expression was elevated in GC and associated with GC progression
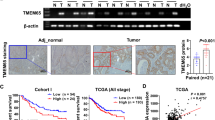
To assess the clinical significance of DRAM1 in GC, The Cancer Genome Atlas (TCGA) database was used to analyze the different DRAM1 expressions in normal gastric tissues and tumors (Fig. 1a). Furthermore, the Kaplan–Meier survival curve indicated that patients with lower DRAM1 mRNA levels had better overall survival than those with higher levels (Fig. 1b). Western blotting was performed on GC and adjacent tissues to detect the expression level of DRAM1, revealing that DRAM1 expression in tumor tissues was significantly higher than that in adjacent tissues (Fig. 1c,d). Subsequently, DRAM1 expression was examined in normal gastric mucosal cells and five GC cell lines. DRAM1 was highly expressed in adenocarcinoma gastric stomach AGS and MGC803 cells and low in HGC27 cells (Fig. 1e,f). Therefore, we constructed stable, low-expression DRAM1 cell lines in AGS cells and stably overexpressed DRAM1 in HGC27 cells. We used quantitative polymerase chain reaction (qPCR) to assess the efficiency of the knockdown and overexpression (Fig. 1g,h). In summary, these results suggest that DRAM1 is inversely associated with the adverse outcomes of GC progression.
DRAM1 was upregulated in gastric cancer (GC) and was associated with the progression of GC. (a) DRAM1 mRNA was significantly higher in GC tissues than the adjacent normal tissues (***: P = 0.001) based on the TCGA dataset (375 GC tissues and 32 adjacent normal tissues). (b) Survival analysis based on the TCGA dataset indicated that higher DRAM1 expression was associated with poor OS in patients with GC (P = 0.039). (c) DRAM1 protein expression was confirmed in four pairs of GC tissues by western blotting. (d) Bar chart was the quantitative analysis of DRAM1 protein in four pairs of GC tissues (***: P < 0.001;**: P = 0.003). (e) Western blot was used to confirm the expression of DRAM1 in gastric cancer cell lines. (f) The histogram showed the expression of DRAM1 mRNA in gastric cancer cells . Compared with normal gastric mucosa, DRAM1 was low expressed in HGC27 cells (**: P = 0.005) and high expression in AGS, MKN45, MGC803, and SGC7901 cells (****: P < 0.001). (g) Bar chart represented the efficiency of DRAM1 knockdown in AGS cells (Ctrl vs shDRAM1#1, **: P = 0.008; Ctrl vs shDRAM1#2, **: P = 0.008). (h) Bar chart represented the efficiency of overexpression of DRAM1 in HGC27 cells (NC vs OE, ***: P < 0.001).
DRAM1 knockdown suppressed the migration, proliferation, and metastasis of GC cells
To elucidate the molecular function of DRAM1, DRAM1 knockdown was performed in AGS cells. DRAM1 knockdown inhibited the migration and proliferation of GC cells. This conclusion was supported by cell scratch and plate clone formation assays (Fig. 2a–d). To investigate the association between epithelial-mesenchymal transition (EMT) and cell migration and invasion, we performed immunoblotting analysis to detect EMT markers. DRAM1 knockdown may contribute to the epithelial marker E-cadherin upregulation and mesenchymal marker N-cadherin downregulation in AGS cells (Fig. 2e). Furthermore, we evaluated the impact of DRAM1 knockdown on the mobility and multiplication of GC cells using cell counting kit-8 (CCK-8), Transwell, and EdU cell proliferation tests. AGS cell proliferation and metastasis were inhibited by suppressing DRAM1 (Fig. 2f–h). These results suggest that DRAM1 promotes the metastasis of GC cells, and EMT may be involved in the role of DRAM1 in promoting GC metastasis.
Knockdown of DRAM1 inhibited the migration, proliferation, and metastasis of gastric cancer cells. (a) The effect of DRAM1 on cell migration was evaluated using wound-healing assay, and the results showed that DRAM1 knockdown inhibited the migration of gastric cancer cell. (b) Bar chart showed the difference in migration area between DRAM1 knocked down GC cells and NC cells (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ***: P < 0.001). (c) Plate cloning experiments confirmed that knockdown of DRAM1 inhibited the proliferation of gastric cancer cells. (d) Bar chart showed the number of knocked down GC cells and NC cells analyzed by plate cloning experiments (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ***: P < 0.001). (e) Western blotting was used to detect EMT pathway-related proteins between DRAM1 knocked down GC cells and NC cells. Compared with NC gastric cancer cells, the expression of E-cadherin in gastric cancer cells with DRAM1 knockdown was up-regulated, and the expression of N-cadherin was down-regulated. (f) CCK8 assay confirmed that knockdown of DRAM1 inhibited the proliferation of gastric cancer cells (Ctrl vs shDRAM1#1, ****: P < 0.001; Ctrl vs shDRAM1#2, ****: P < 0.001). (g) The Transwell assay confirmed that knockdown of DRAM1 inhibited gastric cancer cell migration and metastasis. (h) EDU assay confirmed that knockdown of DRAM1 inhibited the proliferation of gastric cancer cells. Scale bar = 100 μm.
DRAM1 overexpression promoted the migration, proliferation, and metastasis of GC cells
HGC27 cells were used to establish a cell line in which DRAM1 was stably overexpressed. DRAM1 overexpression enhanced the migration and proliferation of HGC27 cells, as demonstrated by cell scratch assay and plate cloning experiments (Fig. 3a–d). Cell proliferation assays were performed using CCK8, Transwell, and EdU. DRAM1 overexpression promoted HGC27 cell proliferation (Fig. 3f–g). Furthermore, DRAM1 overexpression increased the number of cells crossing the polycarbonate membrane compared with that in the control group (Fig. 3e). In HGC27 cells, E-cadherin and N-cadherin expressions were detected using western blotting, revealing that DRAM1 overexpression may contribute to N-cadherin upregulation and E-cadherin downregulation (Fig. 3h).
Overexpression of DRAM1 promoted the migration, proliferation, and metastasis of gastric cancer cells. (a) The effect of DRAM1 on cell migration was evaluated using wound-healing assay, and the results showed that DRAM1 overexpression markedly promoted the migration of gastric cancer cell. (b) Bar chart showed the difference in migration area between DRAM1 overexpressing GC cells and NC cells (NC vs OE, **: P = 0.002). (c) Plate cloning experiments confirmed that DRAM1 overexpression promoted the proliferation of gastric cancer cells. (d) Bar chart showed the number of DRAM1 overexpressing GC cells and NC cells analyzed by plate cloning experiments (NC vs OE, ***: P < 0.001). (e) The Transwell assay confirmed that overexpressing DRAM1 promoted gastric cancer cell migration and metastasis. (f) CCK8 assay confirmed that overexpressing DRAM1 promoted the proliferation of gastric cancer cells (NC vs OE, ****: P < 0.001). (g) EDU assay confirmed that knockdown of overexpressing DRAM1 promoted the proliferation of gastric cancer cells. Scale bar = 100 μm. (h) Western blotting was used to detect EMT pathway-related proteins between DRAM1 overexpressing GC cells and NC cells. Compared with NC gastric cancer cells, the expression of N-cadherin in gastric cancer cells with DRAM1 overexpression was up-regulated, and the expression of E-cadherin was down-regulated.
Suppressing DRAM1 promoted apoptosis of GC cells, while DRAM1 overexpression inhibited apoptosis of GC cells
We determined whether DRAM1 affects apoptosis in GC cells. DRAM1 knockdown increased apoptosis occurrence in AGS cells, whereas DRAM1 overexpression inhibited apoptosis in HGC27 cells (Fig. 4a–d). Western blotting was performed to ascertain the presence of apoptotic proteins in cells. These findings indicated that suppressing DRAM1 increased the expression of the pro-apoptotic protein Bax in AGS cells while decreasing the expression of the anti-apoptotic protein Bcl-2. However, when DRAM1 was overexpressed, the opposite outcome was observed (Fig. 4e–h).
Silencing DRAM1 promoted apoptosis of gastric cancer cells, while overexpression of DRAM1 inhibits apoptosis of gastric cancer cells. (a) The apoptosis of DRAM1 knocked down gastric cancer cells and NC gastric cancer cells was detected by flow cytometry. (b) Bar chart showed that knockdown of DRAM1 promoted apoptosis in gastric cancer cells (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ***: P < 0.001). (c) The apoptosis of DRAM1 overexpressing gastric cancer cells and NC gastric cancer cells was detected by flow cytometry. (d) Bar chart showed that DRAM1 overexpressing inhibited apoptosis in gastric cancer cells (NC vs OE, *: P = 0.013). (e) Western blotting was used to detect apoptotic proteins in DRAM1 knocking gastric cancer cells and NC gastric cancer cells. (f) Protein quantitative analysis showed that knockdown of DRAM1 up-regulated the expression of Bax (Ctrl vs shDRAM1#1, *: P < 0.015; Ctrl vs shDRAM1#2, *: P < 0.031)and down-regulated the expression of Bcl-2 (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ****: P < 0.001). (g) Detecting apoptotic proteins in DRAM1-overexpressing gastric cancer cells and NC gastric cancer cells by Western blotting. (h) Bar chart showed that overexpressed DRAM1 up-regulated Bcl-2 (NC vs OE, ***: P < 0.001) expression and down-regulated Bax (NC vs OE, ****: P < 0.001) by Western blotting analysis.
DRAM1 inhibited GC cell proliferation via the PI3K/AKT/mTOR signaling pathway
Previous experiments showed that suppressing DRAM1 can impede the growth of GC cells. However, the mechanisms underlying these inhibitory effects were unknown. Transcriptome sequencing of untreated and DRAM1-silenced AGS cells was performed. Figure 5a shows the technology roadmap for this experiment. Transcriptomic data were subjected to differential gene expression and enrichment analyses. The differentially expressed genes were visualized using heat maps (Fig. 5b). Differential gene enrichment analysis showed that the mTOR signaling pathway played a significant role (Fig. 5c)13. Subsequently, western blotting was used to detect the expression of proteins related to the mTOR signaling pathway, including PI3K, AKT, mTOR, and p62. Suppressing DRAM1 in AGS cells inhibited the expression of PI3K, AKT, p-AKT, mTOR, p-mTOR, and p62 (Fig. 5d,e). The opposite results were observed in HGC27 cells overexpressing DRAM1 (Fig. 5f,g). In addition, the HGC27 cells and HGC27 cells overexpressing DRAM1 were treated with the PI3K inhibitor LY294002. The CCK-8 assay was used to assess GC cell proliferation. The results indicated that DRAM1 overexpression significantly promoted HGC27 cell proliferation, whereas LY294002 significantly inhibited GC cell proliferation (Fig. 5h). Western blot analysis demonstrated that AKT and mTOR protein levels were consistent with the expected outcomes. LY294002 treatment reduced AKT, p-AKT, mTOR, and p-mTOR levels (Fig. 5i). These experimental findings confirmed that DRAM1 knockdown impedes GC cell proliferation by suppressing the PI3K/AKT/mTOR signaling pathway.
DRAM1 inhibited gastric cancer cell proliferation through the PI3K/AKT/mTOR signaling pathway. (a) Flowchart of RNA sequencing of DRAM1 knocked GC cells and NC GC cells. (b) Heatmap showed DRAM1 knocked down GC cells and NC GC cell differential genes. (c) GSEA-KEGG enrichment analysis showed that the differential genes were significantly enriched in the mTOR signaling pathway. (d) Western blotting was used to detect PI3K-AKT-mTOR signaling pathway-related proteins in silenced DRAM1 gastric cancer cells and NC gastric cancer cells. (e) Protein quantification analysis showed that DRAM1 silencing inhibited the expression of PI3K (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ****: P < 0.001), AKT (Ctrl vs shDRAM1#1, **: P < 0.002; Ctrl vs shDRAM1#2, **: P < 0.005), p-AKT (Ctrl vs shDRAM1#1, ****: P < 0.001; Ctrl vs shDRAM1#2, ****: P < 0.001), mTOR (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ***: P < 0.001), p-mTOR (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ***: P < 0.001) and P62 (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, ***: P < 0.001) in GC cells compared with NC GC cells. (f) Detecting PI3K-AKT-mTOR signaling pathway-related proteins by Western blotting in DRAM1 gastric cancer cells and NC gastric cancer cells. (g) Bar chart showed that compared with NC gastric cancer cells, overexpression of DRAM1 promoted the expression of PI3K (NC vs OE, **: P = 0.007), AKT (NC vs OE, **: P = 0.002), p-AKT (NC vs OE, ***: P < 0.001), mTOR (NC vs OE, **: P = 0.001), p-mTOR (NC vs OE, ***: P < 0.001) and P62 (NC vs OE, **: P = 0.001) in gastric cancer cells. (h) CCK8 assay confirmed that PI3K inhibitor LY294002 (10 μm) significantly inhibited the proliferation of gastric cancer cells overexpressing DRAM1 (NC vs OE, ****: P < 0.001, NC vs OE + LY294002, ****: P < 0.001). (i) Western blotting confirmed that LY294002 (10 μm) significantly inhibited the expression of mTOR, p-mTOR, AKT and p-AKT in gastric cancer cells overexpressing DRAM1.
mTOR in energy metabolism of GC cells is mediated by DRAM1
mTOR is a crucial molecule involved in cellular energy metabolism. We performed the Kyoto Encyclopedia of Genes and Genomes enrichment analysis of the differentially expressed genes in the transcriptome. The analysis revealed significant alterations in the biological processes of glucose metabolism (Fig. 6a–c)13. Hence, we used qPCR to analyze the mRNA expression of genes associated with glucose metabolism in GC cells. DRAM1 knockdown in AGS cells significantly inhibited the expression of energy metabolism-related genes, such as HK2, PFK1, GCK, PKM2, LDHA, SLC1A5, and IDH1 (Fig. 6d,e). Conversely, DRAM1 overexpression in HGC27 cells significantly upregulated the expression of PFK1, GCK, PKM2, LDHA, SLC7A5 and SLC1A5 (Fig. 6f). Furthermore, ATP and lactate production experiments in GC cells demonstrated that suppressing DRAM1 inhibited ATP and lactate production in AGS cells (Fig. 6g,h). In addition, immunofluorescence experiments confirmed that suppressing DRAM1 attenuated the fluorescence intensity of SLC1A5 and LDHA in AGS cells (Fig. S1a–c). Conversely, DRAM1 overexpression enhanced the fluorescence intensities of LDHA and SLC3A2 in HGC27 cells (Fig. S2a,b). mTOR is an important molecule for sensing changes in cellular energy metabolism. To investigate the regulation of energy metabolism in GC cells by DRAM1 via the mTOR signaling pathway, AGS and HGC27 cells were treated with rapamycin, an mTOR inhibitor. Next, we performed western blotting to analyze the expression of genes related to energy metabolism. The results demonstrated that inhibiting mTOR in GC cells led to the downregulation of HK2, PFK1, GCK, PKM2, LDHA, and SLC1A5 expressions (Fig. S3a,b). In conclusion, these results suggest that DRAM1 knockdown can mediate the metabolic reprogramming of GC cells through mTOR.
DRAM1 mediated energy metabolism in gastric cancer cells through mTOR. (a) GO analysis was performed to analyze the differential genes of DRAM1 silencing GC cells and NC GC cells, which were significantly related to energy metabolism pathway. (b) KEGG analysis was conducted to examine the differential genes between DRAM1 silenced gastric cancer cells and non-silenced gastric cancer cells. The results revealed a significant association with the energy metabolism pathway. (c) KEGG was used to analyze the linkages between the metabolic pathways of differentially enriched genes. (d) Knocking down of DRAM1 inhibited the mRNA expression of HK2 (Ctrl vs shDRAM1#1,ns; Ctrl vs shDRAM1#2, *: P = 0.013), PFK1 (Ctrl vs shDRAM1#1, **: P = 0.001; Ctrl vs shDRAM1#2, **: P = 0.002), GCK (Ctrl vs shDRAM1#1, *: P = 0.011; Ctrl vs shDRAM1#2, **: P = 0.004), PKM2 (Ctrl vs shDRAM1#1, *: P = 0.042; Ctrl vs shDRAM1#2,ns), ATP6V1A (Ctrl vs shDRAM1#1, **: P = 0.001; Ctrl vs shDRAM1#2, **: P = 0.002), LDHA (Ctrl vs shDRAM1#1, **: P = 0.004; Ctrl vs shDRAM1#2, **: P = 0.006), c-MYC (Ctrl vs shDRAM1#1, ***: P < 0.001; Ctrl vs shDRAM1#2, **: P = 0.002) and IDH1 (Ctrl vs shDRAM1#1, * : P = 0.038; Ctrl vs shDRAM1#2, **: P = 0.005) in gastric cancer cells by q-PCR analysis. (e) Western blotting confirmed that knockdown of DRAM1 inhibited the expression of PKM2, HK2, GCK, SLC1A5 and LDHA proteins in gastric cancer cells. (f) Western blotting confirmed that overexpression of DRAM1 promoted the expression of PKM2, GCK, SLC1A5, LDHA, PFK1 and SLC7A5 proteins in gastric cancer cells. (g) Knockdown of DRAM1 inhibit ATP production in gastric cancer cells (Ctrl vs shDRAM1#1, **: P = 0.006; Ctrl vs shDRAM1#2, **: P = 0.008). (h) Knockdown of DRAM1 inhibited lactate production in gastric cancer cells (Ctrl vs shDRAM1#1, **: P = 0.002; Ctrl vs shDRAM1#2, **: P = 0.002).
Suppressing DRAM1 reduced the growth of subcutaneous tumors in nude mice
To confirm the in vivo effects of DRAM1, AGS, and DRAM1-knockdown AGS cells were injected into the ventral lateral aspect of male nude mice to create xenograft models. The mice were observed for 4–6 weeks. Compared with control tumors, DRAM1 knockdown significantly reduced the growth rate, size, and weight of AGS cells in mice (Fig. 7a–d). Subsequently, western blot analysis was performed on mouse tumor tissues, and DRAM1 knockdown inhibited mTOR, p-mTOR, AKT, and p-AKT proteins (Fig. 7e). In addition, we stained mouse tissues with hematoxylin and eosin (H&E), Ki-67, and TUNEL. DRAM1 knockdown significantly inhibited tumor growth in vivo and increased tumor apoptosis in vivo (Fig. 7f–i).
Knockdown of DRAM1 inhibited the growth of subcutaneous tumors in nude mice. (a) Knocking down of DRAM1 inhibited subcutaneous growth in nude mice through vivo experiments (n = 3). (b) Knockdown of DRAM1 significantly inhibited tumor size in nude mice (n = 3). (c) Knockdown of DRAM1 significantly inhibits the volume of tumors in nude mice by measuring mouse volume (Ctrl vs shDRAM1#1, **: P = 0.002). (d) Knocking down DRAM1 remarkablely inhibited tumor weight in nude mice (Ctrl vs shDRAM1#1, ****: P < 0.001). (e) Western blot analysis of nude mouse tumor tissues showed that knockdown of DRAM1 significantly inhibited the expression of p-AKT and p-mTOR. (f) HE staining of DRAM1 knockdown tumor tissues and control tissues. Scale bar = 100 μm. (g) DRAM1 expression in tumor tissues and control tissues with DRAM1 knockdown was detected by IF. Scale bar = 100 μm. (h) Immunofluorescence assay confirmed that knockdown of DRAM1 significantly inhibited the expression of ki-67 in tumor tissues. Scale bar = 100 μm. (i) Tunel staining confirmed that knockdown of DRAM1 promotes apoptosis in nude mice. Scale bar = 100 μm.
p53 regulates the expression of DRAM1
Our findings indicate that DRAM1 regulates the proliferation, migration, and metastasis of gastric cancer cells via the PI3K/AKT/mTOR pathway. To better understand the upstream regulation of DRAM1, we reviewed the relevant literature and identified DRAM1 as a direct downstream target of the p53 gene. Consequently, we investigated whether p53 regulates DRAM1 expression in gastric cancer cells. In both HGC27 and AGS cells, we found that p53 knockdown reduced DRAM1 mRNA and protein expression levels (Fig. 8a,b).
p53 regulates the expression of DRAM1. (a) In HGC27 and AGS cells, knocking down the p53 gene can inhibit the mRNA levels of DRAM1 (AGS:NC vs sip53-1, ***: p < 0.001, NC vs sip53-2, ***: p < 0.001;HGC27: NC vs sip53-1, **: p = 0.002, NC vs sip53-2, **: p = 0.002). (b) In HGC27 and AGS cells, knocking down the p53 gene can inhibit the protein expression levels of DRAM1.
Discussion
The role and mechanism of DRAM1 have been reported in several types of tumors but are rarely reported in GC. DRAM1 acted as a tumor suppressor in lung cancer by promoting the assembly of lysosomal v-ATPase and the lysosomal degradation of EGFR. In renal clear cell carcinoma, DRAM1 promoteed apoptosis of cancer cells by inhibiting the degradation of BAX, while also suppressing proliferation, migration, and invasion through the Akt signaling pathway. Additionally, DRAM1 participated in regulating the migration and invasion of HepG2 cells via the autophagy-EMT pathway.This study aimed to elucidate the molecular functions and underlying mechanisms of DRAM1 in GC. Suppressing DRAM1 expression inhibits glucose metabolism in GC by modulating the PI3K/AKT/mTOR signaling pathway. Therefore, targeting DRAM1 may be a novel approach for treating patients with GC.
DRAM1 is a downstream target of p53 and plays an important role in autophagy4. DRAM1 regulates autophagy and cell proliferation by inhibiting the PI3K/AKT/mTOR/rpS6 pathway14. DRAM1 regulates autophagy by regulating lysosomal degradation pathways. Furthermore, DRAM1 has a significant impact on regulating tumor cell migration and invasion via the autophagy-EMT pathway15,16. Autophagy and metabolic reprogramming are closely linked. Tumor metabolism plays a crucial role in signal transduction during tumorigenesis, drives immune evasion, and hinders immune surveillance through metabolites. This has broad implications for regulating antitumor immune responses17,18,19. Macrophages, followed by T cells and tumor cells, absorb most glucose in tumor tissues. Tumor cells have the highest glutamine absorption rate20.
Abnormal glucose metabolism can affect various physiological processes, including cancer cell proliferation, the cell cycle, drug resistance, DNA damage repair induction, autophagy enhancement, tumor microenvironment alteration, and increased exosome secretion21,22,23,24. Glucose metabolism includes anaerobic glycolysis, aerobic oxidation, and the PPP25. Benzylserine, an inhibitor of ASCT2, can significantly inhibit GC progression26.
Macrophages are the most prevalent inflammatory cells in tumors. Polarization may occur in the presence of cytokines and metabolites27,28,29. M1 macrophages have antitumor effects, whereas M2 macrophages have pro-tumor effects. The glycolytic metabolite lactate and glutamine metabolite α-ketoglutarate can promote macrophage M2 polarization30,31. This study suggested that suppressing DRAM1 not only inhibited glycolysis in GC cells but also suppressed the expression of crucial enzymes involved in glutamine metabolism of the GC cells. In summary, the results of this study corroborate the potential of targeted DRAM1-mediated metabolic reprogramming as a novel therapeutic strategy against GC while highlighting its potential involvement in developing chemotherapeutic resistance. This study also suggests that suppressing DRAM1 not only inhibits glycolysis in GC cells but also suppresses the expression of crucial enzymes involved in glutamine metabolism of the GC cells.
Malignant tumors are the primary cause of death globally, with their occurrence and fatality rates rising rapidly1. However, developing new drugs for treating malignant tumors is time-consuming and expensive32. Drug repurposing for cancer treatment has been in practice for several years. Atorvastatin is a widely used statin medication, primarily prescribed to reduce cholesterol levels and prevent cardiovascular diseases.Statins are potent competitive inhibitors of 3-hydroxy-3-methylglutaryl-CoA reductase and are commonly used as lipid-lowering drugs. Statins exhibit pleiotropic effects that are mediated by epigenetic mechanisms33. However, recent study of statins in tumors has garnered significant attention34. For example, statins modulate the inflammatory tumor microenvironment and improve the effectiveness of anti-PD-1 immunotherapy in non-small-cell lung cancer. This is achieved by transcriptional PD-L1 expression inhibition and inducing ferroptosis35. Statins overcome chemoresistance in small-cell lung cancer via the mevalonate-geranylgeranyl diphosphate pathway36. Statins reduced the incidence of GC and improved the survival rate of patients with GC37. The anticancer mechanism of statins involves inhibiting the absorption of proteins by tumor cells. Therefore, glucose is not the only nutrient that stimulates cancer cells, and amino acids are crucial for the sustenance of cancer cells38. Additionally, this study confirmed that atorvastatin could induce apoptosis in GC cells, with a more significant effect observed in cells in which DRAM1 was suppressed (Figures S4a–e). Therefore, combining targeted inhibition of DRAM1 and atorvastatin may significantly impact the treatment and prognosis of patients with GC.
This study revealed significant DRAM1 upregulation in GC, establishing a negative correlation with patient prognosis. Through meticulous exploration, we elucidated the intricate involvement of DRAM1 in GC onset and progression via the PI3K/AKT/mTOR signaling pathway. In a xenograft tumor mouse model, DRAM1 suppression effectively curbed tumor size, volume, and weight. This groundbreaking discovery highlights the potential of DRAM1 as an innovative target for future targeted therapies in GC.
Research limitations
This study mainly focuses on the role of DRAM1 in gastric cancer, and although some progress has been made, there are still certain limitations. First, the research is limited to the PI3K/AKT/mTOR signaling pathway, and future studies should expand to other potential pathways to gain a more comprehensive understanding of DRAM1’s function in gastric cancer. Second, the study found that DRAM1 affects the energy metabolism of gastric cancer cells, but the specific regulatory mechanisms were not explored in depth. Future research should investigate these regulatory mechanisms to clarify DRAM1’s specific role in energy metabolism.
Materials and methods
Cell lines, cell culture and cell transfection
Human gastric cancer cell lines GES-1, AGS, HGC27, MKN45, MGC803, and SGC7901 were obtained from the Shanghai Institute of Cell Research, Chinese Academy of Sciences. These cell lines were cultured in Roswell Park Memorial Institute 1640 (Biochannel, Nanjing, P.R. China) supplemented with 10% fetal bovine serum (FBS; Wisent, Nanjing, P.R. China) and 1% penicillin/streptomycin (Thermo Fisher Scientific, MA, USA). DRAM1 knockdown lentivirus was purchased from GENECHEM (Shanghai, P. R. China). DRAM1 overexpression lentivirus was purchased from Corues Biotechnology (Nanjing, P.R. China). The cells were evenly distributed in 6-well plates at a density of 100,000 cells/well for lentiviral cell transfection experiments. A mixture of 1 ml of complete medium, HitransG A/P infection enhancement solution, and lentivirus was added to the cells. The infection multiplicity was 20, and the virus titer was 7.5 × 108. After 24 h, the medium was replaced, and stable cell lines were screened with puromycin after 72 h. The cell lines were placed in a humidified environment with 5% CO2 and incubated at 37℃ for cultivation.
Tissue samples
We collected 4 gastric cancer samples and their corresponding normal tissues from Nanjing Drum Tower Hospital. None of the patients had undergone radiation or chemotherapy prior to surgery. This study received approval from the Ethics Committee of Drum Tower Hospital, Nanjing University Medical School, and informed consent was obtained from all patients’ guardians. Clinical tissue samples were managed in accordance with the ethical standards outlined in the Declaration of Helsinki.
Bioinformatics
TCGA (https://www.cancer.gov/ccg) was used for gene set enrichment analysis to analyze DRAM1 expression in patients with GC. The impact of DRAM1 on clinical survival was analyzed using the Kaplan–Meier plotting method (https://www.kmplot.com/).KEGG database (https://www.kegg.jp/kegg/kegg1.html) was used to analyze the pathways and metabolism-related features of differential genes.
RNA isolation, reverse transcription (RT), and qPCR
The total RNA was extracted from tissues and cells using the TRIzol® reagent (Kangwei Century, Changzhou, P.R. China). Furthermore, a TRUEscript RT kit (Vazyme Biotech, Nanjing, P.R. China) was used to reverse transcribe 1000 ng of total RNA into cDNA, with a final reaction volume of 100 μL. Commercially synthesized specific primers (GenScript Biotech, Nanjing, P.R. China)were used for qPCR. SYBR Green qPCR Supermix (Vazyne Biotech, Nanjing, P.R. China) and a 7500 real-time PCR system (Applied Biosystems, MA, USA)were used for qPCR analysis. The relative mRNA expression was normalized to the expression of β-actin and calculated using the 2-ΔΔCT method. The primer sequences used were as follows: Human-β-actin F: GCGTGACATTAAGGAGAAG, R: GAAGGAAGGCTGGAAGAG; human-DRAM1 F: TCAACCCCTTCCTCCCGTAT, R: TCGTGGCTGCACCAAGAAAT; human-SLC1A5 F: TCCAGCCCTCGGGAGTAAATA, R: TCACCCTGGTTCCGGTGATA; human-PKM2 F: ATGTCGAAGCCCCATAGTGAA, R: TGGGTGGTGAATCAATGTCCA; human-SLC7A5 F: GGAAGGGTGATGTGTCCAATC, R: TAATGCCAGCACAATGTTCCC; human-SLC3A2 F: TGAATGAGTTAGAGCCCGAGA, R: GTCTTCCGCCACCTTGATCTT; human-c-myc F: GTCAAGAGGCGAACACACAAC, R: TTGGACGGACAGGATGTATGC; human-LDHA F: ATGGCAACTCTAAAGGATCAGC, R: CCAACCCCAACAACTGTAATCT; human-IDH1 F: CACCAAATGGCACCATACGAA, R: CCCCATAAGCATGACGACCTAT; human-GOT1 F: ATTTCTTAGCGCGTTGGTACA, R: ACACAGCATTGTGATTCTCCC; human-GOT2 F: GCCTGGGTGATTTGCTCTTTT, R: CCTTTAGTGCAGTGGTGAACTT; human-HK2 F: GAGCCACCACTCACCCTACT, R: CCAGGCATTCGGCAATGTG; human-PFK1 F: AGCGTTTCGATGATGCTTCAG, R: GGAGTCGTCCTTCTCGTTCC; human-GCK F: GCAGAAGGGAACAATGTCGTG, R: CGTAGTAGCAGGAGATCATCGT; and human-ATP6V1A F: GAGATCCTGTACTTCGCACTGG, R: GGGGATGTAGATGCTTTGGGT.
Western blotting assay
GC tumor tissues and cells were harvested. RIPA lysis buffer (P0013B, Beyotime, P.R. China) was placed on ice for 30 min, followed by protein quantification using the bicinchoninic acid assay (KeyGEN Biotech, Nanjing, P.R. China) and western blotting. The following primary antibodies were used: β-actin (proteintech; 66009-1-immunoglobin [Ig]; 1:3000), DRAM1 (Abcam; Ab208160; 1:1000), SLC1A5 (proteintech; 20350-1-AP; 1:2000), SLC7A5 (proteintech; 28670-1-AP; 1:2000), mTOR (proteintech; 66888-1-Ig; 1:2000), p-mTOR (proteintech; 67778-1-Ig; 1:2000), P62 (proteintech; 18420-1-AP; 1:2000), p-AKT (proteintech; 66444-1-Ig; 1:2000), E-cadherin (proteintech; 20874-1-AP; 1:2000), N-cadherin (proteintech; 22018-1-AP; 1:2000), Bax (Cell Signaling Technology; 2772S; 1:1000), Bcl-2 (Cell Signaling Technology; 15071S; 1:1000), PI3K (Wanleibio; WL03380; 1:1000), AKT (Cell Signaling Technology; 9272S; 1:1000), HK2 (Abways; AB3194; 1:1000), PFK1 (Abways; CY8047; 1:1000), GCK (Abways; CY8036; 1:1000), LDHA (bioworld; BS6179;1:1000), PKM2 (Cell Signaling Technology; 4053S; 1:1000), p53 (proteintech; 60283-22-Ig; 1:2000),LC3 (proteintech; 14600-1-AP; 1:2000), anti-rabbit antibody horseradish peroxidase (HRP) rabbit (Cell Signaling Technology; 7074S; 1:3000), and anti-mouse antibody HRP mouse (Cell Signaling Technology; 7076S; 1:3000).
RNA sequencing
Inoculated AGS-negative cells and AGS cells with DRAM1 knocked down into 10 cm culture dishes, and after they have grown to confluence, collect the cells for RNA sequencing.
Cell proliferation assay (CCK-8 method)
HGC27 control cells, HGC27 cells overexpressing DRAM1, AGS-negative cells, and AGS cells with DRAM1 knockdown were seeded in 96-well plates at a density of 3,000 cells/well. Cell proliferation was assessed by adding 10 μL of CCK-8 solution (Target Mol, Boston, MA, USA) to each well at 24, 48, 72, and 96 h, respectively.
EdU proliferation
HGC27 control cells, HGC27 cells overexpressing DRAM1, AGS-negative cells, and AGS cells with DRAM1 knockdown were seeded in 24-well plates, and appropriate concentrations of EdU reagent (C0075S, Beyotime, P.R. China) were added for 3 h. The cells were washed twice with phosphate-buffered saline (PBS) for 5 min each, followed by fixation with 4% paraformaldehyde for 30 min. After washing twice with PBS, the cells were permeabilized with 0.3% Triton X-100 for 15 min and stained with the reaction solution. A thin microscope was used to capture images at 20 × magnification.
TUNEL staining
Frozen sections were immersed in PBS for 20 min. A histochemistry pen was used to draw a small circle around the periphery of the tissue. Next, 100 μL of Proteinase K working solution (Pinuofei Biological, Wuhan, P.R. China) was added to the sample and incubated for 10 min at room temperature. PBS was used for three washes, followed by 50 μL of equilibration buffer. The cells were incubated at room temperature for 10 min, washed thrice with PBS, and incubated with TDT for 1 h at room temperature. Finally, nuclear staining was performed, and images were collected at 20 × magnification using a thunder microscope.
Immunofluorescence
The cells were washed thrice with PBS, treated with 4% paraformaldehyde (Servicebio, Wuhan, P.R. China) for 15 min, and incubated with 0.5% Triton X100 for another 15 min. Subsequently, they were immersed in a 5% FBS solution for 1 h at room temperature and incubated overnight with the primary antibody. The following primary antibodies were used: SLC1A5 (proteintech; 20350-1-AP; 1:200), DRAM1 (Abcam; Ab208160; 1:200), LDHA (bioworld; BS6179; 1:200), and SLC3A2 (proteintech; 15193-1-AP; 1:200). The following day, the cells were washed thrice with PBST for 5 min each. The cells were then incubated with a secondary antibody labeled with a CoraLite488-conjugated Goat Anti-Rabbit antibody (1:200, Proteintech, China) for 1 h at room temperature. The cells were then rinsed thrice with PBST for 5 min each. Finally, the nuclei were stained with DAPI (10 μg/ml) (KeyGEN biotech, Nanjing, P.R. China) for 10 min at room temperature. We used Thunder to capture photos and obtain images.
Colony formation assay
Cells were seeded at a density of 1000 cells/well in 6-well plates and subsequently cultured in a complete medium for 10–14 days. Colonies were treated with 4% paraformaldehyde (Servicebio, Wuhan, P.R. China) at room temperature for 20 min and stained with 0.1% crystal violet solution for 20 min.
Wound healing assays
The cells were seeded in 6-well plated at a density of 6 × 105 cells/well. After the cells reached confluence, the monolayer was damaged using a 200 μL pipette tip, and the detached cells with PBS were removed. Subsequently, the cells were cultured in a serum-free medium to inhibit growth. Images were captured at 0 and 24 h, and the wound size was assessed using ImageJ software.
Cell migration and invasion assays
The migratory and invasive abilities of the cells were assessed using a transwell assay. For the migration experiments, a suspension of 3 × 104 cells in a serum-free medium (250 μL) was added to the upper chamber, while the lower chamber was filled with 10% FBS medium (500 μL) to stimulate cell migration through the membrane. After 24–48 h of incubation, the chambers were removed and fixed with 4% paraformaldehyde for 20 min. The upper cells were cleaned with a cotton swab, and a 0.1% crystal violet solution was applied for 20 min to stain them. To conduct invasion experiments, Matrigel (GENEBAY, Nanjing, P.R. China) was added to the upper chamber at a dilution of 1:8. The procedure remained unchanged, as previously mentioned. Cells were counted in five random fields at × 100 magnification.
Flow cytometry analysis of apoptosis
An Annexin V-FITC/PI apoptosis assay kit (Vazyme Biotech, Nanjing, P.R. China) was used to detect apoptosis. Briefly, 6 × 105 cells were seeded into 6-well plates. After 24 h of incubation, the cells were collected and resuspended in the buffer. The cells were stained with Annexin V-FITC (50 μg/mL) and propidium iodide (10 μg/mL) for 15 min in a light-protected environment before flow cytometry (BD Biosciences, San Diego, CA, U.S.A.).
ATP determination
Cells from different treatment groups were collected into 1.5 mL EP tubes, and a centrifuged cell pellet was obtained. After washing the cells with PBS buffer, 200 μL of lysis buffer from the ATP kit (Beyotime, S0026, P.R. China) was added, and the cells were then lysed following the provided instructions. The lysate was centrifuged at 12,000 × g for 5 min at 4 °C. The liquid above the sediment was transferred into a fresh 1.5 mL EP tube, and ATP testing was conducted using the ATP assay kit.
Lactate determination
Cells from the various treatment groups (1 × 106) were seeded in 6-well plates and cultured overnight. Subsequently, the medium was aspirated, and the cells were rinsed twice with pre-chilled PBS. After a 24-h incubation period, the cells were collected. Then, 200 μL of lactate assay buffer was added, and the cells were thoroughly shaken and mixed. Finally, the collected cells were centrifuged at 12,000g for 5 min at 4 °C, and the supernatant was removed for lactate analysis. The assay was prepared following the CheKine™ Lactate Assay Kit (KTB1100, Abbkine, Wuhan, P.R. China). After 30 min, a microplate reader was used to measure the optical density value at 450 nm, followed by the analysis of lactate metabolism levels.
Mouse xenograft models
Four-week-old female BALB/c-nu and male nude mice were obtained from Nanjing Huachuang Biotechnology Co., Ltd. and housed in a specific pathogen-free facility with a 12/12-h light/dark cycle, providing them with ad libitum access to food and water. This animal study was approved by the Experimental Animal Management Ethics Committee of Drum Tower Hospital, Nanjing University Medical School (2023AE01054).Animals were euthanized if their body weight loss exceeded 20%–25% or if the weight of the tumor exceeded 10% of their body weight. AGS (1 × 107) and DRAM1-silencing AGS cells were separately suspended in 0.1 mL PBS for in vivo tumor growth studies. Subsequently, cells were inoculated into the flanks of nude mice via subcutaneous injection. Tumor volume was calculated using the formula 1/2 (length × width2), with measurements taken every three days. After 21 days, the mice were euthanized, and a small amount of subcutaneous tumor tissue was collected for Western blot analysis. The remaining tissue was placed in a 4% paraformaldehyde solution and embedded in OCT, and frozen sections were prepared for analysis using H&E, ki-67, and TUNEL staining. All animal experiments were conducted in accordance with the ARRIVE guidelines.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism (Version 9.0). Quantitative data was presented as mean ± standard deviation. The difference in means between two groups was analyzed using the Student’s t-test, and the difference in means between three or more groups was analyzed using One-way analysis of variance. The Kaplan–Meier test was used to calculate survival rates, whereas the log-rank test was used to assess differences in survival between the two groups. All statistical tests were two-tailed exact tests, and statistical significance was set at P < 0.05.
Data availability
All information produced or analyzed for this inquiry is contained in this article. The RNA sequencing (RNA-seq) data used in this study are publicly available in the NCBI database under the BioProject accession number PRJNA1182365.
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Chen, J. Q. & Russo, J. Dysregulation of glucose transport, glycolysis, TCA cycle and glutaminolysis by oncogenes and tumor suppressors in cancer cells. Biochem. Biophys. Acta. 370–384, 2012. https://doi.org/10.1016/j.bbcan.2012.06.004 (1826).
Crighton, D. et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 126, 121–134. https://doi.org/10.1016/j.cell.2006.05.034 (2006).
Beaumatin, F. et al. mTORC1 activation requires DRAM-1 by facilitating lysosomal amino acid efflux. Mol. Cell 76, 163-176.e168. https://doi.org/10.1016/j.molcel.2019.07.021 (2019).
Tan, J. et al. DRAM1 increases the secretion of PKM2-enriched EVs from hepatocytes to promote macrophage activation and disease progression in ALD. Mol. Ther. Nucleic Acids 27, 375–389. https://doi.org/10.1016/j.omtn.2021.12.017 (2022).
Wu, X. et al. Increased expression of DRAM1 confers myocardial protection against ischemia via restoring autophagy flux. J. Mol. Cell. Cardiol. 124, 70–82. https://doi.org/10.1016/j.yjmcc.2018.08.018 (2018).
Zhang, Y. et al. DNA damage-regulated autophagy modulator 1 (DRAM1) mediates autophagy and apoptosis of intestinal epithelial cells in inflammatory bowel disease. Digest. Dis. Sci. 66, 3375–3390. https://doi.org/10.1007/s10620-020-06697-2 (2021).
Qi, Y. et al. ARHGAP4 promotes leukemogenesis in acute myeloid leukemia by inhibiting DRAM1 signaling. Oncogene 42, 2547–2557. https://doi.org/10.1038/s41388-023-02770-y (2023).
Zhang, X. et al. Long non-coding RNA LINC00511 accelerates proliferation and invasion in cervical cancer through targeting miR-324-5p/DRAM1 axis. OncoTargets Ther. 13, 10245–10256. https://doi.org/10.2147/ott.S255067 (2020).
Geng, J. et al. DRAM1 plays a tumor suppressor role in NSCLC cells by promoting lysosomal degradation of EGFR. Cell Death Dis. 11, 768. https://doi.org/10.1038/s41419-020-02979-9 (2020).
Guan, J. J. et al. DRAM1 regulates apoptosis through increasing protein levels and lysosomal localization of BAX. Cell Death Dis. 6, e1624. https://doi.org/10.1038/cddis.2014.546 (2015).
Banducci-Karp, A. et al. DRAM1 promotes lysosomal delivery of mycobacterium marinum in macrophages. Cells 12. https://doi.org/10.3390/cells12060828 (2023).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587-d592. https://doi.org/10.1093/nar/gkac963 (2023).
Lu, T. et al. DRAM1 regulates autophagy and cell proliferation via inhibition of the phosphoinositide 3-kinase-Akt-mTOR-ribosomal protein S6 pathway. Cell Commun. Signal. CCS 17, 28. https://doi.org/10.1186/s12964-019-0341-7 (2019).
Zhang, X. D., Qi, L., Wu, J. C. & Qin, Z. H. DRAM1 regulates autophagy flux through lysosomes. PloS one 8, e63245. https://doi.org/10.1371/journal.pone.0063245 (2013).
Chen, C. et al. DRAM1 regulates the migration and invasion of hepatoblastoma cells via autophagy-EMT pathway. Oncol. Lett. 16, 2427–2433. https://doi.org/10.3892/ol.2018.8937 (2018).
Sun, L., Zhang, H. & Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 13, 877–919. https://doi.org/10.1007/s13238-021-00846-7 (2022).
Xia, L. et al. The cancer metabolic reprogramming and immune response. Mol. Cancer 20, 28. https://doi.org/10.1186/s12943-021-01316-8 (2021).
Li, Z., Sun, C. & Qin, Z. Metabolic reprogramming of cancer-associated fibroblasts and its effect on cancer cell reprogramming. Theranostics 11, 8322–8336. https://doi.org/10.7150/thno.62378 (2021).
Reinfeld, B. I. et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288. https://doi.org/10.1038/s41586-021-03442-1 (2021).
Lin, J. et al. The roles of glucose metabolic reprogramming in chemo- and radio-resistance. J. Exp. Clin. Cancer Res. CR 38, 218. https://doi.org/10.1186/s13046-019-1214-z (2019).
Vallée, A., Lecarpentier, Y., Guillevin, R. & Vallée, J. N. Demyelination in multiple sclerosis: Reprogramming energy metabolism and potential PPARγ agonist treatment approaches. Int. J. Mol. Sci. 19. https://doi.org/10.3390/ijms19041212 (2018).
Kim, J. Regulation of immune cell functions by metabolic reprogramming. J. Immunol. Res. 2018, 8605471. https://doi.org/10.1155/2018/8605471 (2018).
Su, B. et al. Diallyl disulfide inhibits TGF-β1-induced upregulation of Rac1 and β-catenin in epithelial-mesenchymal transition and tumor growth of gastric cancer. Oncol. Rep. 39, 2797–2806. https://doi.org/10.3892/or.2018.6345 (2018).
Kang, S. W., Lee, S. & Lee, E. K. ROS and energy metabolism in cancer cells: alliance for fast growth. Arch. Pharm. Res. 38, 338–345. https://doi.org/10.1007/s12272-015-0550-6 (2015).
Ye, J. et al. Targeting of glutamine transporter ASCT2 and glutamine synthetase suppresses gastric cancer cell growth. J. Cancer Res. Clin. Oncol. 144, 821–833. https://doi.org/10.1007/s00432-018-2605-9 (2018).
Murray, P. J. Macrophage polarization. Annu. Rev. Physiol. 79, 541–566. https://doi.org/10.1146/annurev-physiol-022516-034339 (2017).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832–844. https://doi.org/10.1016/j.cell.2014.04.016 (2014).
Piao, H. et al. A positive feedback loop between gastric cancer cells and tumor-associated macrophage induces malignancy progression. J. Exp. Clin. Cancer Res. CR 41, 174. https://doi.org/10.1186/s13046-022-02366-6 (2022).
Zhang, L. & Li, S. Lactic acid promotes macrophage polarization through MCT-HIF1α signaling in gastric cancer. Exp. Cell Res. 388, 111846. https://doi.org/10.1016/j.yexcr.2020.111846 (2020).
Liu, P. S. et al. α-ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994. https://doi.org/10.1038/ni.3796 (2017).
Morgan, S., Grootendorst, P., Lexchin, J., Cunningham, C. & Greyson, D. The cost of drug development: A systematic review. Health Policy (Amsterdam, Netherlands) 100, 4–17. https://doi.org/10.1016/j.healthpol.2010.12.002 (2011).
Mohammadzadeh, N. et al. Statins: Epidrugs with effects on endothelial health?. Eur. J. Clin. Invest. 50, e13388. https://doi.org/10.1111/eci.13388 (2020).
Jiang, W., Hu, J. W., He, X. R., Jin, W. L. & He, X. Y. Statins: a repurposed drug to fight cancer. J. Exp. Clin. Cancer Res. CR 40, 241. https://doi.org/10.1186/s13046-021-02041-2 (2021).
Mao, W. et al. Statin shapes inflamed tumor microenvironment and enhances immune checkpoint blockade in non-small cell lung cancer. JCI Insight 7. https://doi.org/10.1172/jci.insight.161940 (2022).
Guo, C. et al. Therapeutic targeting of the mevalonate-geranylgeranyl diphosphate pathway with statins overcomes chemotherapy resistance in small cell lung cancer. Nature Cancer 3, 614–628. https://doi.org/10.1038/s43018-022-00358-1 (2022).
Lou, D., Fu, R., Gu, L., Su, H. & Guan, L. Association between statins’ exposure with incidence and prognosis of gastric cancer: An updated meta-analysis. Expert Rev. Clin. Pharmacol. 15, 1127–1138. https://doi.org/10.1080/17512433.2022.2112178 (2022).
Jiao, Z. et al. Statin-induced GGPP depletion blocks macropinocytosis and starves cells with oncogenic defects. Proc. Natl. Acad. Sci. USA 117, 4158–4168. https://doi.org/10.1073/pnas.1917938117 (2020).
Acknowledgements
The work was supported by the Jiangsu Provincial Medical Science and Technology Development Plan Project (ZA983-2020), the Clinical Research Projects of Nanjing Drum Tower Hospital (HA9830202101, HA9830202201, EA9830202301) and Taikang Youth Medical Research Initiation Fund (No.2022001).
Author information
Authors and Affiliations
Contributions
Xiaotan Dou, Xiaoping Zou, and Min Chen designed the study. Xinrong Wu, Weiwei Wang, and Yifan Li performed the in vitro experiments. Xinrong Wu, Bei Zhao, Wenqi Sun, and Jiale Xu conducted the in vivo experiments. Dan Ge and Longying Xiong analyzed the data. Xinrong Wu wrote the manuscript. Min Chen and Lei Wang reviewed the manuscript. All the authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Morality and ethics
The Medical Ethics Committee of Drum Tower Hospital, Nanjing University Medical School approved this study. The feasibility of tissue sampling in the experiment is strictly adhered to and standardized ethically according to the Helsinki Declaration. Each patient signed a document requesting their informed consent, ensuring their understanding and agreement. Animal experiments were conducted in Nanjing, and all animal experimental methods were carried out in accordance with the ARRIVE principles.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, X., Li, Y., Wang, W. et al. DRAM1 enhances the proliferation and metastasis of gastric cancer through the PI3K/AKT/mTOR signaling pathway and energy metabolism. Sci Rep 15, 3542 (2025). https://doi.org/10.1038/s41598-025-87389-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-87389-7