Abstract
Identifying DNA-binding proteins and their binding residues is critical for understanding diverse biological processes, but conventional experimental approaches are slow and costly. Existing machine learning methods, while faster, often lack accuracy and struggle with data imbalance, relying heavily on evolutionary profiles like PSSMs and HMMs derived from multiple sequence alignments (MSAs). These dependencies make them unsuitable for orphan proteins or those that evolve rapidly. To address these challenges, we introduce TransBind, an alignment-free deep learning framework that predicts DNA-binding proteins and residues directly from a single primary sequence, eliminating the need for MSAs. By leveraging features from pre-trained protein language models, TransBind effectively handles the issue of data imbalance and achieves superior performance. Extensive evaluations using diverse experimental datasets and case studies demonstrate that TransBind significantly outperforms state-of-the-art methods in terms of both accuracy and computational efficiency. TransBind is available as a web server at https://trans-bind-web-server-frontend.vercel.app/.
Similar content being viewed by others
Introduction
DNA-protein interactions play a pivotal role in gene regulation and major cell cycle events, including DNA replication, transcription, and translation1,2,3,4. Certain amino acid residues inside a DNA-binding protein tend to bind with DNA, which are called DNA-protein binding residues (or sites). Micro-array-based experimental approaches, such as protein microarray assays5, protein binding microarray6, and ChIP-seq7 are widely used for the precise identification of DNA binding residues. Although highly precise, these experimental approaches are costly and time- and labor-intensive. Therefore, considerable effort is being put into developing computational methods for predicting DNA-binding proteins and DNA-binding protein residues.
Given the growing availability of protein databases and rapid advances in machine learning (ML) methods (especially, deep learning techniques), the application of ML techniques to leverage the available data in the accurate prediction of DNA-binding proteins and sites has gained significant attention1,8,9,10,11,12,13,14. DNA-protein interaction prediction generally deals with two types of tasks. One is predicting whether a protein sequence is DNA-binding, which means that it will interact with a DNA8,9,10,11, and the second task deals with identifying specific amino acids within a protein sequence that will bind to a DNA1,12. Although the input for both these tasks is the same– a protein sequence, from a prediction model’s perspective, the first is a binary classification task for the input protein sequence and the latter is a binary classification on each of the residues of the protein sequence. Most of the existing approaches address either of these two problems, except for some which deal with both of these tasks13,14. TransBind is designed to address both these problems with high precision and sensitivity.
Most of the existing DNA-binding prediction methods primarily use evolutionary features, such as position specific scoring matrix (PSSM), and hidden Markov models (HMM) profile derived from MSAs and structural information (e.g., secondary structures, torsion angles, etc.). Similar to many other problems in computational proteomics, PSSMs and HMMs are among the most widely used features in DNA-binding protein prediction15,16,17,18,19.
However, MSA-based evolutionary feature extraction is a time-consuming task, making it a major bottleneck for using evolutionary features. Moreover, MSAs of homologous proteins are not always available, such as with orphan proteins (proteins with few or no homologs20,21 as they do not belong to any functionally characterized protein family) and antibodies that tend to have noisy MSAs due to fast evolution. As a result, identifying the feature sets, which are less computationally demanding and alignment-free, yet effective for highly accurate prediction of DNA-binding proteins and DNA-binding residues is of great interest.
Particularly, with the advancement of Natural Language Processing (NLP), pretrained language models have become popular to extract features that can be used for building predictive models. In 2021, ProtTrans22 was published, a transformer-like protein language model trained on 393 billion amino acids from 2.1 billion protein sequences, which can generate feature sets from amino acids capable of performing very well while taking significantly less time to generate compared to PSSM. TransBind presents a successful utilization of pretrained protein models in DNA-binding protein and residue prediction. Recently, some methods have attempted to use language model-based features for predicting DNA-binding proteins and their respective binding sites23,24,25. For instance, Wei et al.26 utilized the ESM-1b27 pre-trained language model to identify DNA-binding proteins. However, their approach lacks the capability to detect specific DNA-binding sites within proteins, meaning it cannot identify DNA-binding protein residues. In another study, Littmann et al.28 employed a generalized framework based on language models to predict binding residues for various ligand classes, including metal ions, nucleic acids, or small molecules. However, the accuracy of predicting DNA/RNA binding sites using this generalized framework was notably low. Furthermore, a number of methods have been proposed to identify DNA binding protein residues which require structural information in addition to language model-based features and evolutionary features23,24,29,30,31,32,33,34,35,36. While leveraging 3D structure can enhance performance, experimentally determined structures are often unavailable for newly discovered sequences.
In this paper, we propose TransBind (Transfer Learning-based Binding Prediction), a sequence-only approach for precisely identifying DNA-binding proteins and residues without relying on MSA-based evolutionary features or structural information. TransBind circumvents some of the important challenges in the field. It uses the “global” features generated by the pretrained protein model ProtTrans22, to generate a residue level feature embedding for each amino acid residue by incorporating the global context of the protein sequence with a self-attention mechanism. To effectively capture the local features within each amino acid residue, we employ a convolution network based approach—namely, the inception V237 architecture. Thus, the features generated by ProtTrans are subsequently processed by a stacked layer of inception-based “local” feature extractor to leverage the inter-relationship among the features generated by protTrans.
TransBind effectively tackles the class-imbalance problem where the positive sample is the minority class (i.e., the number of binding sites is significantly smaller than the non-binding sites) by leveraging a class-weighted training scheme. We report, on an extensive evaluation study, the performance of TransBind for both the DNA-binding protein prediction and the DNA-protein binding residue prediction tasks. In addition to greatly increasing computational efficiency, the inception network-based feature extraction from ProtTrans features coupled with deep learning allows TransBind to achieve remarkable improvements over the state-of-the-art methods on a wide range of benchmark datasets and for a diverse array of evaluation metrics. We present additional results to enhance our understanding of the comparative performances of different methods. Specifically, we analyze the performance of various methods in 20 amino acid classes individually, allowing for a more detailed assessment. We also assessed the performance of TransBind on different model conditions with varying levels of homologous data, showcasing its applicability on orphan proteins or those with a limited number of homologs. Furthermore, we include case studies that involve visual inspection of predicted DNA-binding residues. Importantly, TransBind is a sequence-only method which does not rely on MSA-based evolutionary features and structural information, making it suitable for orphan proteins that do not belong to any functionally characterized protein family and rapidly evolving proteins.
Results
We first briefly present our proposed method TransBind. Next, we report, on an extensive evaluation study, the performance of TransBind compared to the state-of-the-art DNA protein binding prediction methods on a collection of widely used benchmark datasets, spanning a wide range of prediction difficulty levels. We primarily assess the performance of TransBind in predicting DNA-protein binding residues. Furthermore, we demonstrate that the TransBind architecture performs well in predicting DNA binding proteins as well.
The TransBind framework
TransBind takes a sequence of amino acid X = {x1, x2, . . . , xn} as an input where xi represents the ith residue. For each residue xi, TransBind determines whether this residue corresponds to a DNA binding residue or not. In contrast to deriving MSA-based features (e.g., PSSM and HMM) for embedding the protein8,9,38, we used the ProtTrans22 language model.
The ProtT5-XL-UniRef50 model uses the BERT39 architecture and is pretrained to produce an embedding of length 1024 for each amino acid residue within a protein. Thus, ProtT5-XL-UniRef50 generates a sequence of embedding vectors q = {q1, q2, q3, …, qn}, \({q}_{i}\in {{\mathbb{R}}}^{d}\,(d=1024)\), where qi denotes the features generated for each amino acid within the protein sequence. Therefore, for each protein of length n, input to the TransBind model is a sequence of amino acid residues of length n, each of which is encoded into a 1024-dimensional feature vector using ProtT5-XL-UniRef50. This IRN×1024 feature set is passed through our proposed pipeline within the TransBind architecture, which produces an output of dimension IRN×1—indicating whether a residue is DNA-binding or not.
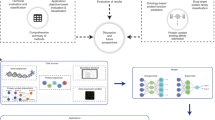
The overall architecture of TransBind is shown in Fig. 1. Instead of passing the whole encoded sequence through the classifier, TransBind ensures the global feature representation with self-attention, and then processes each nucleotide individually with the local feature extractor network. This residue-level granularity allows us to design a weighted class training to effectively handle data imbalance. We describe different components of TransBind in the “Methods” section.
a For an input protein sequence, its amino acid sequence is used as the input to the masked language model (Prot-T5 in TransBind). The language model provides positional encoding for each of the amino acid tokens. b Understanding global features with self attention: within the self-attention block of the transformer, each amino acid attends to all other amino acids in the protein sequence. Hence, it provides a global understanding of the sequence to all the amino acids. c Separation of amino acids: instead of processing amino acids in parallel, each one is isolated and passed through the local feature extraction network individually. This approach enhances the ability to understand the feature space of individual amino acids more effectively compared to processing the entire sequence at once. d Local feature extraction using stacked inception v2 modules: the local feature extraction component works on each amino acid separately. Here, stacked layers of inception V2 module is used as the local feature extractor. Each block consists of six convolutional layers and two pooling layers. \({{{{\bf{d}}}}}^{{\prime} }\) Training with weighted loss distribution for tackling class imbalance: during the training of the local feature extractor, we implement a weighted class loss which provides greater loss value to the incorrectly predicted amino acids of the minority class, which allows to tackle the class imbalance issue. e The prediction head provides binary classification over all the amino acids associated with the given protein in two classes: (i) amino acids that bind to nucleic acid and (ii) those that do not bind to nucleic acid.
DNA-protein binding residue prediction results
The validation accuracies (using 10-fold cross-validation) of TransBind and other competing methods on the PDNA-224 dataset are shown in Table 1. TransBind remarkably outperformed other competing methods across all evaluation metrics.
For example, TransBind achieved an impressive MCC score of 0.82, significantly outperforming the previous best reported MCC score of 0.48, indicating a notable improvement of 70.8%. Furthermore, TransBind exhibited substantial improvements in other performance metrics such as accuracy, sensitivity, and specificity. These improvements signify TransBind’s ability to make more accurate predictions, identify a higher proportion of true binding residues, and achieve better discrimination between binding and non-binding residues. Moreover, TransBind demonstrates remarkably high AUC and AUPR values (Tables 1 and 2), highlighting its consistent and balanced performance despite the presence of substantial amounts of imbalance in the benchmark datasets.
Similar trends were observed in the PDNA-316 dataset (Table 3). TransBind outperformed other competing methods in all evaluation metrics, except for specificity where Saber et al.14 is slightly (less than 1%) better than TransBind, and this small improvement is not statistically significant (p value > 0.05). Moreover, Saber et al.14 is considerably less sensitive than TransBind as the sensitivity of Saber et al.14 and TransBind are 66.91 and 85.00, respectively. Notably, the improvements of TransBind in accuracy, sensitivity and MCC score over other competing methods are remarkable. Moreover, TransBind achieved remarkably high AUC and AUPR scores of 0.965 and 0.951, respectively.
The results on PDNA-543 dataset are shown in Table 4. Saber et al.14 and TransBind showed comparable performance, and both of them are substantially better than the other alternative methods.
Overall, TransBind achieved dramatic improvements in the validation accuracy over other methods on these three benchmark datasets. While other methods tend to achieve high specificity at the cost of sensitivity (meaning that they tend to produce a substantial amount of false negative DNA-binding residue predictions), TransBind achieves higher and more balanced sensitivity and specificity scores, resulting in notably higher MCC scores. Moreover, TransBind achieved impressive AUC and AUPR values across all the datasets.
Finally, we assessed the performance of TransBind and other competing methods in predicting DNA-binding protein residues on the PDNA-41 independent test set. Note that, unlike other methods, we have evaluated and reported the performance of TransBind on the 37 non-redundant sequences, after removing the four sequences with ≥30% sequence similarity, as discussed in section “Test datasets”. Remarkably, the overall performance of TransBind is significantly better than other methods (Table 5). Specifically, TransBind achieved a substantially higher MCC score than all other methods and was 38.7% better than the previous best method, iProDNA. Although ProteDNA had better specificity and accuracy than TransBind, its sensitivity is miserably low (only 4.77%), meaning that ProteDNA tends to classify most residues as non-binding, resulting in a large number of false negative predictions. ProteDNA fails to detect a substantial number of binding sites and incorrectly labels the residues that are actually binding sites as non-binding. However, since non-binding residues are the majority class (i.e., the number of non-binding sites is substantially higher than that of binding sites), it has obtained high accuracy and specificity. But these metrics alone do not suffice to evaluate its overall effectiveness as a predictive model. The low sensitivity score (4.77) underscores ProteDNA’s limitation in capturing true binding sites, which is a critical aspect of binding site prediction. Our method, on the other hand, produces comparable accuracy and specificity, but achieves a substantially higher sensitivity score—making it the best method in terms of the MCC score.
Similarly, other methods (e.g., DNABind, TargetDNA, EC-RUS) that achieved higher sensitivity than TransBind had lower accuracy and specificity, making them worse than TransBind in terms of the MCC score. Note that, in the presence of data imbalance, the MCC score is a more appropriate evaluation metric than accuracy, sensitivity, and specificity. Our results demonstrate that the overall performance of TransBind is notably improved and balanced compared to other methods, which showcases the efficacy of TransBind in detecting the binding residues and achieving better discrimination between binding and non-binding residues. Moreover, TransBind achieved an AUC score of 0.851 and an AUPR of 0.844, highlighting its accurate and balanced predictive performance.
Performance analysis on recent nucleic acid binding datasets
In recent years, a diverse array of methods have emerged that combine structural information with sequence- and MSA-based features to enhance the prediction of protein attributes, especially in the context of protein-nucleic acid interactions.
Additionally, newer datasets such as DNA-129 and RNA-117 have been introduced and are now widely utilized for benchmarking protein-nucleic acid binding prediction models23.
Methods such as GraphBind23, CLAPE-DB25, NucBind29, DNAPred30, targetDNA40, NABind31, and DNABind41 have been benchmarked on these datasets. Some of these methods use structural information along with the MSA- and sequence-based features. For example, NucBind, DNAPred, NABind, DNABind, and GraphBind leverage three-dimensional protein structural embeddings in conjunction with primary sequence data to predict nucleic acid-protein interactions. The availability of structural information serves as a valuable indicator for identifying interacting residues. Several studies, such as42,43, have demonstrated that insights into the surface topology of a protein provide significant information about its binding residues. This is primarily because binding pockets are predominantly located on the protein’s surface42. Consequently, structural data helps address the issue of data imbalance by naturally deprioritizing residues buried within the protein folds when identifying nucleic acid-binding residues.
However, These approaches rely on the availability of the 3D structural information, which is often lacking for newly discovered proteins. As a result, methods like GraphSite24, and GLMSite36 depend on inferring 3D structures using predictive tools such as AlphaFold44 or ESM-fold27, a process that is both time-consuming and resource-intensive.
In contrast, TransBind operates independently of MSA and structural ___domain information. Yet, we compared its performance to the methods that use these information for a more comprehensive assessment of our method. Notably, as shown in Table 6, TransBind outperforms TargetDNA, DNAPred, SVMNuc, COACH-D, NucBind, and DNABind across all metrics. NABind31 achieves the highest scores across all metrics, with the exception of precision on the RNA 117 dataset, where TransBind delivers the best performance. TransBind outperforms GraphBind in terms of precision on both DNA-129 and RNA-117 datasets, while GraphBind is slightly better than TransBind in other metrics. Overall, TransBind remains highly competitive with both NABind and GraphBind, with only marginal differences in performance. It is important to highlight that NABind leverages a combination of language-based features, MSA-derived evolutionary features (e.g., PSSMs, HMM), and structural information. Remarkably, among the sequence only models (indicated by ‘*’ in Table 6), TransBind achieved the best performance, significantly outperforming other methods in this category.
Structure-based methods require known PDB structure data, and analyzing 3D structural data is computationally more expensive than sequence-based analysis. For instance, GraphBind employs a multi-stage graph neural network to aggregate structural features into its prediction head, necessitating both structural information and substantial computational resources. These results indicate that TransBind provides fast and reliable nucleic-acid binding predictions without relying on MSA and structural data, offering a practical solution for computational proteomics. It operates efficiently even with limited resources, meeting the growing demand to assess protein functionality as new sequences are regularly discovered.
Performance analysis based on homologous sequence similarity
The quality of evolutionary features such as HMM or PSSM profiles heavily depends on the quality of multiple sequence alignments (MSAs). Orphan proteins, which do not have homologous sequences, or those with a limited number of homologs, fail to produce meaningful MSA features. Consequently, methods reliant on evolutionary features may work very well when proteins under consideration have a large amount of homologous information in the public sequence databases, but their efficacy diminishes for proteins lacking sufficient homologous data. In contrast, TransBind utilizes language model-generated features that do not rely on homology searches, providing the flexibility to generate meaningful predictions even for proteins with low sequence similarity.
In this experiment, we have assessed the impact of available homologous information on the performance of TransBind and those that use evolutionary features. To quantitatively identify orphan proteins or those with low homology, we used the NEFF score (Number of Effective Sequences in the MSA). The NEFF score, ranging from 1 to 20, serves as an indicator of homology—proteins with low NEFF values are considered to have limited sequence similarity and potential uniqueness within the dataset45. We performed a homology search using HHblits, using the official package from HHsuite (https://github.com/soedinglab/hh-suite), on the PDNA-41 dataset. For this search, we used the latest protein database (https://wwwuser.gwdguser.de/compbiol/uniclust/2023_02) available in HHsuite.
We calculated the NEFF values for each of the 41 proteins in the PDNA-41 dataset and sorted them in ascending order. These NEFF scores, ranging between 3 to 12, were then grouped into nine bins (b1, b2, …, b9), with b1 containing proteins with the lowest NEFF scores and b9 containing those with the highest. Figure 2a illustrates the distribution of proteins in these nine bins, and Fig. 2b displays the average MCC scores of TransBind and iProDNA (an MSA-based method accessible via a web server) for each bin. The results clearly show that the improvement of TransBind over iProDNA increases significantly with decreasing levels of NEFF values. There is no significant difference between them on b9 (the bin corresponding to the highest level of NEFF score, i.e., the highest level of homologous information), but as we decrease the level of NEFF scores, TransBind becomes significantly more accurate than iProDNA and attains the highest level of improvement on b1 (the bin with the lowest level of NEFF score). This clearly indicates the efficacy and superiority of TransBind for proteins with low homology information compared to the methods that rely on evolutionary features.
Ablation study
We conducted three types of ablation studies to assess the contributions of different components in TransBind. First, we assessed the performance of a simple Multilayer Perceptron (MLP) using only global features from ProtTrans, without incorporating any inception blocks. As shown in Table 7, relying solely on the language model-based global features with a basic neural network (without the inception blocks of the local extractor) did not yield satisfactory performance. This highlights the necessity and effectiveness of using both local and global feature extractors in TransBind. Next, we experimented with different numbers of inception blocks in the local feature extraction module. As shown in Table 7, increasing the number of inception blocks from two to three yielded the best performance. However, adding more blocks did not enhance performance and instead increased network complexity, likely due to over-fitting in the local feature extractor. Consequently, we opted to use three inception blocks. Finally, we examined the effect of including a weighted loss function. We observed a significant decline in the MCC score when the weighted loss function was omitted. This underscores the importance of our implemented loss function based on class weights to effectively address the data imbalance problem, ensuring a highly accurate and balanced performance.
Impact of different protein language models
We assessed the impact of different language models (ProtTrans and ESM) on the performance of TransBind using two recent benchmark datasets: DNA-129 and RNA-117. Our analysis, presented in Table 8, shows that ProtTrans-based embeddings outperform ESM-based embeddings. This finding is consistent with recent comparative analysis studies46, which demonstrate that ProtT5-XL-UniRef50 performs better in four of five downstream protein tasks compared to ESM-based models of similar size.
Although ESM models offer larger language models, such as the 15B model of esm2_t48_15B_UR50D or ESM3 with 98B parameters47, these are not feasible for inference or prediction tasks on local machines or typical standalone web servers. Given the competitive or superior performance of ProtTrans, we chose to use ProtT5-XL-UniRef50. It provides better performance than ESM-based embeddings within a similar model size and structure.
Performance on different amino acids
To further elucidate the predictive capabilities of our model on DNA-binding residues and enhance the interpretability of the results, we conducted an analysis of TransBind’s performance across 20 different amino acids using the PDNA-224 and PDNA-316 datasets. The DNA-binding amino acid composition (i.e., the percentage of 20 different amino acids) within a protein sequence is highly skewed, with certain DNA-binding amino acids being significantly less prevalent (underrepresented) than others (see Fig. 3a). For instance, Cysteine (C), Isoleucine (I), Leucine (L), and Methionine (M) are underrepresented in the datasets we examined. This distribution introduces an additional layer of class imbalance across 20 types/classes of amino acids in residue-level prediction methods, beyond the existing imbalance between interacting and non interacting residues. Our findings, presented in Fig. 3b, c, reflect this challenge, highlighting the complexities faced when predicting residue interactions across diverse amino acid classes. We compared TransBind’s results with those from iProDNA. Recognizing the challenges posed by imbalanced datasets, we focused on sensitivity as a key metric, given its importance in minimizing false negatives. Our analysis highlights a significant variance in prediction difficulty across different amino acids. Figure 3b, c shows the average sensitivity scores achieved by TransBind and iProDNA across 20 different amino acid types on two benchmark datasets. iProDNA achieves a perfect sensitivity score for certain residues but fails to identify the DNA-binding residues that are under-represented such as Cysteine (C), Isoleucine (I), Leucine (L), and Methionine (M), resulting in a sensitivity score of zero. It shows that existing methods are not suitable to handle the imbalance in the amino acid compositions. In contrast, TransBind consistently delivers valid and reasonably high sensitivity scores across all amino acid types, demonstrating robustness even for the underrepresented amino acids where iProDNA struggles. Remarkably, while sensitivity scores of iProDNA on Cysteine (C) and is Leucine (L) are zero on PDNA 224 dataset, TransBind achieves the perfect sensitivity score (100%). We believe that the way we separate amino acids in the global and local extractors within TransBind, combined with the class-weighted training scheme, contributes to the improved capacity of TransBind to identify interacting residues among the underrepresented amino acids. These findings underscore the importance of analyzing model performance at the amino acid level and demonstrate the capability of TransBind to provide reliable predictions across diverse residue types.
DNA-binding protein prediction results
For identifying DNA-binding proteins, we used an appropriately modified version of TransBind for protein-level predictions (see “Classification module” section). PDB-1075 dataset was used for training and validation, and TransBind was assessed for its performance on a widely used independent test set PDB-186.
Table 9 presents the validation performance of various methods on the training and validation dataset PDB-1075. While TransBind did not outperform other competing methods on the validation dataset, a notable advantage of TransBind becomes apparent when evaluated on the independent test set, PDB-186 (see Table 10). Specifically, TransBind achieved the highest MCC score of 0.82 and the highest accuracy of 90.86% on the test set, surpassing the performance of DTLM-DBP13, which obtained the highest MCC score of 0.64 and an accuracy of 81.18% on the validation dataset. Furthermore, TransBind demonstrated a significantly higher specificity of 98.92%, outperforming all competing methods in this regard.
Although TransBind achieved a sensitivity of 82.79%, which is lower than the best sensitivity of 94.6% achieved by ref. 10, TransBind substantially outperformed it across the other three metrics. The substantial improvement of TransBind over the state-of-the-art methods on the independent test set compared to the validation set indicates its robustness to unseen data and its ability to mitigate overfitting issues more effectively than other existing methods.
Case studies
TransBind has achieved remarkably better and more balanced (in terms of sensitivity and specificity) performance than the best alternative methods. In order to visually demonstrate the efficacy of TransBind in predicting the DNA-binding residues, we show in Fig. 4 the true and predicted (by TransBind and iProDNA) binding sites on two representative proteins from the PDNA-316 dataset, namely Phage 434 OR2/R1-69 complex and Human PAX3 Homeodomain. It is clear from the figures that iProDNA produces substantially more false positive predictions than TransBind. Moreover, TransBind is not only highly specific (with less numbers of FPs), it is considerably sensitive as all the true binding residues are correctly recognized by TransBind. The specificity of TransBind on these two proteins are 94.11% and 92.15%, respectively which are substantially higher than iProDNA which obtained 66.67% and 56.86% specificity, respectively.
Running time
TransBind significantly reduces the running time for DNA-binding protein and residue prediction by leveraging transfer learning. We assessed the time required to generate features for TransBind and compared it to methods relying on evolutionary features like PSSM, as detailed in Table 11 for various datasets analyzed in this study. We also report the inference time for TransBind. All analyses were conducted on the same machine with an 13th Generation IntelⓇ Core i7-13700HX Processor, 64GB RAM, and an NVIDIA GeForce RTX 4070 (Laptop GPU) (8GB memory). These results indicate that generating ProtTrans-based features is considerably faster than producing evolutionary features. For instance, generating ProtTrans-based features for the PDNA-224 datasets took less than 5 min, with TransBind requiring an additional 30 s for inference. In contrast, generating PSSM features took approximately 37 h. Consequently, methods relying on MSA-based PSSM features, such as iProDNA1, Zhang et al.17, PDRLGB12, NABind31, and GraphBind23 are significantly slower than TransBind.
Discussion
We presented TransBind, a highly accurate and fast method to accurately identify both DNA-binding proteins and DNA-protein binding residues from primary sequences alone. Our study leverages a transformer-based protein language model pretrained on unaligned sequences to predict DNA-binding residues and proteins from protein sequences alone, without relying on MSA-based evolutionary features or structural information. The primary novelty of this study lies in the integration of global and local feature extractors, specifically the separation of amino acids in the local extractor—a concept not previously introduced in the field. Moreover, we have addressed the critical challenge of class imbalance by implementing a carefully designed weighted loss function. TransBind remarkably surpassed the state-of-the-art across a wide variety of data sets and evaluation metrics. Our systematic analyses of the performance of different methods under various model conditions with varying levels of complexities indicate the predictive power and effectiveness of TransBind in detecting binding sites and proteins. This study demonstrates practical strengths of protein language models relative to MSAs in DNA-binding protein prediction. Therefore, we believe that TransBind advances the state-of-the-art in this ___domain and will be considered a useful tool for predicting DNA-binding proteins and residues.
This study can be extended in several directions. We utilized the protein language model ProtTrans22 for feature generation. A more recent and larger language model (ESM-2)48 for proteins. Aminobert20, another transformer-like method, uses an end-to-end large-language model to predict secondary and 3D structures from protein sequences. As an immediate extension of this study, we will explore the performance of these newly released language models in predicting DNA-binding residues and proteins. In addition to DNA-binding proteins, the proteins that bind RNA also play a vital role in various biological processes, including pre-mRNA processing, post-transcriptional gene regulation, and RNA degradation34. There is a promising opportunity to explore the applicability of TransBind in predicting RNA-binding proteins. The existing benchmark datasets in this ___domain do not contain orphan and rapidly evolving proteins for which generating MSA-based features is not feasible. In such cases, TransBind, being an alignment-free and sequence-alone method, holds promise for being more suitable compared to MSA-based methods. We systematically demonstrated the superiority of TransBind over methods that rely on evolutionary features for proteins with low homology information. However, due to the unavailability of suitable datasets, we were unable to comprehensively evaluate the performance of various methods under this specific model condition. Therefore, compiling a benchmark dataset that specifically accounts for orphan and fast-evolving proteins and conducting a comparative study on this specific condition remains an important research direction for future investigations.
The timing of this language model-based method seems appropriate as language models are receiving significant attention from the computational proteomics community, which leads many to speculate on its potential as a practical way of generating useful features from the primary sequence alone. We believe TransBind will continue to evolve with the availability of new protein databases and language models, and in response to new scientific findings—laying a firm foundation for alignment-free and sequence-only prediction of DNA-binding proteins and residues.
Methods
The TransBind architecture
The different components of TransBind (as shown in Fig. 1) are discussed in subsequent sections.
Understanding global features with self attention
This component consists of generating global features for each amino acid in a protein using the protein language model (ProtT5-XL-UniRef50). Here, each amino acid in the protein sequence generates an attention score corresponding to the significance of relation to each other amino acid. As each amino acid gathers information from all other residues with the help of attention mechanism, we call this module as global feature extractor.
In the ProtT5-XL-UniRef50 architecture, the input protein sequences are first passed through the large language model (LLM) as shown in Fig. 1a, which generates an embedding for each of the residues in the protein. For an input protein sequence \(X=\{{x}_{1},{x}_{2},\ldots ,{x}_{{N}_{x}}\}\) with Nx amino acid residues, the LLM generates a vector of dimension \({{\mathbb{R}}}^{{N}_{x}\times {D}_{x}}\), where Dx = 1024. The embedding is then manipulated by the self-attention layer to retrieve the global features for each amino acid residue in a protein sequence, as shown in Fig. 1b.
To incorporate contextual information, the attention score αij between amino acids pi and pj and the context vector ci for each amino acid pi are computed as follows:
The context vector for each amino acid pi is then given by:
Here, \({{{{\bf{W}}}}}_{Q}\in {{\mathbb{R}}}^{{D}_{x}\times {D}_{k}}\), \({{{{\bf{W}}}}}_{K}\in {{\mathbb{R}}}^{{D}_{x}\times {D}_{k}}\), and \({{{{\bf{W}}}}}_{V}\in {{\mathbb{R}}}^{{D}_{x}\times {D}_{v}}\) are the query, key, and value weight matrices, respectively. The vectors ei and ej are the embeddings of the amino acids pi and pj, respectively, each of dimension Dx. The resulting attention score αij determines the influence of amino acid pj on amino acid pi, enabling the construction of a context vector ci that captures relevant information from the sequence.
Separation of amino acids instead of parallel processing
In this step, we separate the amino acids to process them individually by the local feature extractor as shown in Fig. 1c. Let C be the matrix containing the context vectors of all amino acids in a protein sequence of length n:
where \({{{\bf{C}}}}\in {{\mathbb{R}}}^{n\times d}\) and \({{{{\bf{c}}}}}_{i}\in {{\mathbb{R}}}^{d}\) for i = 1, 2, …, n. Processing the matrix C as a whole through the local feature extractor could have been an option, as done in most token-level prediction studies49. However, since our approach already captures the global context of each amino acid relative to other residues through attention mechanisms, we can leverage this by processing each amino acid separately. This ensures that the local feature extraction and prediction network attends to each amino acid individually, thus enhancing the robustness of the learning process. This approach provides a more detailed and focused analysis, ultimately improving the accuracy and reliability of our predictions.
Local feature extraction using stacked inception V2 modules
Utilizing a local feature extractor to gather information from a close neighborhood is a widely adopted approach in protein-related interaction prediction tasks. Different methods employ various types of local feature extractors. For instance, Visual19 uses a sliding window of length seven-capturing features from three residues on either side of the central residue. However, a distinctive aspect of TransBind is that it considers each individual residue as an input to the local feature extractor. For each residue \({{{\bf{C}}}}\in {{\mathbb{R}}}^{d}\), the d-dimensional language model feature is used as the local feature. Such approach allows us to implement an efficient weighted loss training to handle the data imbalance issue of protein-nucleic acid interaction prediction.
We used an assembly of inception V2 modules in three stacked layers to build the local feature extractor (Fig. 1d). The number of inception modules was chosen with an ablation study, which is discussed in the “Ablation study” section. For each amino acid pi in the protein sequence, we apply a stacked inception v2 module to its context vector ci. The inception V2 module operates on the embedding dimension, extracting local features through multiple convolutional operations.
The inception V2 module takes a context vector ci as input and produces local features \({{{{\mathcal{F}}}}}_{{{{\rm{inception}}}}}({{{{\bf{c}}}}}_{i})\) as follows:
Here, \({{{{\mathcal{C}}}}}_{2}^{1\times 1}\) and \({{{{\mathcal{C}}}}}_{3}^{1\times 1}\) are intermediate 1 × 1 convolutions used before the 3 × 3 and 5 × 5 convolutions, respectively, to reduce the dimensionality. The max pooling operation \({{{{\mathcal{P}}}}}^{3\times 3}\) is followed by a 1 × 1 convolution \({{{{\mathcal{C}}}}}_{4}^{1\times 1}\) to maintain the same dimensionality across all branches before concatenation.
Training with weighted loss distribution for tackling class imbalance
In computational proteomics, datasets often exhibit a significant class imbalance, where the number of positive samples are substantially lower than the number of negative samples. This class-imbalance causes machine learning models to underestimate the significance of the minority class. As a result, ML models can often predict the negative samples quite well due to the large amount of negative training samples, but it fails to recognize positive samples, resulting in a relatively low sensitivity score50. As shown in Table 12, the minority to majority ratio is around 15%. While there are many sampling techniques and methods involving artificial data generation to address class imbalance, we did not adopt those approaches due to potential drawbacks such as overfitting and the introduction of synthetic artifacts that may not accurately represent real data51,52. To address this issue, we use a weighted loss function during training, which provides more penalty for incorrectly classified instances of the minority class, compared to incorrect predictions of the majority class, as detailed in Algorithm 1. The weighted class training scheme is shown in Fig. 1d′.
Given the training dataset \(D={\{({{{{\bf{f}}}}}_{i},{y}_{i})\}}_{i = 1}^{N}\), where \({{{{\bf{f}}}}}_{i}\in {{\mathbb{R}}}^{d}\) is the feature vector for the ith amino acid and yi ∈ {0, 1} is its corresponding classification label (assuming binary classification), we first compute the frequency of each class as follows:
where ck denotes the count of class k, and \({\mathbb{I}}(\cdot )\) is the indicator function. We then calculate the weight for each class as the inverse of their frequencies:
We normalize the weights as follows:
Finally, employing a weighted cross-entropy loss function to handle the class imbalance, we implement weighted cross-entropy loss \({{{\mathcal{L}}}}\) as follows:
where \({\hat{y}}_{k}\) is the predicted probability for class k.
Algorithm 1
Training with Weighted Loss Function
1: Input: Training dataset D, model M, learning rate η, number of epochs E
2: Compute class counts \({c}_{k}={\sum }_{(x,y)\in D}{\mathbb{I}}(y=k)\) for k ∈ {0, 1}
3: Compute class weights \({w}_{k}=\frac{1}{{c}_{k}}\) for k ∈ {0, 1}
4: Normalize class weights \({\tilde{w}}_{k}=\frac{{w}_{k}}{{\sum }_{j = 0}^{1}{w}_{j}}\) for k ∈ {0, 1}
5: Define weighted cross-entropy loss
6: function WEIGHTED_CROSS_ENTROPY_LOSSy, y_pred
7: return\(-\mathop{\sum }_{k = 0}^{1}{\tilde{w}}_{k}{\mathbb{I}}(y=k)\log (y\_pred[k])\)
8: end function
9: for epoch in 1 to E do
10: for (x, y) in D do
11: Forward pass: y_pred = M(x)
12: Compute loss: loss = weighted_cross_entropy_loss(y, y_pred)
13: Backward pass and optimization:
14: loss.backward()
15: optimizer.step()
16: optimizer.zero_grad()
17: end for
18: end for
Classification module
After the local feature extraction, we pass the features for each amino acid generated by the stacked inception module to a classification layer. This layer consists of two dense neuron layers with 128 and 32 neurons. We use the Rectified Linear Unit (ReLU) activation function53 for the classification part followed by a dropout layer with a dropout value of 0.2.
From residue level prediction to protein level prediction
In protein-level prediction tasks, such as prediction of the entire sequence interaction14, subcellular ___location54, we need to aggregate the residue-level predictions to make a classification at the sequence level. This process involves combining the predictions for all residues in a given protein and using this combined information for the final protein classification.
Let y = {y1, y2, …, yn} be the set of residue-level predictions for a protein with n residues, where yi denotes the prediction for the ith residue. To aggregate these predictions into a single protein-level prediction, we employ a weighted pooling strategy. In this approach, each residue-level prediction yi is assigned a weight wi that reflects relative importance. The aggregated protein-level prediction \({\hat{y}}_{{{{\rm{protein}}}}}\) is then computed as a weighted sum of these predictions:
Once the aggregated protein-level prediction \({\hat{y}}_{{{{\rm{protein}}}}}\) is obtained, it can be used for the final classification of the protein. Let \({{{\mathcal{C}}}}\) denote the classification function. The final protein classification \({\hat{c}}_{{{{\rm{protein}}}}}\) is given by:
This approach allows us to leverage detailed residue-level information to make accurate predictions at the protein level, enhancing the overall performance of protein-level tasks.
Dataset
We used widely used benchmark datasets to evaluate the performance of TransBind and other competing methods. Table 12 presents a summary of these datasets.
Training and validation datasets
We used widely used benchmark datasets, PDNA-22455, PDNA-31656, PDNA-54340, DNA-57323, and RNA-49523 to train and validate TransBind for predicting DNA-protein binding residues.
These datasets were deposited in PDB before 2016. For a more recent dataset on nucleic acid binding protein site prediction, we used the DNA-573 and RNA-495 datasets. For DNA-binding protein prediction (i.e., protein-level prediction), we used the PDB-1075 dataset57 for training and validation.
PDNA-224
The PDNA-224 dataset was assembled by ref. 55 which consists of 224 protein sequences comprising 3778 DNA-binding residues and 53,570 non-DNA-binding residues.
PDNA-316
The PDNA-316 dataset was originally constructed by ref. 56, which contains 316 protein sequences collected from PDB. There are 72,718 amino acid residues, of which 5609 residues are DNA-binding and 67,109 residues are non-DNA-binding.
PDNA-543
PDNA-543 was assembled by ref. 40 comprising 7186 protein sequences, collected from PDB before October 10, 2014. 543 non-redundant protein sequences were identified using CD-HIT58, where no two sequences had more than 30% sequence similarity. This dataset contains 144,544 amino acid residues, 9549 of which are DNA-binding and 134,995 of which are non DNA-binding.
DNA-573
This DNA-binding protein dataset, compiled and analyzed by ref. 23, contains 573 non-redundant sequences and was collected from BioLiP59 and augmented by transferring binding annotations from similar proteins. Redundancy was minimized using CD-HIT to ensure less than 30% sequence identity23.
RNA-495
This RNA-binding protein dataset, compiled and analyzed by ref. 23, was collected from BioLiP and enhanced by transferring binding annotations from similar proteins. A total of 495 non-redundant sequences were identified using CD-HIT with a 30% sequence identity threshold.
PDB-1075
For the DNA-binding protein prediction task, we use the widely used PDB-1075 dataset which was constructed by ref. 57. This dataset contains 525 DNA-binding protein and 550 non-DNA-binding protein sequences.
Test datasets
We assessed the performance of TransBind and other competing methods on widely used test sets for independent testing on unseen data. We used the PDNA-41 dataset for DNA binding residue classifications and the PDB-186 dataset for DNA binding protein classifications.
PDNA-41
This is the test dataset corresponding to the training sets PDNA-316, PDNA-543, and PDNA-224. This dataset was compiled by ref. 40 that contains 41 independent amino acid sequences used as test data for DNA binding residue classification task. This dataset contains 14,755 amino acid residues, of which 734 are DNA-binding and 14,021 are non DNA-binding residues. We note that no information regarding the sequence similarity between the training datasets (e.g., PDNA-316, PDNA-543, and PDNA-224) and PDNA-41 was reported in ref. 40. Therefore, we performed a CD-HIT analysis on the PDNA-41 dataset and the corresponding training datasets: PDNA-224, PDNA-316, and PDNA-543. We identified 4 sequences (out of the 41 sequences) with more than 30% sequence similarity with the training sets. Consequently, we evaluated TransBind on the entire PDNA-41 dataset (to ensure a fair comparison with prior studies that reported their results on the entire set of 41 sequences) as well as on the 37 non-redundant sequences after removing these four sequences.
DNA-129
This is the independent test dataset, containing 129 non-redundant sequences, corresponding to the DNA-573 dataset. This dataset was compiled and previously analyzed by ref. 23. Redundancy between training and test dataset was removed using 30% sequence similarity threshold.
RNA-117
This is the independent test set corresponding to the training dataset RNA-495, containing 117 non-redundant sequences23. It has ≤30% sequence similarity with the training set.
PDB-186
This is the corresponding test dataset for training set PDB-1075. This is a protein (sequence) level prediction dataset. This dataset60 contains 93 DNA-binding sequences and 93 non-DNA binding protein sequences, and is widely used for the DNA-binding protein prediction task.
Performance evaluation
We compared our proposed method with existing competing methods. We used six evaluation metrics, that are frequently used to assess the performance of various methods, namely accuracy, sensitivity, specificity, AUC (Area Under the Curve), AUPR (Area Under Precision-Recall), and MCC (Matthews Correlation Coefficient) score. The reported results on different performance metrics for the existing methods were obtained from their respective publications, unless otherwise specified:
Note that accuracy is not a meaningful evaluation metric for an unbalanced data set. MCC is a more appropriate performance metric, as predicting DNA binding residues/proteins is an unbalanced learning problem61,62.
Statistics and reproducibility
To ensure the reliability and robustness of the results, we conducted thorough statistical analyses. Specifically, all training and testing datasets were processed to eliminate potential overlaps. Using CD-HIT with a 30% sequence similarity threshold, we verified that no sequences in the training and testing datasets shared similarity beyond this threshold. Number of sequences in each dataset, training sample size, and the test datasets are described in details in the “Dataset” section. While reporting the validation performances, we used ten-fold cross validation and reported the average performance. To promote reproducibility, we provide the complete implementation, including the training and evaluation pipelines, as an open-source codebase.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets underlying this article are available at https://zenodo.org/records/10215073. Numerical source data for Figs. 2a, b and 3a can be found in Supplementary Data 1.
Code availability
TransBind is freely available as an open source project at Zenodo63. A server side application is available at https://trans-bind-web-server-frontend.vercel.app/.
References
Nguyen, B. P., Nguyen, Q. H., Doan-Ngoc, G.-N., Nguyen-Vo, T.-H. & Rahardja, S. iProDNA-CapsNet: identifying protein-DNA binding residues using capsule neural networks. BMC Bioinform 20, 1–12 (2019).
Si, J., Zhao, R. & Wu, R. An overview of the prediction of protein DNA-binding sites. Int. J. Mol. Sci. 16, 5194–5215 (2015).
Shen, C., Ding, Y., Tang, J., Song, J. & Guo, F. Identification of DNA–protein binding sites through multi-scale local average blocks on sequence information. Molecules 22, 2079 (2017).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015).
Ho, S.-W., Jona, G., Chen, C. T. L., Johnston, M. & Snyder, M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc. Natl. Acad. Sci. USA 103, 940–9945 (2006).
Berger, M. F. et al. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 24, 1429–1435 (2006).
Valouev, A. et al. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods 5, 29–834 (2008).
Rahman, M. S., Shatabda, S., Saha, S., Kaykobad, M. & Rahman, M. S. DPP-PseAAC: a DNA-binding protein prediction model using Chou’s general PseAAC. J. Theor. Biol. 452, 22–34 (2018).
Qian, Y., Jiang, L., Ding, Y., Tang, J. & Guo, F. A sequence-based multiple kernel model for identifying DNA-binding proteins. BMC Bioinform 22, 1–18 (2021).
Zou, Y., Ding, Y., Peng, L. & Zou, Q. FTWSVM-SR: DNA-binding proteins identification via fuzzy twin support vector machines on self-representation. Interdiscip. Sci. Comput. Life Sci. 1, 1–13 (2022).
Zhang, Q. et al. StackPDB: predicting DNA-binding proteins based on XGB-RFE feature optimization and stacked ensemble classifier. Appl. Soft Comput. 99, 2021 (2021).
Deng, L. et al. PDRLGB: precise DNA-binding residue prediction using a light gradient boosting machine. BMC Bioinform 19, 135–145 (2018).
Saber, S., Khairuddin, U., Yusof, R. & Madan, A. DTLM-DBP: deep transfer learning models for DNA binding proteins identification. Comput. Mater. Contin. 68, X (2021).
Saber, S., Khairuddin, U. & Yusof, R. DNA-binding-proteins identification based on hybrid features extraction from hidden Markov model. International Conference on Advanced Machine Learning Technologies and Applications 1, 137–147 (2022).
Xu, R. et al. Identifying DNA-binding proteins by combining support vector machine and PSSM distance transformation. BMC systems biology 9, 1–12 (2015).
Lu, W., Song, Z., Ding, Y., Wu, H. & Huang, H. A prediction method of DNA-binding proteins based on evolutionary information. Intelligent Computing Theories and Application: 15th International Conference CIC 2019, Nanchang, China; Volumne: Part II 15 ; Page range: 418-429 (2019).
Zhang, Y. et al. Identification of DNA–protein binding sites by bootstrap multiple convolutional neural networks on sequence information. Eng. Appl. Artif. Intell. 79, 58–66 (2019).
Sharma, R., Kumar, S., Tsunoda, T., Kumarevel, T. & Sharma, A. Single-stranded and double-stranded DNA-binding protein prediction using HMM profiles. Anal. Biochem. 612, 113954 (2021).
Wardah, W. et al. Predicting protein-peptide binding sites with a deep convolutional neural network. J. Theor. Biol. 496, 110278 (2020).
Chowdhury, R. et al. Single-sequence protein structure prediction using a language model and deep learning. Nat. Biotechnol. 40, 1617–1623 (2022).
Michaud, J. M., Madani, A. & Fraser, J. S. A language model beats alphafold2 on orphans. Nat. Biotechnol. 40, 1576–1577 (2022).
Elnaggar, A. et al. Prottrans: toward understanding the language of life through self-supervised learning. IEEE Trans. Pattern Anal. Mach. Intell. 44, 7112–7127 (2021).
Xia, Y., Xia, C.-Q., Pan, X. & Shen, H.-B. GraphBind: protein structural context embedded rules learned by hierarchical graph neural networks for recognizing nucleic-acid-binding residues. Nucleic Acids Res 49, e51–e51 (2021).
Shi, W. et al. Graphsite: ligand binding site classification with deep graph learning. Biomolecules 12, 1053 (2022).
Liu, Y. & Tian, B. Protein–DNA binding sites prediction based on pre-trained protein language model and contrastive learning. Brief. Bioinform. 25, bbad488 (2024).
Wei, C., Ye, Z., Wang, P. & Lan, W. DBP2Vec: predicting DNA-binding proteins directly using pre-trained protein language model. bioRxiv (2022).
Rives, A. et al. Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences. Proc. Natl. Acad. Sci. USA 118, e2016239118 (2021).
Littmann, M., Heinzinger, M., Dallago, C., Weissenow, K. & Rost, B. Protein embeddings and deep learning predict binding residues for various ligand classes. Sci. Rep. 11, 23916 (2021).
Su, H., Liu, M., Sun, S., Peng, Z. & Yang, J. Improving the prediction of protein–nucleic acids binding residues via multiple sequence profiles and the consensus of complementary methods. Bioinformatics 35, 930–936 (2019).
Yan, J. & Kurgan, L. DRNApred, fast sequence-based method that accurately predicts and discriminates DNA-and RNA-binding residues. Nucleic Acids Res 45, e84–e84 (2017).
Jiang, Z., Shen, Y.-Y. & Liu, R. Structure-based prediction of nucleic acid binding residues by merging deep learning-and template-based approaches. PLoS Comput. Biol. 19, e1011428 (2023).
Kuznetsov, I. B., Gou, Z., Li, R. & Hwang, S. Using evolutionary and structural information to predict DNA-binding sites on DNA-binding proteins. Proteins Struct. Funct. Bioinf. 64, 19–27 (2006).
Suresh, V., Liu, L., Adjeroh, D. & Zhou, X. RPI-Pred: predicting ncRNA-protein interaction using sequence and structural information. Nucleic Acids Res 43, 1370–1379 (2015).
Li, P. & Liu, Z.-P. PST-PRNA: prediction of RNA-binding sites using protein surface topography and deep learning. Bioinformatics 38, 2162–2168 (2022).
Terribilini, M. et al. RNABindR: a server for analyzing and predicting RNA-binding sites in proteins. Nucleic Acids Res 25, W578–W584 (2007).
Song, Y., Yuan, Q., Zhao, H. & Yang, Y. Accurately identifying nucleic-acid-binding sites through geometric graph learning on language model predicted structures. Brief. Bioinform. 24, bbad360 (2023).
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J. & Wojna, Z. Rethinking the inception architecture for computer vision Proceedings of the IEEE conference on computer vision and pattern recognition 2818-2826 (2016).
Zhao, S., Ding, Y., Liu, X. & Su, X. HKAM-MKM: a hybrid kernel alignment maximization-based multiple kernel model for identifying DNA-binding proteins. Comput. Biol. Med. 145, 105395 (2022).
Devlin, J., Chang, M.-W., Lee, K. & Toutanova, K. Bert: pre-training of deep bidirectional transformers for language understanding. Proceedings of naacL-HLT Vol. 1, (2018).
Hu, J. et al. Predicting protein-DNA binding residues by weightedly combining sequence-based features and boosting multiple SVMs. IEEE/ACM Trans. Comput. Biol. Bioinform. 14, 1389–1398 (2016).
Liu, R. & Hu, J. DNABind: a hybrid algorithm for structure-based prediction of DNA-binding residues by combining machine learning-and template-based approaches. Proteins Struct. Funct. Bioinf. 81, 1885–1899 (2013).
Gainza, P. et al. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 17, 184–192 (2020).
Tubiana, J. ScanNet: an interpretable geometric deep learning model for structure-based protein binding site prediction. Nat. Methods 19, 730–739 (2022).
Bryant, P., Pozzati, G. & Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 13, 1265 (2022).
Wang, Q. et al. Prior knowledge facilitates low homologous protein secondary structure prediction with DSM distillation. Bioinformatics 38, 3574–3581 (2022).
Shang, J. et al. Accurate and efficient protein embedding using multi-teacher distillation learning. Bioinformatics 40, btae567 (2024).
Hayes, T. et al. Simulating 500 million years of evolution with a language model. bioRxiv eads0018 (2024).
Lin, Z. et al. Language models of protein sequences at the scale of evolution enable accurate structure prediction. bioRxiv 379, 1123–1130 (2022).
Uddin, M. R., Mahbub, S., Rahman, M. S. & Bayzid, M. S. SAINT: self-attention augmented inception-inside-inception network improves protein secondary structure prediction. Bioinformatics 36, 4599–4608 (2020).
Zhang, Y. et al. How to balance the bioinformatics data: pseudo-negative sampling. BMC Bioinform 20, 1–13 (2019).
Chawla, N. V., Bowyer, K. W., Hall, L. O. & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. J. Artif. Intell. Res. 16, 321–357 (2002).
He, H., Bai, Y., Garcia, E. A. & Li, S. ADASYN: adaptive synthetic sampling approach for imbalanced learning. 2008 IEEE international joint conference on neural networks (IEEE world congress on computational intelligence) 1322-1328 (2008).
Agarap, A. F. Deep learning using rectified linear units (relu). Preprint at arXiv https://doi.org/10.48550/arXiv.1803.08375 (2018).
Wang, J., Horlacher, M., Cheng, L. & Winther, O. DeepLocRNA: an interpretable deep learning model for predicting RNA subcellular localization with ___domain-specific transfer-learning. Bioinformatics 40, btae065 (2024).
Li, T. et al. PreDNA: accurate prediction of DNA-binding sites in proteins by integrating sequence and geometric structure information. Bioinformatics 29, 678–685 (2013).
Si, J., Zhang, Z., Lin, B., Schroeder, M. & Huang, B. MetaDBSite: a meta approach to improve protein DNA-binding sites prediction. BMC Syst. Biol. 5, 1–7 (2011).
Liu, B. et al. iDNA-Prot∣ dis: identifying DNA-binding proteins by incorporating amino acid distance-pairs and reduced alphabet profile into the general pseudo amino acid composition. PLoS ONE 9, e106691 (2014).
Qiao, S. et al. Predicting long-term trajectories of connected vehicles via the prefix-projection technique. IEEE Trans. Intell. Transp. Syst. 19, 2305–2315 (2017).
Yang, J., Roy, A. & Zhang, Y. BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res 41, D1096–D1103 (2012).
Lou, W. et al. Sequence based prediction of DNA-binding proteins based on hybrid feature selection using random forest and Gaussian naive Bayes. PLoS ONE 9, e86703 (2014).
Chicco, D. & Jurman, G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21, 1–13 (2020).
Mahbub, S. & Bayzid, M. S. EGRET: edge aggregated graph attention networks and transfer learning improve protein–protein interaction site prediction. Brief. Bioinform. 23, bbab578 (2022).
Tahmid, M. T. Source Code : TransBind allows precise detection of DNA-binding proteins and residues using language models and deep learning. Zenodo. https://doi.org/10.5281/zenodo.14272818 (2024).
Zhou, J., Lu, Q., Xu, R., He, Y. & Wang, H. EL_PSSM-RT: DNA-binding residue prediction by integrating ensemble learning with PSSM relation transformation. BMC Bioinform 18, 1–16 (2017).
Wang, L. & Brown, S. J. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res 34, W243–W248 (2006).
Hwang, S., Gou, Z. & Kuznetsov, I. B. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics 23, 634–636 (2007).
Chu, W.-Y. et al. ProteDNA: a sequence-based predictor of sequence-specific DNA-binding residues in transcription factors. Nucleic Acids Res. 37, W396-W401 (2009).
Acknowledgements
This work was supported by the Basic Research Grant at Bangladesh University of Engineering and Technology (BUET) and the Research and Innovation Centre for Science and Engineering (RISE) at BUET Internal Research Grant (ID: 2021-01-16).
Author information
Authors and Affiliations
Contributions
M.T.T. and M.S.B. conceived and designed the study, M.T.T., M.S.B. and A.K.M.M.H. developed the algorithm, M.T.T. implemented the proposed solutions, M.T.T. performed the experiments, M.T.T. and A.K.M.M.H. developed the web server, M.T.T. and M.S.B. wrote the manuscript, M.S.B. supervised the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Quang-Thai Ho, and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editors: Chien-Yu Chen and David Favero. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tahmid, M.T., Hasan, A.M. & Bayzid, M.S. TransBind allows precise detection of DNA-binding proteins and residues using language models and deep learning. Commun Biol 8, 568 (2025). https://doi.org/10.1038/s42003-025-07534-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-07534-w