Abstract
Astrocytes promote neuroinflammation and neurodegeneration in multiple sclerosis (MS) through cell-intrinsic activities and their ability to recruit and activate other cell types. In a genome-wide CRISPR-based forward genetic screen investigating regulators of astrocyte proinflammatory responses, we identified the C-type lectin ___domain-containing 16A gene (CLEC16A), linked to MS susceptibility, as a suppressor of nuclear factor-κB (NF-κB) signaling. Gene and small-molecule perturbation studies in mouse primary and human embryonic stem cell-derived astrocytes in combination with multiomic analyses established that CLEC16A promotes mitophagy, limiting mitochondrial dysfunction and the accumulation of mitochondrial products that activate NF-κB, the NLRP3 inflammasome and gasdermin D. Astrocyte-specific Clec16a inactivation increased NF-κB, NLRP3 and gasdermin D activation in vivo, worsening experimental autoimmune encephalomyelitis, a mouse model of MS. Moreover, we detected disrupted mitophagic capacity and gasdermin D activation in astrocytes in samples from individuals with MS. These findings identify CLEC16A as a suppressor of astrocyte pathological responses and a candidate therapeutic target in MS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
27,99 € / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
209,00 € per year
only 17,42 € per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Data supporting the findings of this study are available as supplementary and source data files. All raw and processed deep-sequencing data have been deposited into the Gene Expression Omnibus under SuperSeries accession numbers GSE189030 and GSE283395. Source data are provided with this paper.
Code availability
No custom software code was used in this research.
References
Alvarez, J. I. et al. The Hedgehog pathway promotes blood–brain barrier integrity and CNS immune quiescence. Science 334, 1727–1731 (2011).
Anderson, M. A. et al. Astrocyte scar formation aids central nervous system axon regeneration. Nature 532, 195–200 (2016).
Escartin, C. et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat. Neurosci. 24, 312–325 (2021).
Molofsky, A. V. et al. Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194 (2014).
Nagai, J. et al. Hyperactivity with disrupted attention by activation of an astrocyte synaptogenic cue. Cell 177, 1280–1292 (2019).
Stogsdill, J. A. et al. Astrocytic neuroligins control astrocyte morphogenesis and synaptogenesis. Nature 551, 192–197 (2017).
Anderson, M. A. et al. Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature 561, 396–400 (2018).
Gibson, E. M. et al. Methotrexate chemotherapy induces persistent tri-glial dysregulation that underlies chemotherapy-related cognitive impairment. Cell 176, 43–55 (2019).
Glasgow, S. M. et al. Glia-specific enhancers and chromatin structure regulate NFIA expression and glioma tumorigenesis. Nat. Neurosci. 20, 1520–1528 (2017).
Sanmarco, L. M. et al. Gut-licensed IFNγ+ NK cells drive LAMP1+TRAIL+ anti-inflammatory astrocytes. Nature 590, 473–479 (2021).
Wheeler, M. A. et al. MAFG-driven astrocytes promote CNS inflammation. Nature 578, 593–599 (2020).
Lee, H.-G., Wheeler, M. A. & Quintana, F. J. Function and therapeutic value of astrocytes in neurological diseases. Nat. Rev. Drug Discov. 21, 339–358 (2022).
Sanmarco, L. M., Polonio, C. M., Wheeler, M. A. & Quintana, F. J. Functional immune cell–astrocyte interactions. J. Exp. Med. 218, e20202715 (2021).
Wheeler, M. A. & Quintana, F. J. Regulation of astrocyte functions in multiple sclerosis. Cold Spring Harb. Perspect. Med. 9, a029009 (2019).
Clark, I. C. et al. Barcoded viral tracing of single-cell interactions in central nervous system inflammation. Science 372, eabf1230 (2021).
Ito, M. et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 565, 246–250 (2019).
Rothhammer, V. et al. Microglial control of astrocytes in response to microbial metabolites. Nature 557, 724–728 (2018).
Rothhammer, V. et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 22, 586–597 (2016).
Wheeler, M. A. et al. Environmental control of astrocyte pathogenic activities in CNS inflammation. Cell 176, 581–596 (2019).
Chao, C.-C. et al. Metabolic control of astrocyte pathogenic activity via cPLA2-MAVS. Cell 179, 1483–1498 (2019).
Mayo, L. et al. Regulation of astrocyte activation by glycolipids drives chronic CNS inflammation. Nat. Med. 20, 1147–1156 (2014).
Knott, G. J. & Doudna, J. A. CRISPR–Cas guides the future of genetic engineering. Science 361, 866–869 (2018).
Joung, J. et al. Genome-scale CRISPR–Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR–Cas9. Science 346, 1258096 (2014).
Berge, T., Leikfoss, I. S. & Harbo, H. F. From identification to characterization of the multiple sclerosis susceptibility gene CLEC16A. Int. J. Mol. Sci. 14, 4476–4497 (2013).
International Multiple Sclerosis Genetics Consortium. Multiple sclerosis genomic map implicates peripheral immune cells and microglia in susceptibility. Science 365, eaav7188 (2019).
Liddelow, S. A. & Barres, B. A. Reactive astrocytes: production, function, and therapeutic potential. Immunity 46, 957–967 (2017).
Everhart, M. B. et al. Duration and intensity of NF-κB activity determine the severity of endotoxin-induced acute lung injury. J. Immunol. 176, 4995–5005 (2006).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Kim, S., Naylor, S. A. & DiAntonio, A. Drosophila Golgi membrane protein Ema promotes autophagosomal growth and function. Proc. Natl Acad. Sci. USA 109, E1072–E1081 (2012).
Lee, D.-F. et al. IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 130, 440–455 (2007).
Li, Z. et al. BCL-6 negatively regulates expression of the NF-κB1 p105/p50 subunit. J. Immunol. 174, 205–214 (2005).
Schuster, C. et al. The autoimmunity-associated gene CLEC16A modulates thymic epithelial cell autophagy and alters T cell selection. Immunity 42, 942–952 (2015).
Soleimanpour, S. A. et al. The diabetes susceptibility gene Clec16a regulates mitophagy. Cell 157, 1577–1590 (2014).
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Green, D. R., Galluzzi, L. & Kroemer, G. Mitochondria and the autophagy–inflammation–cell death axis in organismal aging. Science 333, 1109–1112 (2011).
Sidarala, V. et al. Mitophagy protects β cells from inflammatory damage in diabetes. JCI Insight 5, e141138 (2020).
Martorell-Riera, A. et al. Mfn2 downregulation in excitotoxicity causes mitochondrial dysfunction and delayed neuronal death. EMBO J. 33, 2388–2407 (2014).
Chen, Y. & Dorn, G. W. 2nd. PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science 340, 471–475 (2013).
Iwashita, H. et al. Live cell imaging of mitochondrial autophagy with a novel fluorescent small molecule. ACS Chem. Biol. 12, 2546–2551 (2017).
Takahashi, D. et al. AUTACs: cargo-specific degraders using selective autophagy. Mol. Cell 76, 797–810 (2019).
Fang, E. F. et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 22, 401–412 (2019).
Georgakopoulos, N. D., Wells, G. & Campanella, M. The pharmacological regulation of cellular mitophagy. Nat. Chem. Biol. 13, 136–146 (2017).
Motori, E. et al. Inflammation-induced alteration of astrocyte mitochondrial dynamics requires autophagy for mitochondrial network maintenance. Cell Metab. 18, 844–859 (2013).
Azarias, G. et al. Glutamate transport decreases mitochondrial pH and modulates oxidative metabolism in astrocytes. J. Neurosci. 31, 3550–3559 (2011).
Bélanger, M., Allaman, I. & Magistretti, P. J. Brain energy metabolism: focus on astrocyte–neuron metabolic cooperation. Cell Metab. 14, 724–738 (2011).
Jackson, J. G. & Robinson, M. B. Regulation of mitochondrial dynamics in astrocytes: mechanisms, consequences, and unknowns. Glia 66, 1213–1234 (2018).
Zhou, R., Yazdi, A. S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Murphy, M. P. et al. Guidelines for measuring reactive oxygen species and oxidative damage in cells and in vivo. Nat. Metab. 4, 651–662 (2022).
West, A. P. & Shadel, G. S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 17, 363–375 (2017).
Nakahira, K. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 (2011).
Shimada, K. et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 (2012).
Barclay, W. & Shinohara, M. L. Inflammasome activation in multiple sclerosis and experimental autoimmune encephalomyelitis (EAE). Brain Pathol. 27, 213–219 (2017).
Guo, H., Callaway, J. B. & Ting, J. P. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015).
Lieberman, J., Wu, H. & Kagan, J. C. Gasdermin D activity in inflammation and host defense. Sci. Immunol. 4, eaav1447 (2019).
Coll, R. C. et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 (2015).
Inoue, M., Williams, K. L., Gunn, M. D. & Shinohara, M. L. NLRP3 inflammasome induces chemotactic immune cell migration to the CNS in experimental autoimmune encephalomyelitis. Proc. Natl Acad. Sci. USA 109, 10480–10485 (2012).
Paré, A. et al. IL-1β enables CNS access to CCR2hi monocytes and the generation of pathogenic cells through GM-CSF released by CNS endothelial cells. Proc. Natl Acad. Sci. USA 115, E1194–E1203 (2018).
Witte, M. E. et al. Reduced expression of PGC-1α partly underlies mitochondrial changes and correlates with neuronal loss in multiple sclerosis cortex. Acta Neuropathol. 125, 231–243 (2013).
Allen, N. J. & Eroglu, C. Cell biology of astrocyte–synapse interactions. Neuron 96, 697–708 (2017).
Bessou, M. et al. The apoptosis inhibitor Bcl-xL controls breast cancer cell migration through mitochondria-dependent reactive oxygen species production. Oncogene 39, 3056–3074 (2020).
McKenzie, B. A. et al. Caspase-1 inhibition prevents glial inflammasome activation and pyroptosis in models of multiple sclerosis. Proc. Natl Acad. Sci. USA 115, E6065–E6074 (2018).
Linnerbauer, M., Wheeler, M. A. & Quintana, F. J. Astrocyte crosstalk in CNS inflammation. Neuron 108, 608–622 (2020).
Magistretti, P. J. & Allaman, I. Lactate in the brain: from metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 19, 235–249 (2018).
Exner, N., Lutz, A. K., Haass, C. & Winklhofer, K. F. Mitochondrial dysfunction in Parkinson’s disease: molecular mechanisms and pathophysiological consequences. EMBO J. 31, 3038–3062 (2012).
González-Rodríguez, P. et al. Disruption of mitochondrial complex I induces progressive parkinsonism. Nature 599, 650–656 (2021).
Mahad, D. H., Trapp, B. D. & Lassmann, H. Pathological mechanisms in progressive multiple sclerosis. Lancet Neurol. 14, 183–193 (2015).
Fünfschilling, U. et al. Glycolytic oligodendrocytes maintain myelin and long-term axonal integrity. Nature 485, 517–521 (2012).
Giladi, A. et al. Cxcl10+ monocytes define a pathogenic subset in the central nervous system during autoimmune neuroinflammation. Nat. Immunol. 21, 525–534 (2020).
Gruber, R. C. et al. The control of reactive oxygen species production by SHP-1 in oligodendrocytes. Glia 63, 1753–1771 (2015).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Locatelli, G. et al. Mononuclear phagocytes locally specify and adapt their phenotype in a multiple sclerosis model. Nat. Neurosci. 21, 1196–1208 (2018).
Masuda, T. et al. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566, 388–392 (2019).
Mendiola, A. S. et al. Transcriptional profiling and therapeutic targeting of oxidative stress in neuroinflammation. Nat. Immunol. 21, 513–524 (2020).
Harper, J. W., Ordureau, A. & Heo, J.-M. Building and decoding ubiquitin chains for mitophagy. Nat. Rev. Mol. Cell Biol. 19, 93–108 (2018).
Mascanfroni, I. D. et al. IL-27 acts on DCs to suppress the T cell response and autoimmunity by inducing expression of the immunoregulatory molecule CD39. Nat. Immunol. 14, 1054–1063 (2013).
Freeman, L. et al. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J. Exp. Med. 214, 1351–1370 (2017).
Mandolesi, G. et al. Interleukin-1β alters glutamate transmission at Purkinje cell synapses in a mouse model of multiple sclerosis. J. Neurosci. 33, 12105–12121 (2013).
Wallach, D., Kang, T.-B., Dillon, C. P. & Green, D. R. Programmed necrosis in inflammation: toward identification of the effector molecules. Science 352, aaf2154 (2016).
Humphries, F. et al. Succination inactivates gasdermin D and blocks pyroptosis. Science 369, 1633–1637 (2020).
Li, S. et al. Gasdermin D in peripheral myeloid cells drives neuroinflammation in experimental autoimmune encephalomyelitis. J. Exp. Med. 216, 2562–2581 (2019).
Fransen, N. L. et al. Post-mortem multiple sclerosis lesion pathology is influenced by single nucleotide polymorphisms. Brain Pathol. 30, 106–119 (2020).
Frischer, J. M. et al. Clinical and pathological insights into the dynamic nature of the white matter multiple sclerosis plaque. Ann. Neurol. 78, 710–721 (2015).
Luchetti, S. et al. Progressive multiple sclerosis patients show substantial lesion activity that correlates with clinical disease severity and sex: a retrospective autopsy cohort analysis. Acta Neuropathol. 135, 511–528 (2018).
Malhotra, S. et al. NLRP3 inflammasome as prognostic factor and therapeutic target in primary progressive multiple sclerosis patients. Brain 143, 1414–1430 (2020).
Kadowaki, A. & Quintana, F. J. The NLRP3 inflammasome in progressive multiple sclerosis. Brain 143, 1286–1288 (2020).
Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015).
Mouton-Liger, F. et al. Parkin deficiency modulates NLRP3 inflammasome activation by attenuating an A20-dependent negative feedback loop. Glia 66, 1736–1751 (2018).
Sorrentino, V. et al. Enhancing mitochondrial proteostasis reduces amyloid-β proteotoxicity. Nature 552, 187–193 (2017).
Ising, C. et al. NLRP3 inflammasome activation drives tau pathology. Nature 575, 669–673 (2019).
Pan, D. et al. A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359, 770–775 (2018).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Kadowaki, A. et al. Gut environment-induced intraepithelial autoreactive CD4+ T cells suppress central nervous system autoimmunity via LAG-3. Nat. Commun. 7, 11639 (2016).
Kadowaki, A., Saga, R., Lin, Y., Sato, W. & Yamamura, T. Gut microbiota-dependent CCR9+CD4+ T cells are altered in secondary progressive multiple sclerosis. Brain 142, 916–931 (2019).
Tcw, J. et al. An efficient platform for astrocyte differentiation from human induced pluripotent stem cells. Stem Cell Rep. 9, 600–614 (2017).
Trombetta, J. J. et al. Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 107, 4.22.1–4.22.17 (2014).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Soneson, C., Love, M. I. & Robinson, M. D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Rath, S. et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 49, D1541–D1547 (2021).
Pagliarini, D. J. et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008).
Polman, C. H. et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 69, 292–302 (2011).
Kuhlmann, T. et al. An updated histological classification system for multiple sclerosis lesions. Acta Neuropathol. 133, 13–24 (2017).
Dhaeze, T. et al. CD70 defines a subset of proinflammatory and CNS-pathogenic TH1/TH17 lymphocytes and is overexpressed in multiple sclerosis. Cell. Mol. Immunol. 16, 652–665 (2019).
Broux, B. et al. Interleukin-26, preferentially produced by TH17 lymphocytes, regulates CNS barrier function. Neurol. Neuroimmunol. Neuroinflamm. 7, e870 (2020).
Acknowledgements
A.K. was supported by a Uehara Memorial Foundation Overseas Research Fellowship, the Japan Society for the Promotion of Science (JSPS) Overseas Research Fellowship, the JSPS PD Research Fellowship and Grant-in-Aids for JSPS fellows (grant title: ‘Investigation of the role of mitophagy-regulating molecules in astrocytic CNS inflammation’). This work was supported by grants NS102807, NS087867, ES02530, AI126880 and AI093903 from the National Institutes of Health (NIH), RG4111A1 from the National MS Society (NMSS) and PA-1604-08459 from the International Progressive MS Alliance and the Chan Zuckerberg Initiative to F.J.Q. M.A.W. was supported by R00NS114111, R01MH130458 and R01MH132632. B.M.A. was supported by the Training Program in Nervous System Tumors (K12CA090354) from the National Cancer Institute (NCI)/NIH, the Career Enhancement Program (CEP) for the SPORE at the Harvard Cancer Center (P50CA165962) from the NCI/NIH and the Postdoctoral Fellowship in Translational Medicine from the PhRMA Foundation. H.-G.L. was supported by the Basic Science Research Program through the National Research Foundation of Korea (2021R1A6A3A14039088). J.-H.L. was supported by the Basic Science Research Program funded by the National Research Foundation of Korea (2022R1A6A3A03071157) and a long-term postdoctoral fellowship funded by the Human Frontier Science Program (LT0015/2023-L). S.A.S. was supported by the JDRF (CDA-2016-189, COE-2019-861), the NIH (R01 DK108921, U01 DK127747) and the Department of Veterans Affairs (I01 BX004444). A.N. was supported by a Max Kade Fellowship. I.C. was supported by the NIH (5R01DK127257, 5R01AI130019), Burroughs Wellcome Trust, Chan Zuckerberg Initiative and Kenneth Rainin Foundation. A.P. holds a T1 Canada Research Chair in MS and was supported by the Canada Institute of Health Research, MS Canada, the NMSS and the Canadian Foundation for Innovation. V.K. is a New York Stem Cell Foundation Robertson Stem Cell Investigator, and this work was additionally supported by NIH grant NS109209-01A1. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. We thank all members of the Quintana laboratory for helpful advice and discussions, as well as L. Ding for assistance with live-cell imaging and quantification, T. Yamamura (National Institute of Neuroscience, Tokyo, Japan) and H. Mochizuki (Osaka University, Osaka, Japan) for support, and R. Saga (BWH) for advice on histology and support.
Author information
Authors and Affiliations
Contributions
A.K. conceptualized and designed the study, performed most of the studies, analyzed and interpreted data, wrote the manuscript and secured funding. M.A.W. designed and performed the CRISPR screen and helped with single-cell data analysis. S.E.J.Z., W.K. and A.P. provided human brain tissues and analyzed them by IHC. A.K., H.B., D.N., M.B. and I.C. performed IHC analysis of mouse CLEC16A, LC3 and GSDMD. A.N. generated hES cell-derived astrocytes and helped with related studies under the supervision of V.K. C.-C.C. offered technical advice, discussion and data interpretation. J.V.M. assisted with EAE and histology. Z.L. performed bioinformatics analyses. B.M.A. assisted with single-cell and mtDNA studies. H.-G.L. quantified IL-1β in EAE. T.I. helped with mitophagy-related protein analysis. J.-H.L. helped with live-cell imaging. G.L. and L.S. kept mouse colonies and assisted with astrocyte cultures. S.A.S. provided Clec16afl/fl mice and CLEC16A-specific antibodies. V.R. participated in the early project stages. F.J.Q. conceptualized and designed the study, interpreted data, wrote the manuscript, secured funding and supervised the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Lars Fugger and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Analysis of the mitophagy-related and mitochondrial proteins.
(a) PINK1 expression in Clec16a siRNA KD primary astrocytes stimulated with TNF/IL-1β. Representative of 3 experiments. (b) Parkin and mitophagy-specific receptor expression in Clec16a siRNA KD primary astrocytes. Representative blots and quantification of mean fold change from baseline (time 0) of normalized mitophagy-specific receptor expression from 3 experiments are shown. *p < 0.05 by paired t-test, two-sided (Calococo2 16Hours p 0.0131). (c) Expression of mitochondrial protein TOM20. Representative blots and quantification from 3 experiments are shown. Mean and s.e.m. p < 0.05 by two-way ANOVA, Sidak post-test (TNFα/IL-1β p 0.0123).
Extended Data Fig. 2 Role of CLEC16A in the control of mitophagy in astrocytes.
(a) Live cell imaging of siRNA KD FCCP or TNF/IL-1β-stimulated murine astrocytes, stained using Mitophagy dye (red) and Lyso-dye (green). Merged area indicate mitochondria undergoing mitophagy. Scale bars 100 μm. Regions of interest are highlighted with white squares and displayed in Fig. 2c. Representative of two experiments. (b, c) Live cell imaging of Control, Atg7 or Clec16a siRNA KD and TNFα/IL-1β-stimulated murine astrocytes, stained using Mitophagy dye (red) and Lyso-dye (green). The astrocytes were also pre-stimulated with TNFα/IL-1β for 13 hrs. (b) Representative images are shown. High Mitophagy dye intensity area generated from the original image by cutting off the low intensity signal area is also shown. Scale bars 30 μm. (c) Quantification of the overlay puncta numbers or area size of the high mitophagy-dye intensity area and lyso-dye are calculated from 9 splitted fields per each time point in each condition. Mean and s.e.m. p** < 0.01, p*** < 0001, p**** < 0.0001 by One-way ANOVA Dunnett’s post-test. (No. overlay puncta siControl versus siAtg7 P = 0.0090, overlay puncta area size siControl versus siAtg7 P = 0.0002).
Extended Data Fig. 3 Cytoplasmic mitochondrial DNA in unstimulated CLEC16A KD astrocytes.
qPCR quantification of mitochondrial gene mt-Co1 DNA extracted from the cytoplasmic fractions of Clec16a KD unstimulated or TNFα/IL-1β-stimulated murine astrocytes. Relative copy numbers to control are displayed. n = 3 samples per condition. Representative of two experiments. Mean and s.e.m. *p < 0.05 by two-way ANOVA, Sidak post-test (TNFα/IL-1β p 0.0189).
Extended Data Fig. 4 Nlrp3 expression in CLEC16A KD astrocytes.
qPCR in Clec16a KD astrocytes. Combination of two experiments, each containing 3 samples per condition, n = 6 in total. Mean and s.e.m. ** p < 0.01 by two-way ANOVA, Sidak post-test (TNFα/IL-1β p 0.0010).
Extended Data Fig. 5 RNA-seq of astrocytes and microglia from CLEC16AGFAP EAE mice.
(a) Clec16a and Ppargc1a expression determined by qPCR in astrocytes and microglia from Control (n = 13) and CLEC16AGFAP (n = 12) EAE mice (day 30). Sorting strategy in Supplementary Fig. 3. Mean and s.e.m. **p < 0.01, *p < 0.05 by two-way ANOVA Sidak post-test (Ppargc1a astrocytes p 0.0032, Clec16a astrocytes p 0.0006). (b) IPA of RNA-Seq data of astrocytes from Control (n = 5) and CLEC16AGFAP (n = 6) EAE mice (day 30). P values were determined using Fisher’s exact test. (c) Differentially expressed genes (DEGs) detected by RNA-seq analysis of microglia from Control (n = 5) or CLEC16AGFAP (n = 6) EAE mice on day 30. (d) IPA of RNA-Seq data of microglia from Control (n = 5) and CLEC16AGFAP (n = 6) EAE mice (day 30). P values were determined using Fisher’s exact test.
Extended Data Fig. 6 Astrocyte-specific CLEC16A deletion in CLEC16AGFAP mice.
(a,b) IHC analysis of CLEC16A (Green), GFAP (purple) in spinal cord sections from Control EAE or CLEC16AGFAP EAE mice (day 21); representative maximum intensity images of Z-stacks are shown. Scale bars 200 μm (upper images) and 30 μm (lower images). (c) Quantification of CLEC16A expression in astrocytes (GFAP), neurons (Neurofilament-L), and microglia (IBA1) are shown. Each condition is quantified from 6 images, containing 2 independent images of whole spinal cord sections from 3 mice. **** p < 0.0001, n.s. not significant by Sidak’s test.
Extended Data Fig. 7 Peripheral immune cells in CLEC16AGFAP EAE mice.
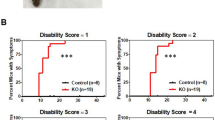
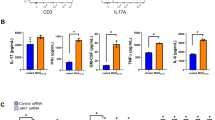
(a) Flow cytometry analysis of CD4+ T cells (CD4+CD3+) and monocytes (CD45hiCD11b+Ly6chi) in CNS and spleen of Control and CLEC16AGFAP mice 30 days after EAE induction. (b) Flow cytometry analysis of IL-17A for Th17 cells, IFNγ for Th1 cells, Foxp3 for regulatory T cells, and IL-10 for type 1 regulatory T cells in CNS and spleen of EAE mice (day 30). (c) Recall cytokine response of splenocytes from Control and CLEC16AGFAP mice (day 30) to MOG(35-55). (a-c) Mean and s.e.m. ***p < 0.001, **p < 0.01, *p < 0.05 by Mann Whiney test, two-sided (CD4+ T cells p 0.0022, monocytes p 0.0022) (a) and two-way ANOVA Sidak post-test (IL-17 + IFNγ- p 0.0007, IL-17-IFNγ + p 0.0353) (b). Representative of two experiments. (a, b) In the box plots, the data are presented as the median and interquartile range and whiskers represent minimum to maximum.
Extended Data Fig. 8 Role of CLEC16A in autophagy in vivo.
(a) Immunohistochemical analysis of LC3 (Green) and GFAP (red) and merged images in spinal cord (SC) sections from Control EAE or CLEC16AGFAP EAE mice (day 21); representative maximum intensity images of Z-stacks are shown. (b) Region 1, 2, and 3 (Fig. 5e) in (a) are depicted in larger images. White scale bars 100 μm, gray scale bars 25 μm. Representative of two experiments.
Extended Data Fig. 9 Proinflammatory gene expression in non-immunized CLEC16AGFAP mice.
(a) qPCR performed in sorted astrocytes from non-immunized Control (n = 6) and CLEC16AGFAP mice (n = 6). Comibination of two experiments. Mean and s.e.m. n.s. not significant by unpaired t-test, two-sided. (b) scRNA-seq analysis of astrocytes in naïve Control and CLEC16AGFAP mice; ingenuity pathway analysis shown. P values were determined using Fisher’s exact test.
Extended Data Fig. 10 scRNA-seq analysis of astrocytes in CLEC16AGFAP EAE mice.
(a) Feature plots depicting astrocyte marker genes. (b) Gene set enrichment analysis (GSEA) was performed in cluster 2 astrocytes. Indicated gene sets were used, normalized enrichment score (NES) and p values are depicted. Nominal p-value was one-tailed test on the appropriate side of the null distribution. (c) IPA analysis of cluster 1 and 3 astrocytes. Representative pathways are shown. P values were determined using Fisher’s exact test.
Supplementary information
Supplementary Information
Supplementary Figs. 1–5 and unprocessed western blots of Supplementary Fig. 2.
Supplementary Tables
Supplementary Table 1. Genes detected in the astrocyte CRISPR–Cas9 screen using NF-κB–EGFP reporter mice. Genes are ranked according to log(fold change), which was determined using MAGeCK to compare the number of reads enriched in the EGFP+ astrocyte fraction versus the EGFP− astrocyte fraction, signifying negative regulators of NF-κB (EGFP) activation. P value was determined by MAGeCK and corresponds to the statistical enrichment of a particular guide within its corresponding fraction. Supplementary Table 2. DEGs detected by RNA-seq in astrocytes from control (n = 5) or CLEC16AGFAP (n = 6) EAE mice on day 30 after EAE induction. Genes are ordered in accordance with the multiplicity-adjusted P value (Padj). Genes with a P value smaller than 0.05 are considered DEGs. Supplementary Table 3. Demographic features of patients with MS and control individuals studied in Fig. 8d–g. PM, postmortem; NA, not applicable.
Supplementary Data
Marker gene lists of astrocyte clusters (1–10) in scRNA-seq experiments (a total of ten Excel files). Files are named ‘Cluster.(# of cluster).markers’.
Source data
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Unprocessed western blots.
Source Data Fig. 4
Unprocessed western blots.
Source Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kadowaki, A., Wheeler, M.A., Li, Z. et al. CLEC16A in astrocytes promotes mitophagy and limits pathology in a multiple sclerosis mouse model. Nat Neurosci 28, 470–486 (2025). https://doi.org/10.1038/s41593-025-01875-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41593-025-01875-9
This article is cited by
-
Ubiquitination-mediated upregulation of glycolytic enzyme MCT4 in promoting astrocyte reactivity during neuroinflammation
Journal of Neuroinflammation (2025)