Abstract
Ubiquitination is a post-translational modification (PTM), which is critical to maintain cell homeostasis. Ubiquitin-specific protease 24 (USP24) plays roles in various diseases, the mechanisms by which USP24 regulates hepatocellular carcinoma (HCC) remain poorly understood. In this study, USP24 is found to be significantly downregulated in HCC. Knocking down USP24 promotes HCC proliferation and migration, whereas USP24 overexpression inhibits HCC in vitro and in vivo. The endogenous interaction between USP24 and Beclin1 is confirmed. Mechanically, USP24 delays Beclin1 degradation by reducing its K48-linked ubiquitination, the effects of overexpressing USP24 on HCC proliferation can be partially reversed by silencing Beclin1. We find that increased autophagy is accompanied by ferroptosis in USP24 overexpressed HCC cells and USP24 increases the susceptibility of HCC to sorafenib. Collectively, this study highlights the critical role of USP24 in regulating autophagy-dependent ferroptosis by decreasing Beclin1 ubiquitination, suggesting that targeting USP24 may be a strategy for treating HCC.
Similar content being viewed by others
Introduction
Primary liver cancer is one of the most prevalent malignant tumors, accounting for 4.7% of new cancer cases, and ranking as the third leading cause of cancer-related deaths1. Hepatocellular carcinoma (HCC) is the most common pathological type, constituting approximately 75%–85% of all primary liver cancers. At present, hepatitis B virus and hepatitis C virus still remain the most important causal risk factors for HCC2. Despite the availability of several treatment modalities, such as surgery, transcatheter arterial chemoembolization (TACE), radiofrequency ablation, targeted therapy, and immunotherapy, therapeutic outcomes are often unsatisfactory3,4,5. Consequently, gaining a deeper understanding of the underlying mechanisms of HCC is crucial for developing therapeutic targets.
Post-translational modifications (PTMs) play a fundamental role in regulating protein functions, and participate in various cell biological processes by regulating the subcellular localization and activity of proteins6. Ubiquitination modification is a type of PTMs that is mediated by ubiquitin ligases and deubiquitinases (DUBs), which are essential for protein stability and function7. Ubiquitination is a reversible process, and the imbalance between ubiquitination and deubiquitination can lead to abnormal protein accumulation or degradation, thus leading to the formation of cancer8. Recent studies have demonstrated that modification of protein ubiquitination occurs in various cancers, and small molecule inhibitors targeting this process can be used as a treatment for tumors9,10. Ubiquitin-specific peptidases (USPs) are a subclass of DUBs that have been found to be involved in the regulation of cancer progression11,12,13. Ubiquitin-specific protease 24 (USP24) is a member of the USPs, whose gene is located at the PARK10 locus on the short arm of chromosome 114. Previous studies have found that USP24 is closely related to susceptibility to developing Parkinson’s disease14,15,16. Additionally, USP24 plays a role in regulating cancers. Studies have found that USP24 is linked to the progression of lung cancer17 and can accelerate the occurrence of drug resistance18. As a counter mechanism, USP24 also acts as a cancer suppressor. USP24 was found to inhibit the progression of neuroblastoma by stabilizing CRMP219. However, the mechanism by which USP24 regulates HCC progression is unclear. Consequently, whether USP24 is an effective molecular target in HCC is worthy of further study.
Unrestricted cell proliferation and reduced cell death are responsible for cancer development. Understanding the modes of cancer cell death is helpful for exploring new treatment strategies. Ferroptosis, a programmed cell death first defined decades ago20, plays pluralistic roles in tumor formation and treatment. Ferroptosis is characterized by the accumulation of reactive oxygen species (ROS) dependent on iron overload. In fact, a study has established that ferroptosis is a type of autophagic cell death21, known as ferritinophagy. During ferritinophagy, the level of autophagy is upregulated, with nuclear receptor coactivator 4 (NCOA4) playing a leading role in this process22. In contrast to the cancer-promoting effects of autophagy in tumors23,24, autophagy plays a role in killing tumor cells during ferritinophagy25,26. Understanding the crosstalk between ferroptosis and autophagy, as well as the mechanisms underlying these two types of cell death, is essential. Previous studies have demonstrated the involvement of ubiquitination in multiple aspects of autophagy27. The ability of USP24 to regulate autophagy and ferroptosis needs further exploration.
From the current investigation, we found that USP24 played an important role in preventing HCC progression. Endogenous USP24 and Beclin1 were demonstrated to interact with each other, and USP24 increased Beclin1 stability by decreasing its K48-linked ubiquitination. More interestingly, the upregulation of autophagy promoted the occurrence of autophagy-dependent ferroptosis in HCC, and USP24 could also increase the sensitivity of HCC to sorafenib. In summary, data from this study provided further insights into the mechanisms by which USP24 regulates HCC and is expected to serve as a target for HCC treatment.
Results
Significantly low expression of USP24 in HCC patients correlates with their prognosis
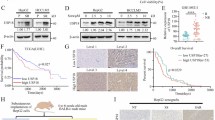
To investigate the role of USP24 in HCC, USP24 expression levels in normal hepatic cells and several HCC cell lines were compared using qRT-PCR and western blot assays. The expression of USP24 was first detected by qRT-PCR, which indicated that the mRNA level of USP24 was lower in HCC cell lines compared to normal hepatic cells (Fig. 1A). This pattern of expression corresponded with the protein level of USP24 measured with western blot (Fig. 1B). The SMMC-7721 and HCCLM3 cell lines were selected for further experimentation. To investigate the clinical significance of USP24 in HCC, human HCC tissue arrays were used to detect the protein expression of USP24 by immunohistochemistry (IHC) staining. USP24 was found to be highly expressed in paracancerous tissues compared with carcinoma tissues with different expression levels of USP24 (Fig. 1C, D). The patients were separated into two groups based on their immunological risk score (IRS): USP24 high-expression and low-expression groups. The Kaplan-Meier analysis demonstrated that patients with high USP24 protein expression had better overall survival than patients with low USP24 protein expression (Fig. 1E). These results indicated that USP24 is low-expressed in HCC and higher expression of USP24 protein indicates better prognosis.
A The mRNA level of USP24 in normal liver cell and HCC cell lines was measured by qRT-PCR (n = 3). B The expression level of USP24 protein in normal liver cell and HCC cell lines was measured by western blot. C Immunohistochemistry assay was performed in paracancerous tissue and paired carcinoma tissue microarray (n = 70). Representative images of USP24 in normal tissues, HCC tissues with different levels of USP24 are shown. Scale bar, 20 μm. D Quantification of the USP24 expression in paracancerous tissues and carcinoma tissues. E Kaplan-Meier curves from patients with HCC expressing low (n = 39) and high (n = 31) USP24 from the tissue microarray. *P < 0.05, **P < 0.01, ***P < 0.001.
USP24 silencing promotes HCC cell proliferation and migration in vitro
To investigate the biological function of USP24 in HCC progression, small interfering RNA (siRNA) was utilized to reduce the expression of USP24. The efficiency of USP24 knockdown was determined using qRT-PCR and western blot assays (Fig. 2A, B). Two siRNAs (si-USP24-1 and si-USP24-3) were utilized for subsequent experiments. As illustrated in Fig. 2C, the CCK-8 assay demonstrated that knocking down USP24 increased the proliferation capacity of SMMC-7721 and HCCLM3 cells. Colony formation assay also indicated that the long-term proliferation ability was increased after silencing USP24 (Fig. 2D, E). Next, the effect of USP24 on the migration capacity of HCC cells was explored. The results showed that knocking down USP24 promoted the wound-healing ability of HCC in vitro (Fig. 2F and Supplementary Fig. 1B, C). Moreover, the transwell experiment was conducted to assess the migration capacity, and the results demonstrated that the migration capacity of HCC cells was increased by knocking down USP24 (Fig. 2G, H). Collectively, these results suggested that the knockdown of USP24 promotes the proliferation and migration of HCC cells in vitro.
A and B The knockdown efficiency of USP24 by siRNA was measured by qRT-PCR and western blot (n = 3). C CCK-8 assays were conducted to determine the viability of SMMC-7721 and HCCLM3 cells after knockdown of USP24 (n = 3). D–E Colony formation assays of HCC cells treated with si-USP24 or si-NC were performed to evaluate the ability of proliferation. The clone number was counted using ImageJ (n = 3). F Migration abilities of SMMC-7721 and HCCLM3 cells after knockdown of USP24 were evaluated with wound healing assay. Scale bar, 1 mm. G–H Transwell assays were performed to detect the ability of migration. The migration rate was calculated (n = 3). Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
USP24 increases the level of autophagy in HCC cells
The above results have elucidated the role of USP24 in regulating HCC progression, however, the exact mechanism by which USP24 performs its function remains elusive. Previous studies have demonstrated the reciprocal cross-talk between ubiquitination and autophagy28. Considering the role of USP24 as a deubiquitinating enzyme, we explored whether USP24 participates in the regulation of autophagy. First, the correlation between USP24 and critical autophagic molecules was analyzed using Gene Expression Profiling Interactive Analysis 2 (GEPIA2). The results revealed that USP24 was positively associated with autophagic markers (Fig. 3A). Our qRT-PCR assay found that the mRNA expressions of ULK1 and Atg5 were decreased after USP24 was silenced, whereas USP24 overexpression increased the mRNA expression of these genes. However, no significant changes in Beclin1 mRNA expression were observed (Fig. 3B, C and Supplementary Fig. 1D–G). Next, western blot was conducted to detect the expression of autophagic proteins. As illustrated in Fig. 3D, E, knocking down USP24 inhibited the expression of ULK1, Atg5, and Beclin1 proteins, whereas overexpression of USP24 increased the protein expression of autophagic molecules as well as LC3B-II, but decreased the expression of p62. Surprisingly, USP24 affected the expression of Beclin1 protein but not its mRNA. The inconsistencies between mRNA and protein expression of Beclin1 caught our attention. Therefore, we hypothesized that there may exist a post-translational modification of Beclin1. To further verify that the level of autophagy increased after USP24 overexpression, the micromorphology of USP24 overexpressed cells was observed by a transmission electron microscopy (TEM). Indeed, results demonstrated an increased number of autophagic vesicles after overexpressing USP24 (Fig. 3F, G and Supplementary Fig. 1H, I). Overall, these evaluations indicated that USP24 increased the level of autophagy in HCC cells.
A The relationship of USP24 expression with autophagy markers was investigated via Gene Expression Profiling Interactive Analysis (GEPIA). B qRT-PCR was performed to measure the mRNA expression of ULK1, Atg5 and Beclin1 after silencing USP24 (n = 3). C qRT-PCR was performed to measure the mRNA expression of ULK1, Atg5 and Beclin1 after overexpressing USP24 (n = 3). D The expression of ULK1, Atg5, Beclin1, LC3B-II and p62 proteins in USP24 knockdown HCC cells were measured by western blot. E The expression of autophagy related proteins after overexpressing USP24 was detected by western blot. F Transmission electron microscopy (TEM) was used to detect the formation of autophagic structures. Red arrows denote representative autolysosome. Scale bar, 5 μm (left), 500 nm (right). G The number of autophagic vesicles was counted and presented in the image (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
USP24 interacts with Beclin1
To explore the exact mechanism by which USP24 induces autophagy, Co-IP was first carried out to identify the potential substrate of USP24. It was found that Beclin1 could be co-immunoprecipitated by USP24 (Fig. 4A), but no other autophagy proteins, such as ULK1 and Atg5 (Supplementary Fig. 2A), indicating that USP24 could interact with Beclin1. Next, a reciprocal Co-IP assay was conducted to further confirm the interaction between USP24 and Beclin1 (Fig. 4B). Additionally, cellular double immunofluorescence assay provided morphological evidence of the interaction between USP24 and Beclin1 protein (Fig. 4C and Supplementary Fig. 2B) and the colocalization coefficients increased after USP24 overexpression (Fig. 4D and Supplementary Fig. 2C). These results collectively confirmed the interaction between USP24 and Beclin1.
A, B Reciprocal Co-IP experiments were performed to detect the interaction between USP24 and Beclin1. An IgG antibody was used as the control. C The cell double immunofluorescence assay was carried out using anti-USP24 and anti-Beclin1 antibodies. The colocalization of USP24 (red) and Beclin1 (green) was observed under a confocal microscope (n = 3). Scale bar, 10 μm. D The colocalization was evaluated by Mander’s colocalization coefficients in ten regions of interest (n = 10). E SMMC-7721 cells were treated with 10 μg/ml MG132 before harvesting. An anti-Beclin1 antibody was used to immunoprecipitate the Beclin1 protein, then an anti-Ubi antibody was used for western blot. F, G The levels of K48 and K63-linked ubiquitination were measured by western blot. H–K Control and USP24 overexpressed HCCLM3 and SMMC-7721 cells were treated with CHX (100 μg/ml) for indicated time. The expression level of Beclin1 was determined by western blot (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
USP24 stabilizes Beclin1 by decreasing its ubiquitination
Previous studies have demonstrated that the protein level of Beclin1 was altered after USP24 inhibition or overexpression, however, the mRNA level of Beclin1 remained unchanged (Fig. 3B–E and Supplementary Fig. 1D–G), suggesting that post-translational modification may be involved in the regulation of Beclin1. Considering the function of USP24 in deubiquitinating proteins, it is proposed that USP24 may regulate the ubiquitination of Beclin1. To confirm this, Beclin1 was immunoprecipitated using an anti-Beclin1 antibody, followed by an anti-Ubi antibody for immunoblot assays. The results demonstrated that the ubiquitination level of Beclin1 significantly decreased after USP24 overexpression (Fig. 4E and Supplementary Fig. 2D). Various connections are made between ubiquitin, primarily through seven lysine residues (K6, K11, K27, K29, K33, K48, and K63) or the first methionine (M1), different types of ubiquitin linkages execute distinct functions29. Among these linkages, K48 and K63-linked polyubiquitin are the two most common types. To explore which type of ubiquitination modification of Beclin1 was affected by USP24, the level of K48 or K63-linked ubiquitination was measured. Our results indicated that USP24 predominantly reduced the K48-linked ubiquitination (Fig. 4F and Supplementary Fig. 2E). Although K63-linked ubiquitination was also reduced after overexpressing USP24 (Supplementary Fig. 2F), it was less pronounced than the reduction in K48-linked ubiquitination, particularly in SMMC-7721 cell line (Fig. 4G). Overall, these results suggested that USP24 inhibits the ubiquitination of Beclin1 mainly by removing the K48-linked polyubiquitylation.
On another hand, the most common consequence of K48-linked polyubiquitylation is proteasome-mediated degradation, whereas K63-linked polyubiquitylation is associated with some cellular processes30. Since the K48-linked polyubiquitylation decreased after USP24 overexpression, it was speculated that USP24 may affect the stability of Beclin1. To confirm this hypothesis, the protein half-life assay was conducted by treating the cells with cycloheximide (CHX) at varied durations. As shown in Fig. 4H–K, the overexpression of USP24 prolonged the half-life of Beclin1. Altogether, these results determined that USP24 inhibits K48-linked polyubiquitination and increases the stability of Beclin1.
USP24 inhibits HCC progression dependent on Beclin1
To explore the mechanism by which USP24 influences the phenotypes of HCC, we overexpressed USP24 and performed rescue experiments by silencing Beclin1. First, the efficiency of overexpression was measured by qRT-PCR and western blot (Fig. 5A, B). It was found that overexpression of USP24 inhibited the proliferation of HCC cells, as indicated by the decrease in both cell viability (Fig. 5C) and colony formation (Fig. 5D, E). Next, wound healing experiments and transwell assays were performed. The results indicated that USP24 overexpression decreased wound-healing ability (Fig. 5F and Supplementary Fig. 3A, B) and inhibited the migration of HCC cells (Fig. 5G, H). To explore whether USP24 plays its role by regulating Beclin1, Bafilomycin A1 was used to inhibit autophagy firstly. The reduced cell viability caused by overexpressing USP24 was partially reversed by inhibiting autophagy (Fig. 6A). The transwell assay was also conducted to evaluate the migration ability and results indicated that the migration ability was increased by treating with BafA1 (Fig. 6B, C). Next, Beclin1 was silenced using siRNA based on the overexpression of USP24 (Fig. 6D). The CCK-8 assay indicated that cell viability could be reversed by silencing Beclin1 (Fig. 6E). Similar results were observed in the colony formation experiments (Supplementary Fig. 3C–E). Additionally, transwell assay demonstrated that decreased migration ability caused by overexpressing USP24 could also be reversed by silencing Beclin1 (Figs. 6F and G). Collectively, these results demonstrated that USP24 inhibits the progression of HCC cells dependent on Beclin1.
A, B The overexpression efficiency of USP24 by lentivirus was measured by western blot and qRT-PCR (n = 3). C CCK-8 assays were conducted to determine the viability of SMMC-7721 and HCCLM3 cells after overexpressing USP24 (n = 3). D–E Colony formation assays of HCC cells treated with LV-CON or LV-USP24 were performed to evaluate the ability of proliferation. The clone number was counted (n = 3). F Migration abilities of SMMC-7721 and HCCLM3 cells after overexpression of USP24 were evaluated by wound healing assay. Scale bar, 1 mm. G-H Transwell assays were performed to detect the ability of migration, and the migration rate was calculated (n = 3). Scale bar, 100 μm. **P < 0.01, ***P < 0.001, ****P < 0.0001.
A–C The cells overexpressed USP24 were treated with Bafilomycin A1 (10 nM) for 24 h. A CCK8 assay was used to detect the cell viability (n = 5). B–C Transwell assay was performed to evaluate the migration ability of HCC cells, and the migration rate was calculated (n = 3). D The efficiency of Beclin1 knockdown was measured by western blotting. E CCK8 assay was performed to determine the cell viability after USP24 overexpression or Beclin-1 knockdown (n = 3). F–G Transwell assays of HCC cells with USP24 overexpression or Beclin1 knockdown were performed to evaluate the ability of migration, and the migration rate was calculated (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
USP24 promotes ferritinophagy and increases HCC sensitivity to sorafenib
Previous studies have demonstrated that USP24 inhibited proliferation and migration of HCC, and our findings also indicated that USP24 increased cell death in HCC cells (Supplementary Fig. 4A). However, the specific mechanism underlying increased cell death following USP24 overexpression remains unclear. Selective inhibitors of cell death pathways were used to explore the exact mechanism responsible for increased cell death after USP24 overexpression. The results revealed that only ferrostatin-1 (Fer-1, inhibitor of ferroptosis) could reverse cell death partially (Fig. 7A), but not other inhibitors, such as zVAD-fmk (inhibitor of apoptosis) and Necrostatin-1 (Ner-1, inhibitor of necroptosis). Erastin was then used to induce ferroptosis, and it was found that cell death was more visible in the USP24 overexpressed group, which can be partially reversed by Fer-1 (Fig. 7B). Additionally, the expression of GPX4, an inhibitory factor of ferroptosis, was observed to decrease after USP24 overexpression (Fig. 7F, G and Supplementary Fig. 4E, F). These results suggested that USP24 promotes ferroptosis of HCC cells.
A USP24 overexpressed SMMC-7721 and HCCLM3 cells were treated with Fer-1 (2 μM), Nec-1 (1 μM) and z-VAD-fmk (10 μM) separately. The cell viability was determined by CCK8 assay (n = 3). B Erastin (20 μM) was used to induce ferroptosis for 24 h, simultaneously treated with Fer-1 (2 μM), Nec-1 (1 μM) and z-VAD-fmk (10 μM). Cell viability was determined by CCK8 assay (n = 3). C Intracellular Fe2+ was detected with the FerroOrange probe (1 μM) under an inverted fluorescence microscope. Scale bar, 100 μm. D The Fe2+ fluorescence intensity was quantified by ImageJ (n = 3). E The intracellular Fe2+ was measured by colorimetric method (n = 3). F–G The mRNA and protein expression of NCOA4, TfR, FPN, ferritin and GPX4 after overexpressing USP24 was measured by qRT-PCR and WB (n = 3). H The level of GSH in HCC cells transfected with LV-CON or LV-USP24 vectors (n = 3). I The level of MDA in HCC cells transfected with LV-CON or LV-USP24 vectors (n = 3). J Control and USP24 overexpressed SMMC-7721 cells were treated with Sorafenib (0, 5, 10, 15, 20 and 25 μM) for 48 h, the cell viabilities were determined by CCK8 assay (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Taking into account that ferroptosis is a type of autophagic cell death, which we called ferritinophagy21, the Fe2+ level was measured using a colorimetric assay and a fluorescence probe. The results indicated that the Fe2+ was found to be elevated after the overexpression of USP24 (Fig. 7C–E and Supplementary Fig. 4B–D). Next, the expression levels of TfR, ferritin, FPN and NCOA4 were also measured, and it was found that NCOA4 and TfR were increased while FPN was decreased after USP24 overexpression (Fig. 7F, G and Supplementary Fig. 4E, F). Since NCOA4 is an essential protein involved in ferritinophagy, responsible for the degradation of ferritin, while TfR and FPN are accountable for the transportation of iron, the regulation of their expression due to USP24 overexpression led to the increased production and decreased exportation of iron in HCC cells. To our surprise, the expression of ferritin was unexpectedly increased, possibly as a compensatory response to the increased labile iron pool. Additionally, the levels of glutathione (GSH) and malondialdehyde (MDA) were measured. The results indicated that overexpression of USP24 decreased GSH while increased the level of MDA in HCC cells (Fig. 7H, I and Supplementary Fig. 4G, H), further confirming that USP24 promotes ferroptosis in HCC cells. To determine whether USP24 could increase the sensitivity of HCC cells to sorafenib, HCC cells were treated with different doses of sorafenib. The CCK-8 assay reveled that cell viability decreased more quickly in the USP24 overexpressed group after treatment with sorafenib (Fig. 7J and Supplementary Fig. 4I). This result suggested that USP24 overexpression combined with sorafenib may synergistically inhibit HCC progression. Altogether, these results confirmed that USP24 promotes the autophagy-dependent ferroptosis and increases the sensitivity of HCC cells to sorafenib.
USP24 inhibits HCC growth in mice xenograft model
Based on the results from in vitro studies, we further investigated the function of USP24 in vivo by establishing the xenograft model in nude mice. The SMMC-7721 cell line that stably overexpressed USP24 was injected subcutaneously (Fig. 8A). It was demonstrated that USP24 overexpression inhibited tumor growth compared with the control group (Fig. 8B–E). While no significant difference in body weight was noted between the two groups (Fig. 8F). Tumor tissues were then microscopically observed after hematoxylin and eosin (H&E) staining (Fig. 8G). IHC staining was conducted to confirm the expression of USP24 in tumor tissues, meanwhile, the Ki-67 was also stained. We found that the percentage of Ki-67 positive areas decreased in the USP24 overexpressed group (Fig. 8H, I). Furthermore, western blot assays were also conducted to evaluate the expression of proteins associated with autophagy and ferroptosis in xenograft tumors, and this presented the same results as the cellular experiments (Fig. 8J). Collectively, these results indicated that USP24 inhibits tumor growth in vivo, further supporting its role as a potential therapeutic target for HCC.
A The process of constructing subcutaneous tumor models in nude mice. B–C Image of subcutaneous tumors in control and USP24 overexpressed groups (n = 5). D The tumor weight is presented as the mean ± standard deviation (n = 5). E Tumor volumes were calculated every 3 days after injection. F The body weight of nude mice during this experiment (n = 5). G Representative images of H&E stained tumor tissues. H Representative images of IHC stained USP24 and Ki-67 in control tissues or USP24 overexpressed xenograft tumor tissues. I The quantification of USP24 and Ki-67 positive cells (n = 5). J Western blot was performed to detect the expression of proteins associated with autophagy and ferroptosis. K Graphical summary of USP24 regulated autophagy-dependent ferroptosis in HCC. ****P < 0.0001.
Discussion
In the present study, ubiquitin-specific peptidase USP24 was found to be downregulated in HCC and correlated with patients’ prognosis. To the best of our knowledge, this is the first evidence demonstrating that USP24 interacts with Beclin1, primarily by inhibiting its K48-linked ubiquitination. The stabilized Beclin1 increases the level of autophagy and thus promotes the occurrence of ferritinophagy. Our results confirmed that USP24 promotes autophagy-dependent ferroptosis by stabilizing Beclin1 (Fig. 8K).
As a member of the ubiquitin-specific peptidases, USP24 is rarely investigated in the field of tumors. Previous research about USP24 mainly focused on its role in Parkinson’s disease16,31,32,33 and inflammatory diseases34,35. Although some researchers have clarified its function in regulating tumor progression in recent years, there still exist different views. Some studies have demonstrated that USP24 is an oncogene in malignant diseases, including lung, bladder, and gastric cancers17,18,36,37,38. On the contrary, USP24 also acts as a tumor suppressor in neuroblastoma19. However, little is known regarding the specific role and clear mechanism of USP24 in HCC. In the current investigation, we found that the protein expression of USP24 is lower in HCC tissues compared with adjacent normal liver tissues and that high expression of USP24 protein is correlated with better prognosis in HCC patients. Although the high expression of USP24 mRNA indicated poor prognosis according to TCGA analysis, there may be widespread data errors in public databases owing to tumor heterogeneity and contamination by immune cells or normal cells39. To address this issue, the mRNA expression levels of USP24 in different cells from HCC tissues in multiple datasets were analyzed by online public tool TISCH (tisch.comp-genomics.org/home/)40. Indeed, it was found that USP24 expression was low in malignant tumor cells, but high in immune cells such as T cells (Supplementary Fig. 1A). Bulk RNA sequencing results from TCGA may interfere with the expression of USP24 in tumor cells, however, the high expression of USP24 in HCC tissues obtained from TCGA cannot be determined whether it is in tumor cells or immune cells. Therefore, the IHC results of our tissue arrays can represent the expression level of USP24 protein in tumor cells to the greatest extent and is more meaningful for analyzing the prognosis of HCC patients. Contrary to previous findings41, our results indicated that the knockdown of USP24 increased HCC proliferation in vitro, while USP24 overexpression inhibited these processes in vitro and in vivo. It is important to consider that different cell lines may yield different results, and there is heterogeneity among tumor cells. More importantly, a previous study was conducted in cells that developed resistance to sorafenib after induction, which differed from the normal HCC cells used in this investigation. Overall, our results here suggested that USP24 may function as a tumor suppressor in HCC.
Ubiquitination modification is a type of post-translational modification that can be reversed by deubiquitination modification. Ubiquitin-specific peptidases (USPs) belong to a class of deubiquitinases, that mainly exert their effects by regulating the function or activity of substrate proteins. Regulation of the stability and function of oncoprotein or tumor suppressor proteins by deubiquitination modification is a universal phenomenon in HCC development. It was previously reported that USP5 promotes HCC progression by stabilizing SLUG42, while USP39 interacts with ZEB1 and regulates HCC proliferation43. Although USP24 interacts with GSDMB37 and PLK138 in bladder and gastric cancers respectively, the substrate protein that interacts with USP24 in HCC remains largely unknown. In our study, USP24 was found to induce autophagy in HCC. This study revealed that USP24 increased Beclin1 stability mainly by decreasing its K48-linked ubiquitination. It is worth mentioning that K48-linked ubiquitination leads to the degradation of proteins, while K63-linked ubiquitination is associated with some cellular processes30. Interestingly, a previous study has demonstrated that Beclin1 can be deubiquitinated by USP11, reducing K48-linked ubiquitination44, whereas another research found that USP14 inhibited the K63-linked ubiquitination of Beclin145. Whereby in our study, it was confirmed that USP24 increased the stability of Beclin1 by decreasing the K48-linked ubiquitination. Indeed, it was also found that K63-linked ubiquitination of Beclin1 was decreased in USP24 overexpressed HCCLM3 cells, however, this was not as pronounced as the change observed in K48-linked ubiquitination. More importantly, the subsequent CHX chase assay further confirmed that the degradation of Beclin1 was slower after USP24 overexpression, which supported the above results more convincingly. Altogether, our study first confirmed that USP24 suppressed the K48-linked ubiquitination of Beclin1.
Beclin1 plays a crucial role in regulating autophagy, and its post-translational modification is a key mechanism for this process. A previous study has shown that the ubiquitination level of Beclin1, which is regulated by USP15, is associated with the level of autophagy46. In line with our results, we found that USP24 initiated autophagy by increasing the level of Beclin1. Moreover, we found that USP24 affected the expression of other autophagy proteins, such as ULK1 and Atg5, although this study could not confirm direct interactions between USP24 and these proteins. This suggests that USP24 may indirectly regulate the expression of ULK1 and Atg5, it was hypothesized that another mechanism may exist.
Autophagy plays a dual role in cancer, and its exact impact on tumor progression remains controversial47. Although autophagy is essential for maintaining cellular homeostasis, excessive autophagy can lead to cell damage and even cell death. This study revealed that increased autophagy driven by USP24 overexpression was associated with decreased cell viability and increased ferroptosis. A recent study has demonstrated that ferroptosis is a type of autophagic cell death21. Interestingly, we found that ferroptosis induced by USP24 is dependent on autophagy. Consistent with a previous study discovered that USP24 regulated ferritinophagy48, our results demonstrated that USP24 regulated iron metabolism thus promoting ferroptosis, and increased HCC sensitivity to sorafenib. We observed an increase in the labile iron pool was caused by increased iron generation and decreased iron exportation. To our surprise, the expression of ferritin was increased, which seems inconsistent with the occurrence of ferroptosis. It was speculated that the increased ferritin content was a compensatory response to the increased labile iron pool, which warrants further investigation. Additionally, the interaction between autophagy and cell death is intricate, and further investigation should be conducted to better understand the boundary between protective autophagy and autophagic cell death.
In summary, our study demonstrated that USP24 stabilizes Beclin1 by decreasing its K48-linked ubiquitination. The interaction between USP24 and Beclin1 represents a key molecular mechanism that promotes autophagy-dependent ferroptosis. Our results emphasized the significance of the post-translational regulation of Beclin1 by USP24 in HCC. Targeting the USP24-Beclin1 axis may provide a therapeutic strategy for HCC.
Methods
Antibodies and reagents
The primary antibodies used in this study are listed in Supplementary Table 1. MG132 (#HY-13259), Bafilomycin A1 (HY-100558), cycloheximide (#HY-12320), Z-VAD-FMK (#HY-16658B), necrostatin-1 (#HY-15760), ferrostatin-1 (#HY-100579), sorafenib (#HY-10201) and erastin (#HY-15763) were acquired from MedChemExpress (Monmouth Junction, NJ, USA) and dissolved in DMSO.
Cell culture and transfection
Human HCC cells (HepG2, SMMC-7721, Huh7 and HCCLM3) and Transformed Human Liver Epithelial-2 (THLE-2) were obtained from the Procell Life Science & Technology (Wuhan, China). Cells have STR authentication and no mycoplasma contamination tests were performed. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Solarbio, Beijing, China) supplemented with 10% fetal bovine serum (FBS, AC03L055, Shanghai Life-iLab Biotech, China), 1% penicillin-streptomycin and 1% L-Ala-Gln (Beyotime, Jiangsu, China). All cell lines mentioned above were cultured in a humidified incubator at 37 °C with 5% CO2. For transfection, three small interfering RNAs targeting USP24 (si-USP24-1, si-USP24-2, si-USP24-3) and the negative control (si-NC), one small interfering RNA targeting Beclin1 (si-Beclin1) and its negative control were acquired from RiboBio (Guangzhou, China). The transfection was performed using the Lipofectamine 2000 kit (Invitrogen, USA). qRT-PCR was conducted 48 h and western blot was detected 72 h after transfection to evaluate the efficiencies of transfection. The sequences of siRNA are listed in Supplementary Table 2.
CRISPR synergistic activation mediator (SAM) system
To overexpress USP24, the CRISPR/dCas9-VP64/MS2-p65-HSF1 gene activation system was utilized49. Briefly, the plasmids encoding MS2-p65-HSF1 and dCas9-VP64 were obtained from GeneChem (Shanghai, China). The sgRNA sequence (Supplementary Table 3) was designed according to the sequence of the target gene, followed by linking the sgRNA to the sgRNA‐MS2‐P65‐HSF1‐Neo vector. A lentivirus was used to deliver dCAS9‐VP64‐Puro vectors into HCC cells first, after being selected by 2 µg/mL puromycin (Solarbio, Beijing, China), the sgRNA‐MS2‐P65‐HSF1‐Neo vector was delivered into cells, and then selected by 300 µg/mL G418 (Beyotime, Jiangsu, China). All the expression plasmids were confirmed by sequencing.
Cell counting Kit-8 (CCK-8) assay and colony formation
For the CCK8 assay, 5 × 103 transfected cells were seeded into 96-well plates and cultured overnight. Cell viability was measured using the CCK-8 assay after 0, 24, 48, 72, and 96 h. Briefly, 10 µL of CCK-8 reagent (Targetmol) was added to 100 µL of medium per well and incubated at 37 °C for 1 h, then the absorbance was measured at 450 nm using a microplate reader (SpectraMax i3x, Molecular Devices, Shanghai, China). For the colony formation assay, 1500 cells were seeded into 6-well plates and cultured for 2 weeks when the colonies were visualized with 1% crystal violet staining (Solarbio, Beijing, China). All experiments were repeated at least three times.
Calcein AM/propidium Iodide (PI) staining assay
A Calcein/PI staining kit (Beyotime, Jiangsu, China) was used to detect living cells and dead cells. Briefly, 5,000 cells transfected with LV-con or LV-USP24 vectors were seeded into 96-well plate. The cells were then incubated with Calcein AM (living cells) and PI (dead cells) for 30 min at 37 °C and observed under a fluorescence microscope.
Transwell and wound-healing assays
Transwell migration assay was utilized to evaluate the migration capacity of HCC cells. Briefly, 1 × 105 cells in 200 µL of serum-free medium were inoculated in the upper chamber, and medium containing 20% FBS was added into the lower chambers. The cells were then cultured for 24 h. Subsequently, cells that migrated to the bottom chamber were fixed with 4% paraformaldehyde followed by staining with 1% crystal violet solution. For wound-healing assays, 3 × 105 cells were seeded into 12-well plates in medium containing 10% FBS. After the cells were fused into monolayer cells, the middle of each well was scratched with a 10 µL pipette tip, then cultured with serum-free medium. Images were captured of the layer of cells as they grow to close the ‘wound’ in the well. All experiments were repeated at least three times.
Total RNA extraction and quantitative RT-PCR
Total RNA was extracted from cells using the TRIzon reagent (CWBIO, China) according to the manufacturer’s instructions. The concentration and purity of total RNA were measured by the NanoDrop One instrument (Thermo, USA). cDNA synthesis was performed using SweScript All-in-One RT SuperMix (G3337, Servicebio, China) and qRT-PCR was conducted with a SYBR Green qPCR Master Mix (G3321, Servicebio, China) to detect the expression level of targeted genes. β-actin was used as an internal control and the results were analyzed using the 2−ΔΔCT method. The sequences of primers are listed in Supplementary Table 4.
Western blot
Total protein was extracted from cells with RIPA buffer (Solarbio, Beijing, China), and the concentration was quantified by a BCA protein assay kit (EpiZyme, Shanghai, China). About 20 µg of total proteins were separated on SDS-PAGE gels and then transferred onto 0.45 µm PVDF membranes (Millipore, USA). The PVDF membranes were blocked with 5% non-fat milk at room temperature for 2 h and then incubated with the primary antibodies at 4 °C overnight. Membranes were washed with TBST three times for 10 min each time. Subsequently, the membranes were incubated with secondary antibodies at room temperature for 1 h and then washed with TBST. The protein bands were visualized by an ECL chemiluminescent reagent (Millipore, USA).
Co-immunoprecipitation (Co-IP)
Co-IP was performed utilizing a Co-IP kit (#88805, Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. Briefly, cells harvested from 10 cm culture plates were lysed in IP Lysis/Wash buffer on ice for 30 min. The supernatant was collected after centrifugation and protein concentrations were measured. Then, 25 µL Protein A/G magnetic beads were prewashed twice and incubated with 5 µg antibodies for 30 min at room temperature. The antibodies were crosslinked to the beads with DSS for 30 min. Then, 500 µL of protein lysate was added into the crosslinked magnetic beads and incubated overnight at 4 °C. Subsequently, the proteins were eluted with an elution buffer, and the supernatants were subjected to western blot.
In vivo ubiquitination assay
For the in vivo ubiquitination assay, HCC cells were treated with 10 µM MG132 for 8 h before harvesting. The cell lysates extracted from USP24 control or overexpression HCC cells were immunoprecipitated with an anti-Beclin1 antibody as described above. Then the ubiquitination level of Beclin1 was measured with anti-ubiquitin, anti-K48-linkage specific polyubiquitin, and anti-K63-linkage specific polyubiquitin antibodies by western blot.
Protein half-life assay
To detect the stability of Beclin1, the protein half-life was measured. Transfected cells in the exponential growth phase were treated with the protein synthesis inhibitor cycloheximide (CHX, 10 µg/mL) for indicated durations (0, 2, 4, 8, and 12 h). Cells were then harvested, and the lysates were subjected to western blot analysis. Protein degradation curves were plotted according to the protein’s expression levels at different time points.
Immunofluorescence staining
HCC cells were seeded on coverslips in 6-well plates. After the cells were attached to the coverslips, the cells were fixed with 4% paraformaldehyde for 20 min, and subjected to permeabilization with 0.5% Triton X-100 (Solarbio, Beijing, China) for 20 min at room temperature. The cells were blocked with 5% BSA for 30 min at room temperature, then incubated with the primary antibodies (anti-USP24 and anti-Beclin1) overnight at 4 °C. After being washed by PBS three times, the cells were incubated with Dylight 594 (red for USP24) and Dylight 488 (green for Beclin1) conjugated secondary antibodies (Abbkine, Wuhan, China) for 1 h at room temperature. Finally, the nuclei were stained with DAPI (Servicebio, China). A confocal microscope (Nikon Eclipse Ti, Nikon, Japan) was utilized to observe the colocalization of USP24 and Beclin1 proteins.
Transmission electron microscopy (TEM)
The formation of autophagic vesicles was observed using TEM. HCC cells were harvested and centrifuged at 1000 × g for 5 min. The cells were resuspended in 2.5% glutaraldehyde in 0.01 M phosphate buffer (Servicebio, China) overnight at 4 °C and then fixed in 1% osmium tetroxide (Ted Pella Inc, USA) for 2 h at room temperature. The samples were dehydrated using a gradient elution of ethanol and embedded in epoxy resin. The resin blocks were cut into 60-80 nm thinness on the ultra microtome, and the tissues were fished out onto the 150 meshes cuprum grids with formvar film. Sections were stained with 2% uranium acetate saturated alcohol solution for 8 min and 2.6% lead citrate for 8 min. Grids were subjected to a TEM (HT7800, HITACHI, Japan) for the observation of autophagic vesicles.
Quantification of cellular iron content
Cellular ferrous iron (Fe2+) was measured using a Cell Ferrous Iron Colorimetric Assay Kit (Elabscience, Wuhan, China) according to the manufacturer’s instructions. Intracellular Fe2+ was also detected by the FerroOrange probe (DojinDo, Japan). Briefly, 2 × 105 HCC cells transfected with USP24 lentivirus or control vectors were seeded into 6-well plates and incubated overnight. These cells were washed three times with Hank’s balanced salt solution (HBSS) followed by treatment with FerroOrange (1 µM). The cells were then incubated for 30 min at 37 °C and observed under a fluorescence microscope.
Determination of glutathione (GSH) and malondialdehyde (MDA) levels
The levels of GSH in HCC cells were determined using a GSH assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). The concentration of MDA in HCC cells was evaluated using the Lipid Peroxidation MDA Assay Kit (Beyotime, Shanghai, China). Briefly, cells were lysed with cell lysis buffer (Beyotime, Shanghai, China) or 0.1 M PBS, and then managed according to the manufacturer’s instruction. The contents of each sample were measured by a microplate reader (SpectraMax i3x, Molecular Devices, Shanghai, China) at 405 nm for GSH and 532 nm for MDA. The levels were standardized according to the protein content in the cells.
Animals
The in vivo tumor model was constructed using 5-week-old female BALB/c nude mice (Beijing HFK Bioscience Co., Ltd.) for tumor xenograft experiments. The animals were housed in a standard room without specific pathogens at the Department of Zhengzhou University Animal Center and fed with enough water and food. The mice were subcutaneously injected with 1 × 107 SMMC-7721 cells stably overexpressing USP24 or control vectors pre-suspended in 100 µL PBS. Tumor size and body weight were measured every three days. The maximum length of the tumor should not exceed 2 cm. The volume formula (length × width2/2) was used to calculate tumor volume. After 18 days, mice were sacrificed by cervical dislocation, then the mice were photographed and the tumors were surgically excised. The isolated tumor tissues were immediately fixed in 4% polyformaldehyde for subsequent experiments. All animal procedures were approved by the Institutional Animal Care and Use Committee of Zhengzhou University (ZZU-LAC2023070401), and we have complied with all relevant ethical regulations for animal use.
Human HCC tissue array, immunohistochemical (IHC) and hematoxylin & eosin (H&E) staining
The human HCC tissue array was acquired from SHANGHAI OUTDO BIOTECH (Shanghai, China). SMMC-7721 xenografts were fixed in 4% polyformaldehyde and embedded in paraffin. The tissues were sliced into 4 µm thick sections used for IHC and H&E staining. The expression of USP24 protein was detected by IHC staining. Briefly, sodium citrate buffer was used for antigen retrieval, then the tissues were incubated with 3% hydrogen peroxide for 25 min and blocked by 3% BSA for 30 min. The sections were incubated with primary antibodies (USP24, 1:200, HUABIO; Ki-67, 1:1000, Servicebio) overnight at 4 °C, and then incubated with HRP-labeled goat anti-rabbit secondary antibody (GB23303, Servicebio, China) for 60 min at room temperature. After staining with DAB substrate solution (Servicebio, China), hematoxylin (Servicebio, China) was used for counterstaining. All slices were observed and the images were taken with a microscope (OLYMPUS, Japan).
Statistics and reproducibility
Experimental data were analyzed using GraphPad Prism 9.0 software. Data that conformed to the normal distribution were analyzed using the student’s t-tests or one-way ANOVA. The Kaplan–Meier method was used for survival analysis. Mander’s correlation analysis was used to evaluate the correlations of the protein expression between USP24 and Beclin1. All statistical data were presented as mean ± standard deviation (SD) or standard error of the mean values. All statistical tests were two-sided, and a P value of <0.05 was statistically significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All data were collected from at least three independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The source data underlying the graphs can be found in Supplementary Data. The original blot images are included in the Supplementary fig. 5. Please contact the corresponding author for all other data requests.
References
Sung, H. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 71, 209–249 (2021).
McGlynn, K. A., Petrick, J. L. & El-Serag, H. B. Epidemiology of Hepatocellular Carcinoma. Hepatol. (Baltim., Md.) 73, 4–13 (2021).
Forner, A., Reig, M. & Bruix, J. Hepatocellular carcinoma. Lancet 391, 1301–1314 (2018).
Llovet, J. M. et al. Immunotherapies for hepatocellular carcinoma. Nat. Rev. Clin. Onco l 19, 151–172 (2022).
Huang, A., Yang, X. R., Chung, W. Y., Dennison, A. R. & Zhou, J. Targeted therapy for hepatocellular carcinoma. Signal Transduct. Target Ther. 5, 146 (2020).
Chakraborty, J., Basso, V. & Ziviani, E. Post translational modification of Parkin. Biol. Direct 12, 6 (2017).
Wilkinson, K. D. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev. Biol. 11, 141–148 (2000).
Qi, J. & Ronai, Z. A. Dysregulation of ubiquitin ligases in cancer. Drug Resist Updat 23, 1–11 (2015).
Hyer, M. L. et al. A small-molecule inhibitor of the ubiquitin activating enzyme for cancer treatment. Nat. Med 24, 186–193 (2018).
Barghout, S. H. et al. Preclinical evaluation of the selective small-molecule UBA1 inhibitor, TAK-243, in acute myeloid leukemia. Leukemia 33, 37–51 (2019).
Tyagi, A. et al. CRISPR/Cas9-based genome-wide screening for deubiquitinase subfamily identifies USP1 regulating MAST1-driven cisplatin-resistance in cancer cells. Theranostics 12, 5949–5970 (2022).
Xu, G. et al. The deubiquitinase USP16 functions as an oncogenic factor in K-RAS-driven lung tumorigenesis. Oncogene 40, 5482–5494 (2021).
Li, J. T. et al. Diet high in branched-chain amino acid promotes PDAC development by USP1-mediated BCAT2 stabilization. Natl Sci. Rev. 9, nwab212 (2022).
Wan, J. Y. et al. Association mapping of the PARK10 region for Parkinson’s disease susceptibility genes. Parkinsonism Relat. Disord. 20, 93–98 (2014).
Li, Y. et al. Genetic evidence for ubiquitin-specific proteases USP24 and USP40 as candidate genes for late-onset Parkinson disease. Hum. Mutat. 27, 1017–1023 (2006).
Wu, Y. R. et al. Ubiquitin specific proteases USP24 and USP40 and ubiquitin thiolesterase UCHL1 polymorphisms have synergic effect on the risk of Parkinson’s disease among Taiwanese. Clin. Chim. acta; Int. J. Clin. Chem. 411, 955–958 (2010).
Wang, Y. C. et al. Variants of ubiquitin-specific peptidase 24 play a crucial role in lung cancer malignancy. Oncogene 35, 3669–3680 (2016).
Wang, S. A. et al. USP24 promotes drug resistance during cancer therapy. Cell death Differ. 28, 2690–2707 (2021).
Bedekovics, T. et al. USP24 Is a Cancer-Associated Ubiquitin Hydrolase, Novel Tumor Suppressor, and Chromosome Instability Gene Deleted in Neuroblastoma. Cancer Res. 81, 1321–1331 (2021).
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Gao, M. et al. Ferroptosis is an autophagic cell death process. Cell Res 26, 1021–1032 (2016).
Mancias, J. D., Wang, X., Gygi, S. P., Harper, J. W. & Kimmelman, A. C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 (2014).
Yang, A. et al. Autophagy Sustains Pancreatic Cancer Growth through Both Cell-Autonomous and Nonautonomous Mechanisms. Cancer Discov. 8, 276–287 (2018).
Li, Q. et al. HIF-1alpha-induced expression of m6A reader YTHDF1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting ATG2A and ATG14 translation. Signal Transduct. Target Ther. 6, 76 (2021).
Li, K. et al. TRIM7 modulates NCOA4-mediated ferritinophagy and ferroptosis in glioblastoma cells. Redox Biol. 56, 102451 (2022).
Lin, P. L., Tang, H. H., Wu, S. Y., Shaw, N. S. & Su, C. L. Saponin Formosanin C-induced Ferritinophagy and Ferroptosis in Human Hepatocellular Carcinoma Cells. Antioxidants (Basel) 9, https://doi.org/10.3390/antiox9080682 (2020).
Yin, Z., Popelka, H., Lei, Y., Yang, Y. & Klionsky, D. J. The Roles of Ubiquitin in Mediating Autophagy. Cells 9, https://doi.org/10.3390/cells9092025 (2020).
Chen, R. H., Chen, Y. H. & Huang, T. Y. Ubiquitin-mediated regulation of autophagy. J. Biomed. Sci. 26, 80 (2019).
Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res 26, 399–422 (2016).
Mattiroli, F. & Sixma, T. K. Lysine-targeting specificity in ubiquitin and ubiquitin-like modification pathways. Nat. Struct. Mol. Biol. 21, 308–316 (2014).
Oliveira, S. A. et al. Identification of risk and age-at-onset genes on chromosome 1p in Parkinson disease. Am. J. Hum. Genet. 77, 252–264 (2005).
Haugarvoll, K. et al. Fine-mapping and candidate gene investigation within the PARK10 locus. Eur. J. Hum. Genet. : EJHG 17, 336–343 (2009).
Thayer, J. A. et al. The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability. Autophagy 16, 140–153 (2020).
Li, X. et al. USP24-dependent stabilization of Runx2 recruits a p300/NCOA3 complex to transactivate ADAMTS genes and promote degeneration of intervertebral disc in chronic inflammation mice. Biol. Direct 18, 37 (2023).
Zang, L. et al. Ubiquitin-specific protease 24 promotes EV71 infection by restricting K63-linked polyubiquitination of TBK1. Virol. Sin. 38, 75–83 (2023).
Wang, Y. C. et al. USP24 induces IL-6 in tumor-associated microenvironment by stabilizing p300 and beta-TrCP and promotes cancer malignancy. Nat. Commun. 9, 3996 (2018).
He, H. et al. USP24-GSDMB complex promotes bladder cancer proliferation via activation of the STAT3 pathway. Int. J. Biol. Sci. 17, 2417–2429 (2021).
Zhi, X., Jiang, S., Zhang, J. & Qin, J. Ubiquitin-specific peptidase 24 accelerates aerobic glycolysis and tumor progression in gastric carcinoma through stabilizing PLK1 to activate NOTCH1. Cancer Sci. https://doi.org/10.1111/cas.15847 (2023).
Chen, L., Liu, P., Evans, T. C. Jr. & Ettwiller, L. M. DNA damage is a pervasive cause of sequencing errors, directly confou nding variant identification. Science 355, 752–756 (2017).
Sun, D. et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic acids Res. 49, D1420–D1430 (2021).
Hu, Z. et al. MiR-21-5p promotes sorafenib resistance and hepatocellular carcinoma progression by regulating SIRT7 ubiquitination through USP24. Life Sci. 325, 121773 (2023).
Meng, J. et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics 9, 573–587 (2019).
Li, X. et al. Deubiquitinase USP39 and E3 ligase TRIM26 balance the level of ZEB1 ubiquitination and thereby determine the progression of hepatocellular carcinoma. Cell death Differ. 28, 2315–2332 (2021).
Rong, Y. et al. USP11 regulates autophagy-dependent ferroptosis after spinal cord ischemia-reperfusion injury by deubiquitinating Beclin 1. Cell death Differ. 29, 1164–1175 (2022).
Xu, D. et al. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev. 30, 1718–1730 (2016).
Kim, M. J. et al. USP15 negatively regulates lung cancer progression through the TRAF6-BECN1 signaling axis for autophagy induction. Cell death Dis. 13, 348 (2022).
Singh, S. S. et al. Dual role of autophagy in hallmarks of cancer. Oncogene 37, 1142–1158 (2018).
Sun, Y. et al. Human Cytomegalovirus Protein pUL38 Prevents Premature Cell Death by Binding to Ubiquitin-Specific Protease 24 and Regulating Iron Metabolism. J. virol. 92, https://doi.org/10.1128/JVI.00191-18 (2018).
Konermann, S. et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588 (2015).
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (Grant reference numbers: 81870457 and 82172944) and Key science and technology projects of Henan Province (No. 232102311048). We thank (www.home-for-researchers.com) Home for Researchers editorial team for language editing service.
Author information
Authors and Affiliations
Contributions
Jiahui Cao, Shitao Wu and Senfeng Zhao performed the experiments and analyzed data. Jiahui Cao wrote the manuscript. Libo Wang, Yahui Wu and Liming Song conducted the cell experiments. Chenguang Sun, Yin Liu, Zhipu Liu performed the animal experiments. Ruopeng Liang provided necessary writing guidance. Rongtao Zhu, Weijie Wang and Yuling Sun arranged subject ideas and provided theoretical, technical guidance.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical Approval and Consent to participate
The research was conducted in accordance with the principles and guidelines of the Ethics Committee of The First Affiliated Hospital of Zhengzhou University. All animal experiments were approved by the Institutional Animal Care and Use Committee of Zhengzhou University (ZZU-LAC2023070401).
Peer review
Peer review information
Communications Biology thanks Jan-Jong Hung and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Mark Collins and Mengtan Xing.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cao, J., Wu, S., Zhao, S. et al. USP24 promotes autophagy-dependent ferroptosis in hepatocellular carcinoma by reducing the K48-linked ubiquitination of Beclin1. Commun Biol 7, 1279 (2024). https://doi.org/10.1038/s42003-024-06999-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06999-5
This article is cited by
-
USP24 promotes hepatocellular carcinoma progression by deubiquitinating and stabilizing YAP1
Cancer Cell International (2025)
-
Autophagy-based therapy for hepatocellular carcinoma: from standard treatments to combination therapy, oncolytic virotherapy, and targeted nanomedicines
Clinical and Experimental Medicine (2024)